 Trees as Python objects Load, create, traverse, search, prune, or modify hierarchical tree structures with ease using the ETE Python API.
Trees as Python objects Load, create, traverse, search, prune, or modify hierarchical tree structures with ease using the ETE Python API.
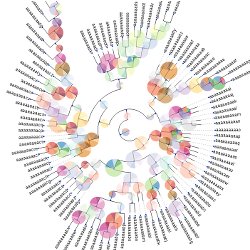 Programmatic tree visualization Get full control of your tree images. Browse them interatively or render SVG, PNG of PDF images.
Programmatic tree visualization Get full control of your tree images. Browse them interatively or render SVG, PNG of PDF images.
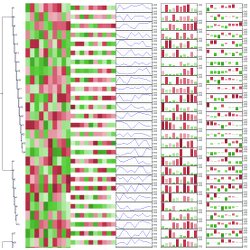 Tree annotation Custom node attributes can be rendered as graphical elements. Choose among external images, charts, symbols, text labels, and more!
Tree annotation Custom node attributes can be rendered as graphical elements. Choose among external images, charts, symbols, text labels, and more!
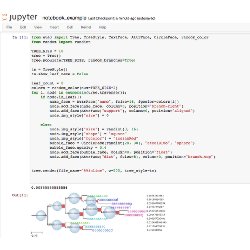 Jupyter notebook support Prototype your methods using the Jupyter notebook framework including inline visualization of trees.
Jupyter notebook support Prototype your methods using the Jupyter notebook framework including inline visualization of trees.
Tools overview Cookbook
 Automated reconstruction of gene and species trees. Run phylogenetic workflows with ease.
Automated reconstruction of gene and species trees. Run phylogenetic workflows with ease.
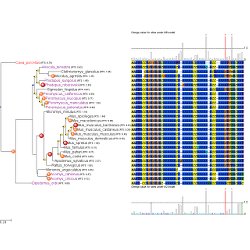 Test evolutionary hypotheses You can run CodeML and SLR easly using ETE, as well as visualizing results.
Test evolutionary hypotheses You can run CodeML and SLR easly using ETE, as well as visualizing results.
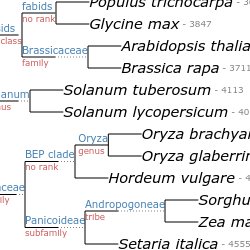 Annotate trees with taxonomic data. Use the NCBI taxonomy database to perform queries efficiently or to annotate your trees.
Annotate trees with taxonomic data. Use the NCBI taxonomy database to perform queries efficiently or to annotate your trees.
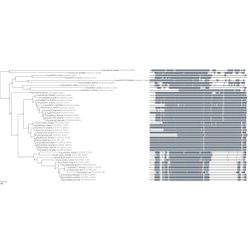
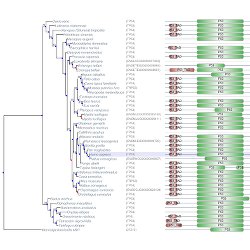 Visualize trees and sequence alignments Choose among detailed view, block-based, domain-based or condensed alignment formats.
Visualize trees and sequence alignments Choose among detailed view, block-based, domain-based or condensed alignment formats.
 Compare tree topologies You can estimate distances between gene and species trees of a different size and with duplicated events.
Compare tree topologies You can estimate distances between gene and species trees of a different size and with duplicated events.
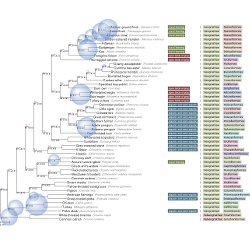
Summarize phylogenetic signal
You can summarize phylogentic signal from multiple gene trees into a single species tree.
 Automated reconstruction of gene and species trees. Run phylogenetic workflows with ease.
Automated reconstruction of gene and species trees. Run phylogenetic workflows with ease. 
 Trees as Python objects Load, create, traverse, search, prune, or modify hierarchical tree structures with ease using the ETE Python API.
Trees as Python objects Load, create, traverse, search, prune, or modify hierarchical tree structures with ease using the ETE Python API.  Programmatic tree visualization Get full control of your tree images. Browse them interatively or render SVG, PNG of PDF images.
Programmatic tree visualization Get full control of your tree images. Browse them interatively or render SVG, PNG of PDF images.  Tree annotation Custom node attributes can be rendered as graphical elements. Choose among external images, charts, symbols, text labels, and more!
Tree annotation Custom node attributes can be rendered as graphical elements. Choose among external images, charts, symbols, text labels, and more!  Jupyter notebook support Prototype your methods using the Jupyter notebook framework including inline visualization of trees.
Jupyter notebook support Prototype your methods using the Jupyter notebook framework including inline visualization of trees.  Test evolutionary hypotheses You can run CodeML and SLR easly using ETE, as well as visualizing results.
Test evolutionary hypotheses You can run CodeML and SLR easly using ETE, as well as visualizing results.  Annotate trees with taxonomic data. Use the NCBI taxonomy database to perform queries efficiently or to annotate your trees.
Annotate trees with taxonomic data. Use the NCBI taxonomy database to perform queries efficiently or to annotate your trees.  Visualize trees and sequence alignments Choose among detailed view, block-based, domain-based or condensed alignment formats.
Visualize trees and sequence alignments Choose among detailed view, block-based, domain-based or condensed alignment formats.