| Version |
5.0 |
| Status |
Expected but not Quantified |
| Creation Date |
2012-09-06 15:16:49 UTC |
| Update Date |
2022-03-07 02:51:34 UTC |
| HMDB ID |
HMDB0014323 |
| Secondary Accession Numbers |
HMDB14323 |
| Metabolite Identification |
|
| Common Name |
Valsartan |
| Description |
Valsartan is an angiotensin-receptor blocker (ARB) that may be used to treat a variety of cardiac conditions including hypertension, diabetic nephropathy and heart failure. Valsartan lowers blood pressure by antagonizing the renin-angiotensin-aldosterone system (RAAS); it competes with angiotensin II for binding to the type-1 angiotensin II receptor (AT1) subtype and prevents the blood pressure increasing effects of angiotensin II. Unlike angiotensin-converting enzyme (ACE) inhibitors, ARBs do not have the adverse effect of dry cough. Valsartan may be used to treat hypertension, isolated systolic hypertension, left ventricular hypertrophy and diabetic nephropathy. It may also be used as an alternative agent for the treatment of heart failure, systolic dysfunction, myocardial infarction and coronary artery disease. |
| Structure |
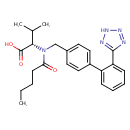 MOL3D MOLSDF3D SDFPDB3D PDBSMILESInChI177 Mrv0541 02231214292D 32 34 0 0 1 0 999 V2000 2.0930 1.2209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -0.0167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 0.8084 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 7.8559 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 9.1909 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.1109 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.9359 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 1.2209 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 3.5220 2.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -2.4917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 16 1 0 0 0 0 2 12 2 0 0 0 0 3 16 2 0 0 0 0 9 4 1 6 0 0 0 4 11 1 0 0 0 0 4 12 1 0 0 0 0 5 7 1 0 0 0 0 5 31 2 0 0 0 0 6 8 2 0 0 0 0 6 31 1 0 0 0 0 7 8 1 0 0 0 0 9 10 1 0 0 0 0 9 16 1 0 0 0 0 10 13 1 0 0 0 0 10 14 1 0 0 0 0 11 17 1 0 0 0 0 12 15 1 0 0 0 0 15 18 1 0 0 0 0 17 20 2 0 0 0 0 17 21 1 0 0 0 0 18 19 1 0 0 0 0 19 25 1 0 0 0 0 20 23 1 0 0 0 0 21 24 2 0 0 0 0 22 23 2 0 0 0 0 22 24 1 0 0 0 0 22 26 1 0 0 0 0 26 27 1 0 0 0 0 26 28 2 0 0 0 0 27 29 2 0 0 0 0 27 31 1 0 0 0 0 28 30 1 0 0 0 0 29 32 1 0 0 0 0 30 32 2 0 0 0 0 M END HMDB0014323 RDKit 3D Valsartan 61 63 0 0 0 0 0 0 0 0999 V2000 -6.2147 -1.9040 -1.0518 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.0355 -2.8131 -0.9456 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.9909 -2.3573 0.0362 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.4229 -1.0028 -0.3172 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.4060 -0.6505 0.7095 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.2138 -1.4268 1.6804 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6351 0.5486 0.6299 N 0 0 0 0 0 0 0 0 0 0 0 0 -0.6676 0.8211 1.6323 C 0 0 0 0 0 0 0 0 0 0 0 0 0.7239 0.4141 1.3796 C 0 0 0 0 0 0 0 0 0 0 0 0 1.1455 -0.8857 1.2415 C 0 0 0 0 0 0 0 0 0 0 0 0 2.4895 -1.2208 1.0227 C 0 0 0 0 0 0 0 0 0 0 0 0 3.4365 -0.2537 0.9384 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8594 -0.4711 0.7246 C 0 0 0 0 0 0 0 0 0 0 0 0 5.7763 -0.1927 1.7553 C 0 0 0 0 0 0 0 0 0 0 0 0 7.1152 -0.3824 1.6223 C 0 0 0 0 0 0 0 0 0 0 0 0 7.6227 -0.8631 0.4484 C 0 0 0 0 0 0 0 0 0 0 0 0 6.7475 -1.1422 -0.5730 C 0 0 0 0 0 0 0 0 0 0 0 0 5.3580 -0.9504 -0.4494 C 0 0 0 0 0 0 0 0 0 0 0 0 4.4759 -1.2505 -1.5492 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8669 -1.7353 -2.7368 N 0 0 0 0 0 0 0 0 0 0 0 0 3.8199 -1.8913 -3.5132 N 0 0 0 0 0 0 0 0 0 0 0 0 2.7481 -1.5156 -2.8517 N 0 0 0 0 0 0 0 0 0 0 0 0 3.1587 -1.1190 -1.6348 N 0 0 0 0 0 0 0 0 0 0 0 0 3.0098 1.0634 1.0781 C 0 0 0 0 0 0 0 0 0 0 0 0 1.6985 1.4189 1.2933 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.9218 1.4452 -0.4675 C 0 0 2 0 0 0 0 0 0 0 0 0 -0.8498 1.5300 -1.4787 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.9414 2.3819 -2.3815 O 0 0 0 0 0 0 0 0 0 0 0 0 0.2273 0.6902 -1.4339 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.3922 2.7957 0.0052 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.6942 3.7419 -1.1081 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.6646 2.6400 0.8265 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.6399 -1.6632 -0.0394 H 0 0 0 0 0 0 0 0 0 0 0 0 -7.0342 -2.4928 -1.5585 H 0 0 0 0 0 0 0 0 0 0 0 0 -6.0590 -1.0185 -1.6701 H 0 0 0 0 0 0 0 0 0 0 0 0 -5.4056 -3.8044 -0.5814 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.6399 -2.9895 -1.9646 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2074 -3.1270 0.0604 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.4626 -2.3597 1.0407 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.9251 -1.0705 -1.3004 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.2246 -0.2413 -0.4069 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.0061 0.3259 2.6037 H 0 0 0 0 0 0 0 0 0 0 0 0 -0.6706 1.9126 1.9457 H 0 0 0 0 0 0 0 0 0 0 0 0 0.4348 -1.6972 1.2996 H 0 0 0 0 0 0 0 0 0 0 0 0 2.7814 -2.2629 0.9184 H 0 0 0 0 0 0 0 0 0 0 0 0 5.4013 0.1909 2.7015 H 0 0 0 0 0 0 0 0 0 0 0 0 7.7801 -0.1454 2.4665 H 0 0 0 0 0 0 0 0 0 0 0 0 8.7020 -1.0122 0.3517 H 0 0 0 0 0 0 0 0 0 0 0 0 7.1111 -1.5263 -1.5219 H 0 0 0 0 0 0 0 0 0 0 0 0 1.7630 -1.5331 -3.2294 H 0 