AIFM1 (apoptosis-inducing factor, mitochondrion-associated, 1) (original) (raw)
DNA/RNA
Note
AIF (Apoptosis-Inducing Factor). Total gene size 36.471 Kb with a transcribed region of 2.215 Kb which codes for 613 amino acids. To date, five isoforms from AIF gene have been described (AIF, AIFexB, AIFsh, AIFsh2, and AIFsh3).
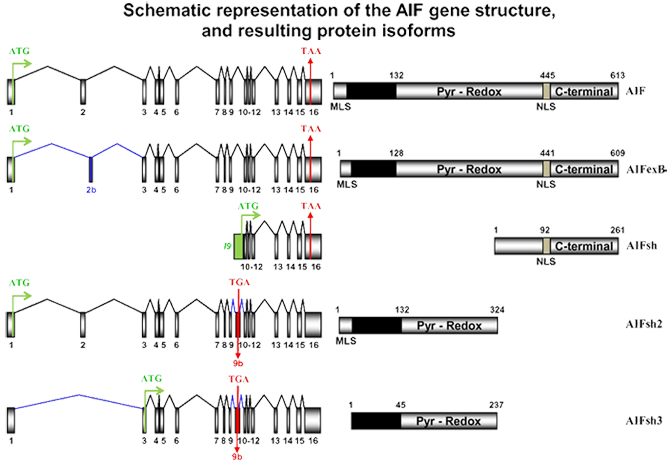
AIF gene structure and known isoforms. Genomic organization of AIF and resulting AIF, AIF-exB, AIFsh, AIFsh2, and AIFsh3 mRNA transcripts (schemas in the left). Translation start (ATG, in green) and stop (TGA/TAA, in red) codons are indicated, and the predicted protein product is shown at the right. Numbers in AIF designate exons (in mRNA transcripts) and amino acids (in the predicted proteins). Mitochondria localization signal (MLS), Pyridoxin-redox (Pyr-Redox), nuclear localization sequence (NLS), and C-terminal domains are indicated. I9 (in green) indicates intron 9. The inclusion of the 203-bp exon 9b (lettering in red) produces AIFsh2 and AIFsh3, which encodes 324- and 237-amino acid proteins, respectively. AIFsh2 contains the MLS and the Pyr-Redox domain, but lacks the C-terminal portion of AIF. AIFsh3 has a similar structure as AIFsh2 with the splicing of exon 2, leading to the loss of MLS. Blue lines indicate the splicing of the different isoforms.
Description
16 exons spanning 36.471 Kb.
Transcription
2,215 bp mRNA.
Proteins
Note
613 amino acids long protein whose structure may be divided into three domains: a FAD-binding domain (residues 128-262 and 401-480), a NADH-binding domain (residues 263-400), and a C-terminal domain (residues 481-608).
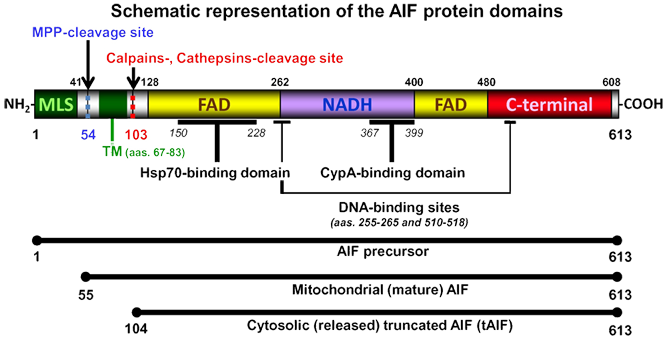
Schematic model representing the three different AIF forms: precursor, mature, and truncated. AIF is a flavoprotein (with an oxidoreductase enzymatic activity) containing a FAD-bipartite domain (yellow, amino-acids 128-262 and 401-480), a NADH-binding motif (violet, amino-acids 263-400), and a C-terminal domain (red, amino-acids 481-608) where the proapoptotic activity of the protein resides. In addition, it has a Mitochondria Localization Sequence (MLS, in green, amino-acids 1-41) placed in its N-terminal region. Between the first-N-terminal FAD motif and the MLS, AIF possesses a potential Transmembrane Domain (TM, in green, amino-acids 67-83). This TM is flanked by two peptidase-processing positions: a Mitochondrial Processing Peptidase (MPP)-cleavage site (in blue, amino-acid 54) and a calpains- and/or cathepsins-cleavage site (in red, amino-acid 103). Hsp70 (Heat Shock Protein-70) and CypA (Cyclophilin A) bind AIF in amino-acids 150-228 and 367-369, respectively. AIF also possesses two DNA-binding sites, which are located in amino-acids 255-265 and 510-518, respectively. AIF precursor protein has 613 amino-acids. The MPP-mediated cleavage generates the mitochondrial mature AIF (amino-acids 55-613). After an apoptotic insult, calpains or cathepsins cleave AIF to produce truncated-AIF (tAIF), which is released from mitochondria to cytosol (amino-acids 104-613).
