ENRG MD for Origami Structure Prediction (original) (raw)
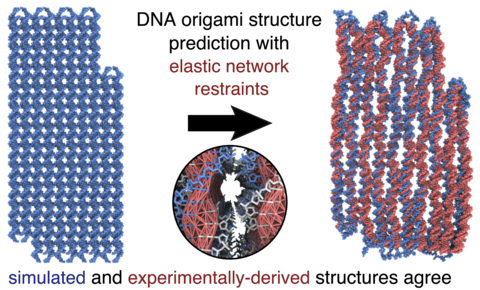
DNA origami is a cost-effective method of controlling matter at the nanoscale that folds a long DNA strand using a custom set of complementary strands. Experimental characterization of DNA origami is essential for accurate design, but has been limited to rather low-resolution techniques. One exception is the high-resolution cryo-electron microscopy (cryo EM) reconstruction of the "pointer" object from the Dietz group.
The need for atomically-detailed computational structure prediction led us to study the pointer object using molecular dynamics simulation (see publication). We found that it is possible to obtain high-quality structures from the idealized configuration using orders of magnitude fewer resources by ommiting solvent and using an elastic-network of restraints to guide the simulation. You can explore these simulations through MDshowcase on NanoHUB.
Simulation setup
This service produces NAMD configuration files needed for an elastic-network guided DNA origami structure prediction simulation. Note that you will have to run the simulations on your own workstation, which can be a time consuming process (overnight to several days), espcially for large systems. It may not be possible to simulate a structure that is initially very far from equilibrium (e.g. having very long bonds in the caDNAno design) out-of-the-box. To gauge how long the simulation might need to run on your hardware, we recommend testing the protocol with a very simple design. The simulation will likely need to run ~2 ns. For a given number of processors, the walltime to run a simulation scales almost linearly with the number of atoms in the system.
Please direct any questions or issues to Chris Maffeo (cmaffeo2@illinois.edu).
Name your job (optional). *
Upload a DNA origami design .json file. Circular strands (without breaks) are not supported.
Select the origami lattice. *
Initial interaxial DNA spacing (Angstrom) *
Select the scaffold sequence. *
m13mp18 (up to 7,249 bases)
Custom
Sequence file
Text file containing nucleotide sequence ('atcg' or 'ATCG'). Pound symbol (#) comments the remainder of a line. Other characters are ignored.
Simulation package *
NAMD (CHARMM FF)
Gromacs (AMBER FF; beta coming soon)
To obtain statistics for our funding agencies, this service collects and stores your IP address, an approximate location derived from the IP address and a timestamp each time a job is submitted. Your email address, if provided, is not associated with other data collected and is deleted from the server once the job completes and an email is sent. Please contact Chris Maffeo (cmaffeo2@illinois.edu) with questions, concerns or requests regarding privacy.