0 0 0 0 0 0 0 0 0 0 0 3.7359 1.8855 1.0178 H 0 0 0 0 0 0 0 0 0 0 0 0 1.3883 2.4594 1.3996 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.7904 1.0165 -1.0741 H 0 0 0 0 0 0 0 0 0 0 0 0 0.6782 0.3828 -2.2955 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.6578 3.2897 0.6760 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.7840 4.3173 -1.3797 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.4028 4.5187 -0.6938 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2174 3.3023 -1.9687 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.6100 1.7410 1.4927 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.5060 2.6018 0.1267 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.7617 3.5370 1.4843 H 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 2 3 1 0 3 4 1 0 4 5 1 0 5 6 2 0 5 7 1 0 7 8 1 0 8 9 1 0 9 10 2 0 10 11 1 0 11 12 2 0 12 13 1 0 13 14 2 0 14 15 1 0 15 16 2 0 16 17 1 0 17 18 2 0 18 19 1 0 19 20 1 0 20 21 2 0 21 22 1 0 22 23 1 0 12 24 1 0 24 25 2 0 7 26 1 0 26 27 1 0 27 28 2 0 27 29 1 0 26 30 1 0 30 31 1 0 30 32 1 0 25 9 1 0 18 13 1 0 23 19 2 0 1 33 1 0 1 34 1 0 1 35 1 0 2 36 1 0 2 37 1 0 3 38 1 0 3 39 1 0 4 40 1 0 4 41 1 0 8 42 1 0 8 43 1 0 10 44 1 0 11 45 1 0 14 46 1 0 15 47 1 0 16 48 1 0 17 49 1 0 22 50 1 0 24 51 1 0 25 52 1 0 26 53 1 6 29 54 1 0 30 55 1 0 31 56 1 0 31 57 1 0 31 58 1 0 32 59 1 0 32 60 1 0 32 61 1 0 M END 177 Mrv0541 02231214292D 32 34 0 0 1 0 999 V2000 2.0930 1.2209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -0.0167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 0.8084 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 7.8559 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 9.1909 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.1109 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.9359 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 1.2209 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 3.5220 2.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -2.4917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 16 1 0 0 0 0 2 12 2 0 0 0 0 3 16 2 0 0 0 0 9 4 1 6 0 0 0 4 11 1 0 0 0 0 4 12 1 0 0 0 0 5 7 1 0 0 0 0 5 31 2 0 0 0 0 6 8 2 0 0 0 0 6 31 1 0 0 0 0 7 8 1 0 0 0 0 9 10 1 0 0 0 0 9 16 1 0 0 0 0 10 13 1 0 0 0 0 10 14 1 0 0 0 0 11 17 1 0 0 0 0 12 15 1 0 0 0 0 15 18 1 0 0 0 0 17 20 2 0 0 0 0 17 21 1 0 0 0 0 18 19 1 0 0 0 0 19 25 1 0 0 0 0 20 23 1 0 0 0 0 21 24 2 0 0 0 0 22 23 2 0 0 0 0 22 24 1 0 0 0 0 22 26 1 0 0 0 0 26 27 1 0 0 0 0 26 28 2 0 0 0 0 27 29 2 0 0 0 0 27 31 1 0 0 0 0 28 30 1 0 0 0 0 29 32 1 0 0 0 0 30 32 2 0 0 0 0 M END > <DATABASE_ID> HMDB0014323 > <DATABASE_NAME> hmdb > CCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)[C@@H](C(C)C)C(O)=O > <INCHI_IDENTIFIER> InChI=1S/C24H29N5O3/c1-4-5-10-21(30)29(22(16(2)3)24(31)32)15-17-11-13-18(14-12-17)19-8-6-7-9-20(19)23-25-27-28-26-23/h6-9,11-14,16,22H,4-5,10,15H2,1-3H3,(H,31,32)(H,25,26,27,28)/t22-/m0/s1 > <INCHI_KEY> ACWBQPMHZXGDFX-QFIPXVFZSA-N > C24H29N5O3 > <MOLECULAR_WEIGHT> 435.5188 > <EXACT_MASS> 435.227039819 > <JCHEM_ACCEPTOR_COUNT> 6 > <JCHEM_AVERAGE_POLARIZABILITY> 47.27314752925239 > <JCHEM_BIOAVAILABILITY> 1 > <JCHEM_DONOR_COUNT> 2 > <JCHEM_FORMAL_CHARGE> 0 > <JCHEM_GHOSE_FILTER> 0 > <JCHEM_IUPAC> (2S)-3-methyl-2-[N-({4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl}methyl)pentanamido]butanoic acid > <ALOGPS_LOGP> 3.68 > <JCHEM_LOGP> 5.2693789780000015 > <ALOGPS_LOGS> -4.27 > <JCHEM_MDDR_LIKE_RULE> 1 > <JCHEM_NUMBER_OF_RINGS> 3 > <JCHEM_PHYSIOLOGICAL_CHARGE> -1 > <JCHEM_PKA> 7.399818067465363 > <JCHEM_PKA_STRONGEST_ACIDIC> 4.366355827724827 > <JCHEM_PKA_STRONGEST_BASIC> -0.11251135131002898 > <JCHEM_POLAR_SURFACE_AREA> 112.07000000000001 > <JCHEM_REFRACTIVITY> 134.7733 > <JCHEM_ROTATABLE_BOND_COUNT> 10 > <JCHEM_RULE_OF_FIVE> 0 > <ALOGPS_SOLUBILITY> 2.34e-02 g/l > <JCHEM_TRADITIONAL_IUPAC> valsartan > <JCHEM_VEBER_RULE> 0 $$ HMDB0014323 RDKit 3D Valsartan 61 63 0 0 0 0 0 0 0 0999 V2000 -6.2147 -1.9040 -1.0518 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.0355 -2.8131 -0.9456 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.9909 -2.3573 0.0362 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.4229 -1.0028 -0.3172 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.4060 -0.6505 0.7095 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.2138 -1.4268 1.6804 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6351 0.5486 0.6299 N 0 0 0 0 0 0 0 0 0 0 0 0 -0.6676 0.8211 1.6323 C 0 0 0 0 0 0 0 0 0 0 0 0 0.7239 0.4141 1.3796 C 0 0 0 0 0 0 0 0 0 0 0 0 1.1455 -0.8857 1.2415 C 0 0 0 0 0 0 0 0 0 0 0 0 2.4895 -1.2208 1.0227 C 0 0 0 0 0 0 0 0 0 0 0 0 3.4365 -0.2537 0.9384 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8594 -0.4711 0.7246 C 0 0 0 0 0 0 0 0 0 0 0 0 5.7763 -0.1927 1.7553 C 0 0 0 0 0 0 0 0 0 0 0 0 7.1152 -0.3824 1.6223 C 0 0 0 0 0 0 0 0 0 0 0 0 7.6227 -0.8631 0.4484 C 0 0 0 0 0 0 0 0 0 0 0 0 6.7475 -1.1422 -0.5730 C 0 0 0 0 0 0 0 0 0 0 0 0 5.3580 -0.9504 -0.4494 C 0 0 0 0 0 0 0 0 0 0 0 0 4.4759 -1.2505 -1.5492 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8669 -1.7353 -2.7368 N 0 0 0 0 0 0 0 0 0 0 0 0 3.8199 -1.8913 -3.5132 N 0 0 0 0 0 0 0 0 0 0 0 0 2.7481 -1.5156 -2.8517 N 0 0 0 0 0 0 0 0 0 0 0 0 3.1587 -1.1190 -1.6348 N 0 0 0 0 0 0 0 0 0 0 0 0 3.0098 1.0634 1.0781 C 0 0 0 0 0 0 0 0 0 0 0 0 1.6985 1.4189 1.2933 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.9218 