Description
AIF was initially identified as a protein released from the mitochondrial intermembrane space during the apoptotic process. First studies showed that upon an apoptotic stimulus AIF translocates from mitochondria to cytosol and further to the nucleus where it triggers caspase-independent programmed cell death. AIF, expressed as a precursor of 67 kDa, is addressed to mitochondria by the two MLS placed within the N-terminal prodomain of the protein. Once in mitochondria, this precursor is processed to a mature form of 62 kDa by a first proteolytic cleavage. In this configuration, AIF is an inner-membrane-anchored protein in which the N-terminus is exposed to the mitochondrial matrix and the C-terminal portion to the mitochondrial intermembrane space. AIF is here required for maintenance or maturation of the mitochondrial respiratory chain complex I. After a cell death insult, the 62 kDa AIF-mitochondrial form is cleaved by activated calpains and/or cathepsins to yield a soluble proapoptotic protein with an apparent molecular weight of 57 kDa tAIF (truncated AIF). tAIF is released from mitochondria to cytosol and nucleus to generate two typical hallmarks of caspase-independent programmed cell death: chromatin condensation and large-scale approximatively 50 kb DNA fragmentation.
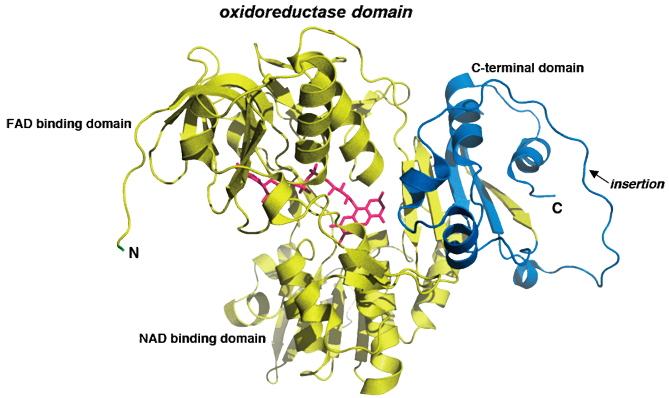
Ribbon structure of mouse AIF in its mature form (pdb id: 1GV4). As depicted here, three domains are present in the protein. The FAD-binding domain and the NAD-binding domain (yellow) are both similar to oxidoreductase domains from members of the glutathione reductase family. In contrast, the C-terminal domain (blue) displays a particular folding with a specific insertion, which includes residues 580 to 610. This picture also includes the AIF cofactor Flavin Adenine Dinucleotide (FAD; magenta).
Expression
Ubiquitously expressed.
Localisation
Mitochondrion.
Function
AIF has a double life/death function.
In its vital role, AIF is required to maintain and/or organize the mitochondrial respiratory complex I, and displays NADH oxidoreductase and peroxide scavenging activities. In addition to this vital function, AIF has been shown to be implicated in programmed cell death (PCD) induction in several experimental models (see bibliography section). In the two most studied AIF-dependent PCD models, AIF death activity is associated with the increase of intracellular Ca2+ (e.g., ischemia/reperfusion injury), or relates with extensive DNA-damage (e.g., treatment with alkylating agents). In the first model, increased intracellular Ca2+ levels trigger depolarization of mitochondrial membrane, subsequent loss of membrane potential, generation of reactive oxygen species (ROS), and AIF mitochondrial release. In the second model, extensive DNA damage, provoked by high doses of alkylating agents such as MNNG or MNU, triggers poly(ADP-ribose) polymerase-1 (PARP-1) over-activation and AIF release from the mitochondrial intermembrane space. This cell death pathway sequentially involves PARP-1, calpains, Bax, and AIF.
Homology
AIF is a highly conserved protein ubiquitously present in all primary kingdoms, Bacteria, Archaea and Eucaryota. The aif gene is inherited from the last universal common ancestor and follows the tree topology with the primary radiation of the archaeo-eukaryotic and bacterial clades. AIF also has a highly significant homology with different families of oxidoreductases, including NADH oxydases, Ascorbate reductases, Glutathione reductases and many NADH-dependent ferredoxin reductases from Archaea and Bacteria to invertebrates and vertebrates.
