CellDesigner (original) (raw)
Note on log4j: CellDesigner uses log4j version 1.2.16 which is not affected by the security vulnerability; https://logging.apache.org/log4j/2.x/security.html
What is CellDesignerTM
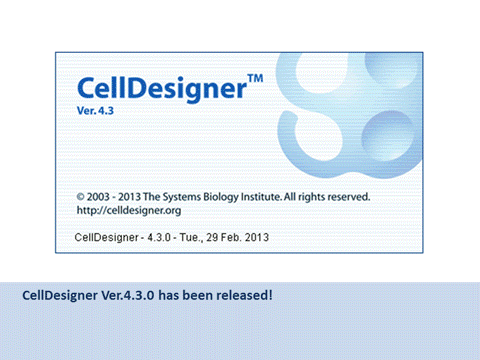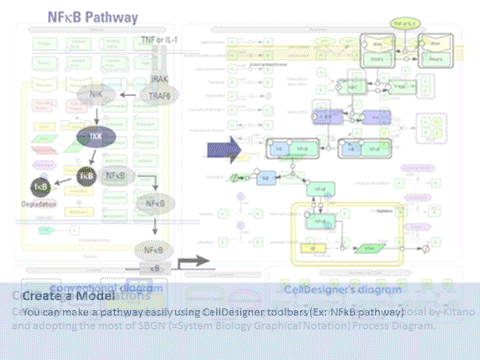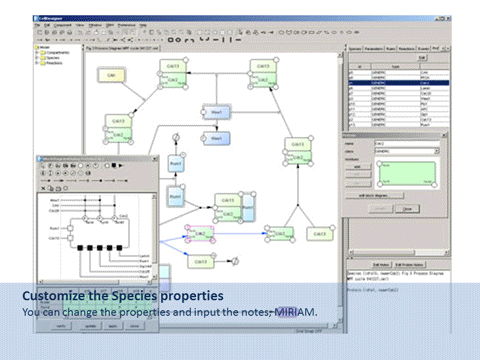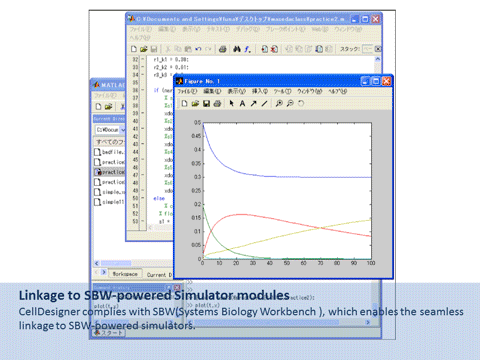
CellDesigner is a structured diagram editor for drawing gene-regulatory and biochemical networks. Networks are drawn based on the process diagram, with graphical notation system proposed by Kitano, and are stored using the Systems Biology Markup Language (SBML), a standard for representing models of biochemical and gene-regulatory networks. Networks are able to link with simulation and other analysis packages through Systems Biology Workbench (SBW). CellDesigner supports simulation and parameter scan by an integration with SBML ODE Solver, SBML Simulation Core and Copasi.
By using CellDesigner, you can browse and modify existing SBML models with references to existing databases, simulate and view the dynamics through an intuitive graphical interface.
CellDesigner is free to use >> License text
How to Cite CellDesigner
- Funahashi, A., Tanimura, N., Morohashi, M., and Kitano, H., CellDesigner: a process diagram editor for gene-regulatory and biochemical networks, BIOSILICO, 1:159-162, 2003 [doi:10.1016/S1478-5382(03)02370-9]
- Funahashi, A.; Matsuoka, Y.; Jouraku, A.; Morohashi, M.; Kikuchi, N.; Kitano, H."CellDesigner 3.5: A Versatile Modeling Tool for Biochemical Networks" Proceedings of the IEEE Volume 96, Issue 8, Aug. 2008 Page(s):1254 - 1265
[doi 10.1109/JPROC.2008.925458] Abstract | Full Text: PDF (1379 KB)
> Graphical Notation
- Kitano, H.et al. "Using process diagram for the graphical representation of biological networks", Nature Biotechnology 23(8), 961-966 (2005)
> Models built with CellDesigner
>> see also Documents.