1.4452 -0.4675 C 0 0 2 0 0 0 0 0 0 0 0 0 -0.8498 1.5300 -1.4787 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.9414 2.3819 -2.3815 O 0 0 0 0 0 0 0 0 0 0 0 0 0.2273 0.6902 -1.4339 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.3922 2.7957 0.0052 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.6942 3.7419 -1.1081 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.6646 2.6400 0.8265 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.6399 -1.6632 -0.0394 H 0 0 0 0 0 0 0 0 0 0 0 0 -7.0342 -2.4928 -1.5585 H 0 0 0 0 0 0 0 0 0 0 0 0 -6.0590 -1.0185 -1.6701 H 0 0 0 0 0 0 0 0 0 0 0 0 -5.4056 -3.8044 -0.5814 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.6399 -2.9895 -1.9646 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2074 -3.1270 0.0604 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.4626 -2.3597 1.0407 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.9251 -1.0705 -1.3004 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.2246 -0.2413 -0.4069 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.0061 0.3259 2.6037 H 0 0 0 0 0 0 0 0 0 0 0 0 -0.6706 1.9126 1.9457 H 0 0 0 0 0 0 0 0 0 0 0 0 0.4348 -1.6972 1.2996 H 0 0 0 0 0 0 0 0 0 0 0 0 2.7814 -2.2629 0.9184 H 0 0 0 0 0 0 0 0 0 0 0 0 5.4013 0.1909 2.7015 H 0 0 0 0 0 0 0 0 0 0 0 0 7.7801 -0.1454 2.4665 H 0 0 0 0 0 0 0 0 0 0 0 0 8.7020 -1.0122 0.3517 H 0 0 0 0 0 0 0 0 0 0 0 0 7.1111 -1.5263 -1.5219 H 0 0 0 0 0 0 0 0 0 0 0 0 1.7630 -1.5331 -3.2294 H 0 0 0 0 0 0 0 0 0 0 0 0 3.7359 1.8855 1.0178 H 0 0 0 0 0 0 0 0 0 0 0 0 1.3883 2.4594 1.3996 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.7904 1.0165 -1.0741 H 0 0 0 0 0 0 0 0 0 0 0 0 0.6782 0.3828 -2.2955 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.6578 3.2897 0.6760 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.7840 4.3173 -1.3797 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.4028 4.5187 -0.6938 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2174 3.3023 -1.9687 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.6100 1.7410 1.4927 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.5060 2.6018 0.1267 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.7617 3.5370 1.4843 H 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 2 3 1 0 3 4 1 0 4 5 1 0 5 6 2 0 5 7 1 0 7 8 1 0 8 9 1 0 9 10 2 0 10 11 1 0 11 12 2 0 12 13 1 0 13 14 2 0 14 15 1 0 15 16 2 0 16 17 1 0 17 18 2 0 18 19 1 0 19 20 1 0 20 21 2 0 21 22 1 0 22 23 1 0 12 24 1 0 24 25 2 0 7 26 1 0 26 27 1 0 27 28 2 0 27 29 1 0 26 30 1 0 30 31 1 0 30 32 1 0 25 9 1 0 18 13 1 0 23 19 2 0 1 33 1 0 1 34 1 0 1 35 1 0 2 36 1 0 2 37 1 0 3 38 1 0 3 39 1 0 4 40 1 0 4 41 1 0 8 42 1 0 8 43 1 0 10 44 1 0 11 45 1 0 14 46 1 0 