Mouse, Rat homology
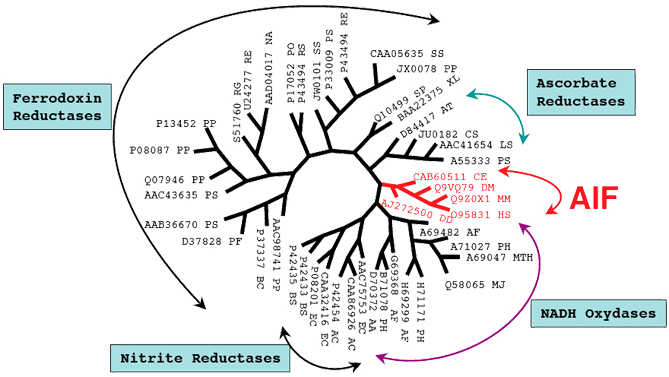
Phylogenetic tree representing the relationship between AIF and other oxidoreductases from different species. Note the proximity of the AIF family (red branch) to the NADH-oxidase family from Archaea. The PIR accession codes are enumerated following the abbreviation of each specie: AA: Aquifex aeolicus; AC: Acinetobacter calcoaceticus; AF: Archaeoglobus fulgidus; AT: Arabidopsis thaliana; BC: Burkholderia cepacia; BS: Bacillus subtilis; CE: Caenorhabditis elegans; DD: Dictyostelium discoideum; DM: Drosophila melanogaster; EC: Escherichia coli; HS: Homo sapiens; LS: Lycopersicon esculentum; MJ: Methanocaldococcus jannaschii; MM: Mus musculus; MTH: Methanobacterium thermoautotrophicum; N A: Novosphingobium aromaticivorans; PF: Pseudomonas fluorescens; PH: Pyrococcus horikoshii; PO: Pseudomonas oleovorans; PP: Pseudomonas putida; PS: Pseudomonas sp.; PSA: Pisum sativum; SP: Schizosaccharomyces pombe; SS: Sphingomonas sp.; RE: Rhodococcus erythropolis; RG: Rhodococcus globerulus; XL: Xenopus laevis.
Mutations
Note
Several polymorphisms have been identified but none of them has shown any association with a disease.
Implicated in
Entity name
Various cancers
Note
Upregulated in cancers (colorectal carcinoma, gastric carcinoma, breast carcinoma and hepatocellular carcinoma, glioblastoma).
AIF expression may play a role in tumor formation and could maintain a transformed state of colon cancer cells involving mitochondrial complex I function.
Disease
AIF has been directly designed as main mediator of cell death in ischemic injuries after overproduction of reactive oxygen species. Indeed, blocking the mitochondrial release of AIF to cytosol and its further nuclear translocation provides protection against neuronal and cardiomyocites cell death. AIF-deficient harlequin mutant mouse presents a significant reduction of neuronal cell death in brain trauma and cerebral ischemia. A similar protective effect was observed in AIF siRNA-treated neurons.
Entity name
Degenerative disorders
Disease
AIF is involved in several degenerative disorders. The elevated production of ROS generated in Amyotrophic Lateral Sclerosis, Alzheimers, or Parkinson diseases concludes in the translocation of AIF. Likewise, AIF release triggered by calpains and cathepsins was observed on in vitro models of Epilepsy and Huntingtons disease. AIF-mediated cell death is involved in the pathogenesis of different retinal affections such as retinal detachment, retinitis pigmentosa, or in models of retinal hypoxia. Moreover, an increase of AIF expression has been reported in patients affected with diabetic retinopathy.
Bibliography
| Pubmed ID | Last Year | Title | Authors |
|---|---|---|---|
| 16636662 | 2006 | CD44 ligation induces caspase-independent cell death via a novel calpain/AIF pathway in human erythroleukemia cells. | Artus C et al |
| 12782632 | 2003 | Cathepsin D triggers Bax activation, resulting in selective apoptosis-inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. | Bidère N et al |
| 17912035 | 2007 | AIF-mediated programmed necrosis: a highly regulated way to die. | Boujrad H et al |
| 14716299 | 2004 | AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. | Candé C et al |
| 16917506 | 2006 | Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. | Cheung EC et al |
| 16644725 | 2006 | Identification and characterization of AIFsh2, a mitochondrial apoptosis-inducing factor (AIF) isoform with NADH oxidase activity. | Delettre C et al |
| 15286713 | 2004 | Apoptosis-inducing factor determines the chemoresistance of non-small-cell lung carcinomas. | Gallego MA et al |
| 14555980 | 2003 | Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. | Gurbuxani S et al |
| 17207087 | 2006 | Immunohistochemical and mutational analysis of apoptosis-inducing factor (AIF) in colorectal carcinomas. | Jeong EG et al |
| 11279485 | 2001 | Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. | Joza N et al |
| 12353028 | 2002 | The harlequin mouse mutation downregulates apoptosis-inducing factor. | Klein JA et al |
| 18180198 | 2007 | Therapeutic potential of AIF-mediated caspase-independent programmed cell death. | Lorenzo HK et al |
| 11967568 | 2002 | The crystal structure of the mouse apoptosis-inducing factor AIF. | Maté MJ et al |
