Computational Diagnostics Group @ Max Planck Institute for Molecular Genetics, Berlin (original) (raw)
 |
Home |
|---|---|
 |
Contact |
 |
Group Members |
 |
Dept. Vingron |
 |
|
 |
Research |
 |
Publications |
 |
Software |
 |
|
 |
Workshops |
 |
Group Seminar |
 |
|
 |
NGFN Microarray Data Analysis Resource |
 |
Collaborators |
adSplit - Annotation-Driven Splits in Microarray Data

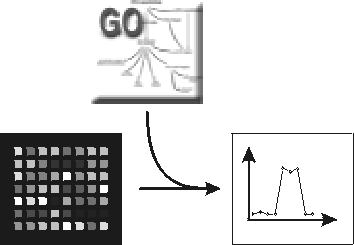 |
adSplit is an R/Bioconductor Package which implements annotation-driven splitting of microarray data into two classes. Functional annotations of genes are used to restrict expression matrices linked to particular annotations. Splits are then computed using 2-means clustering The biological terms used are derived from the Gene Ontology, as well as the KEGG database of pathways. |
|---|
Publication
Annotation-based Distance Measures for Patient Subgroup Discovery in Clinical Microarray Studies
Lottaz C, Toedling J, Spang R
In D. Huson, O. Kohlbacher, A. Lupas, K. Nieselt and A. Zell (eds.)Lecture Notes on Computer Science, Proceedings of the German Conference on Bioinformatics, Tübingen, Germany, Sep 20-22, 2006, 75-91.
Download
Download the tared and gzipped source file, which is compliant toBioconductor. To run the package, you need R and at least packages Biobase, multtest, GO and KEGG from Bioconductor. Furthermore, meta data packages for all microarray you want to analyze are needed. To run the examples, you also need the package golubEsets and vsn.
→ the current source package
→ the current binary package for Windows
Direct installation from the CompDiag R/Bioconductor-repository
Start the R version where you want to install adSplit. Use the following commands on the R prompt to install StAM directly from our repository:
library(reposTools) z <- getReposEntry("http://compdiag.molgen.mpg.de/software/Rrepos") install.packages2("adSplit", z)