| 11278689 | 2001 | NADH oxidase activity of mitochondrial apoptosis-inducing factor. | Miramar MD et al |
| 16621561 | 2006 | Apoptosis-inducing factor: vital and lethal. | Modjtahedi N et al |
| 17470554 | 2007 | Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. | Moubarak RS et al |
| 15775970 | 2005 | Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. | Otera H et al |
| 15590628 | 2005 | Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. | Polster BM et al |
| 11533664 | 2001 | Heat-shock protein 70 antagonizes apoptosis-inducing factor. | Ravagnan L et al |
| 16407317 | 2006 | Distinct hsp70 domains mediate apoptosis-inducing factor release and nuclear accumulation. | Ruchalski K et al |
| 16729031 | 2006 | Regulation of AIF expression by p53. | Stambolsky P et al |
| 9989411 | 1999 | Molecular characterization of mitochondrial apoptosis-inducing factor. | Susin SA et al |
| 16001080 | 2005 | AIF suppresses chemical stress-induced apoptosis and maintains the transformed state of tumor cells. | Urbano A et al |
| 16278674 | 2006 | Physical interaction of apoptosis-inducing factor with DNA and RNA. | Vahsen N et al |
| 12198487 | 2002 | DNA binding is required for the apoptogenic action of apoptosis inducing factor. | Ye H et al |
| 17116881 | 2006 | Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. | Yu SW et al |
| 14678748 | 2003 | Poly(ADP-ribose) polymerase-1 and apoptosis inducing factor in neurotoxicity. | Yu SW et al |
| 15933737 | 2005 | Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. | Yuste VJ et al |
Other Information
Locus ID:
NCBI: 9131
MIM: 310490
HGNC: 8768
Ensembl: ENSG00000156709
Variants:
dbSNP: 9131
ClinVar: 9131
TCGA: ENSG00000156709
COSMIC: AIFM1
RNA/Proteins
Expression (GTEx)
Pathways
| Pathway | Source | External ID |
|---|---|---|
| Apoptosis | KEGG | ko04210 |
| Apoptosis | KEGG | hsa04210 |
Protein levels (Protein atlas)
Not detected
Low
Medium
High
References
| Pubmed ID | Year | Title | Citations |
|---|---|---|---|
| 38043402 | 2024 | NADH improves AIF dimerization and inhibits apoptosis in iPSCs-derived neurons from patients with auditory neuropathy spectrum disorder. | 2 |
| 38449065 | 2024 | AIF translocation into nucleus caused by Aifm1 R450Q mutation: generation and characterization of a mouse model for AUNX1. | 0 |
| 38043402 | 2024 | NADH improves AIF dimerization and inhibits apoptosis in iPSCs-derived neurons from patients with auditory neuropathy spectrum disorder. | 2 |
| 38449065 | 2024 | AIF translocation into nucleus caused by Aifm1 R450Q mutation: generation and characterization of a mouse model for AUNX1. | 0 |
| 36751702 | 2023 | AIFM1 variants associated with auditory neuropathy spectrum disorder cause apoptosis due to impaired apoptosis-inducing factor dimerization. | 3 |
| 36907087 | 2023 | Deep Brain Stimulation for the Management of AIFM1-Related Disabling Tremor: A Case Series. | 1 |
| 37000625 | 2023 | Newly synthesized AIFM1 determines the hypersensitivity of T lymphocytes to STING activation-induced cell apoptosis. | 2 |
| 37365177 | 2023 | Impaired AIF-CHCHD4 interaction and mitochondrial calcium overload contribute to auditory neuropathy spectrum disorder in patient-iPSC-derived neurons with AIFM1 variant. | 1 |
| 36751702 | 2023 | AIFM1 variants associated with auditory neuropathy spectrum disorder cause apoptosis due to impaired apoptosis-inducing factor dimerization. | 3 |
| 36907087 | 2023 | Deep Brain Stimulation for the Management of AIFM1-Related Disabling Tremor: A Case Series. | 1 |
| 37000625 | 2023 | Newly synthesized AIFM1 determines the hypersensitivity of T lymphocytes to STING activation-induced cell apoptosis. | 2 |
| 37365177 | 2023 | Impaired AIF-CHCHD4 interaction and mitochondrial calcium overload contribute to auditory neuropathy spectrum disorder in patient-iPSC-derived neurons with AIFM1 variant. | 1 |
| 34597523 | 2022 | Staurosporine-induced cleavage of apoptosis-inducing factor in human fibrosarcoma cells is independent of matrix metalloproteinase-2. | 0 |
| 34986387 | 2022 | Silencing ESRP1 expression promotes caspase-independent cell death via nuclear translocation of AIF in colon cancer cells. | 5 |
| 35339754 | 2022 | Cyclophosphamide-induced GPX4 degradation triggers parthanatos by activating AIFM1. | 12 |