15 47 1 0 16 48 1 0 17 49 1 0 22 50 1 0 24 51 1 0 25 52 1 0 26 53 1 6 29 54 1 0 30 55 1 0 31 56 1 0 31 57 1 0 31 58 1 0 32 59 1 0 32 60 1 0 32 61 1 0 M END HEADER PROTEIN 23-FEB-12 NONE TITLE NULL COMPND MOLECULE: 177 SOURCE NULL KEYWDS NULL EXPDTA NULL AUTHOR Marvin REVDAT 1 23-FEB-12 0 HETATM 1 O UNK 0 3.907 2.279 0.000 0.00 0.00 O+0 HETATM 2 O UNK 0 9.242 -0.801 0.000 0.00 0.00 O+0 HETATM 3 O UNK 0 5.241 -0.031 0.000 0.00 0.00 O+0 HETATM 4 N UNK 0 7.908 1.509 0.000 0.00 0.00 N+0 HETATM 5 N UNK 0 14.664 2.414 0.000 0.00 0.00 N+0 HETATM 6 N UNK 0 17.156 2.414 0.000 0.00 0.00 N+0 HETATM 7 N UNK 0 15.140 3.879 0.000 0.00 0.00 N+0 HETATM 8 N UNK 0 16.680 3.879 0.000 0.00 0.00 N+0 HETATM 9 C UNK 0 6.574 2.279 0.000 0.00 0.00 C+0 HETATM 10 C UNK 0 6.574 3.819 0.000 0.00 0.00 C+0 HETATM 11 C UNK 0 9.242 2.279 0.000 0.00 0.00 C+0 HETATM 12 C UNK 0 7.908 -0.031 0.000 0.00 0.00 C+0 HETATM 13 C UNK 0 5.241 4.589 0.000 0.00 0.00 C+0 HETATM 14 C UNK 0 7.908 4.589 0.000 0.00 0.00 C+0 HETATM 15 C UNK 0 6.574 -0.801 0.000 0.00 0.00 C+0 HETATM 16 C UNK 0 5.241 1.509 0.000 0.00 0.00 C+0 HETATM 17 C UNK 0 10.576 1.509 0.000 0.00 0.00 C+0 HETATM 18 C UNK 0 6.574 -2.341 0.000 0.00 0.00 C+0 HETATM 19 C UNK 0 5.241 -3.111 0.000 0.00 0.00 C+0 HETATM 20 C UNK 0 11.909 2.279 0.000 0.00 0.00 C+0 HETATM 21 C UNK 0 10.576 -0.031 0.000 0.00 0.00 C+0 HETATM 22 C UNK 0 13.243 -0.031 0.000 0.00 0.00 C+0 HETATM 23 C UNK 0 13.243 1.509 0.000 0.00 0.00 C+0 HETATM 24 C UNK 0 11.909 -0.801 0.000 0.00 0.00 C+0 HETATM 25 C UNK 0 5.241 -4.651 0.000 0.00 0.00 C+0 HETATM 26 C UNK 0 14.577 -0.801 0.000 0.00 0.00 C+0 HETATM 27 C UNK 0 15.910 -0.031 0.000 0.00 0.00 C+0 HETATM 28 C UNK 0 14.577 -2.341 0.000 0.00 0.00 C+0 HETATM 29 C UNK 0 17.244 -0.801 0.000 0.00 0.00 C+0 HETATM 30 C UNK 0 15.910 -3.111 0.000 0.00 0.00 C+0 HETATM 31 C UNK 0 15.910 1.509 0.000 0.00 0.00 C+0 HETATM 32 C UNK 0 17.244 -2.341 0.000 0.00 0.00 C+0 CONECT 1 16 CONECT 2 12 CONECT 3 16 CONECT 4 9 11 12 CONECT 5 7 31 CONECT 6 8 31 CONECT 7 5 8 CONECT 8 6 7 CONECT 9 4 10 16 CONECT 10 9 13 14 CONECT 11 4 17 CONECT 12 2 4 15 CONECT 13 10 CONECT 14 10 CONECT 15 12 18 CONECT 16 1 3 9 CONECT 17 11 20 21 CONECT 18 15 19 CONECT 19 18 25 CONECT 20 17 23 CONECT 21 17 24 CONECT 22 23 24 26 CONECT 23 20 22 CONECT 24 21 22 CONECT 25 19 CONECT 26 22 27 28 CONECT 27 26 29 31 CONECT 28 26 30 CONECT 29 27 32 CONECT 30 28 32 CONECT 31 5 6 27 CONECT 32 29 30 MASTER 0 0 0 0 0 0 0 0 32 0 68 0 END COMPND HMDB0014323 HETATM 1 C1 UNL 1 -6.215 -1.904 -1.052 1.00 0.00 C HETATM 2 C2 UNL 1 -5.036 -2.813 -0.946 1.00 0.00 C HETATM 3 C3 UNL 1 -3.991 -2.357 0.036 1.00 0.00 C HETATM 4 C4 UNL 1 -3.423 -1.003 -0.317 1.00 0.00 C HETATM 5 C5 UNL 1 -2.406 -0.651 0.710 1.00 0.00 C HETATM 6 O1 UNL 1 -2.214 -1.427 1.680 1.00 0.00 O HETATM 7 N1 UNL 1 -1.635 0.549 0.630 1.00 0.00 N HETATM 8 C6 UNL 1 -0.668 0.821 1.632 1.00 0.00 C HETATM 9 C7 UNL 1 0.724 0.414 1.380 1.00 0.00 C HETATM 10 C8 UNL 1 1.145 -0.886 1.241 1.00 0.00 C HETATM 11 C9 UNL 1 2.489 -1.221 1.023 1.00 0.00 C HETATM 12 C10 UNL 1 3.436 -0.254 0.938 1.00 0.00 C HETATM 13 C11 UNL 1 4.859 -0.471 0.725 1.00 0.00 C HETATM 14 C12 UNL 1 5.776 -0.193 1.755 1.00 0.00 C HETATM 15 C13 UNL 1 7.115 -0.382 1.622 1.00 0.00 C HETATM 16 C14 UNL 1 7.623 -0.863 0.448 1.00 0.00 C HETATM 17 C15 UNL 1 6.748 -1.142 -0.573 1.00 0.00 C HETATM 18 C16 UNL 1 5.358 -0.950 -0.449 1.00 0.00 C HETATM 19 C17 UNL 1 4.476 -1.250 -1.549 1.00 0.00 C HETATM 20 N2 UNL 1 4.867 -1.735 -2.737 1.00 0.00 N HETATM 21 N3 UNL 1 3.820 -1.891 -3.513 1.00 0.00 N HETATM 22 N4 UNL 1 2.748 -1.516 -2.852 1.00 0.00 N HETATM 23 N5 UNL 1 3.159 -1.119 -1.635 1.00 0.00 N HETATM 24 C18 UNL 1 3.010 1.063 1.078 1.00 0.00 C HETATM 25 C19 UNL 1 1.699 1.419 1.293 1.00 0.00 C HETATM 26 C20 UNL 1 -1.922 1.445 -0.467 1.00 0.00 C HETATM 27 C21 UNL 1 -0.850 1.530 -1.479 1.00 0.00 C HETATM 28 O2 UNL 1 -0.941 2.382 -2.382 1.00 0.00 O HETATM 29 O3 UNL 1 0.227 0.690 -1.434 1.00 0.00 O HETATM 30 C22 UNL 1 -2.392 2.796 0.005 1.00 0.00 C HETATM 31 C23 UNL 1 -2.694 3.742 -1.108 1.00 0.00 C HETATM 32 C24 UNL 1 -3.665 2.640 0.826 1.00 0.00 C HETATM 33 H1 UNL 1 -6.640 -1.663 -0.039 1.00 0.00 H HETATM 34 H2 UNL 1 -7.034 -2.493 -1.559 1.00 0.00 H HETATM 35 H3 UNL 1 -6.059 -1.018 -1.670 1.00 0.00 H HETATM 36 H4 UNL 1 -5.406 -3.804 -0.581 1.00 0.00 H HETATM 37 H5 UNL 1 -4.640 -2.990 -1.965 1.00 0.00 H HETATM 38 H6 UNL 1 -3.207 -3.127 0.060 1.00 0.00 H HETATM 39 H7 UNL 1 -4.463 -2.360 1.041 1.00 0.00 H HETATM 40 H8 UNL 1 -2.925 -1.070 -1.300 1.00 0.00 H HETATM 41 H9 UNL 1 -4.225 -0.241 -0.407 1.00 0.00 H HETATM 42 H10 UNL 1 -1.006 0.326 2.604 1.00 0.00 H HETATM 43 H11 UNL 1 -0.671 1.913 1.946 1.00 0.00 H HETATM 44 H12 UNL 1 0.435 -1.697 1.300 1.00 0.00 H HETATM 45 H13 UNL 1 2.781 -2.263 0.918 1.00 0.00 H HETATM 46 H14 UNL 1 5.401 0.191 2.701 1.00 0.00 H HETATM 47 H15 UNL 1 7.780 -0.145 2.466 1.00 0.00 H HETATM 48 H16 UNL 1 8.702 -1.012 0.352 1.00 0.00 H HETATM 49 H17 UNL 1 7.111 -1.526 -1.522 1.00 0.00 H HETATM 50 H18 UNL 1 1.763 -1.533 -3.229 1.00 0.00 H HETATM 51 H19 UNL 1 3.736 1.885 1.018 1.00 0.00 H HETATM 52 H20 UNL 1 1.388 2.459 1.400 1.00 0.00 H HETATM 53 H21 UNL 1 -2.790 1.017 -1.074 1.00 0.00 H HETATM 54 H22 UNL 1 0.678 0.383 -2.296 1.00 0.00 H HETATM 55 H23 UNL 1 -1.658 3.290 0.676 1.00 0.00 H HETATM 56 H24 UNL 1 -1.784 4.317 -1.380 1.00 0.00 H HETATM 57 H25 UNL 1 -3.403 4.519 -0.694 1.00 0.00 H HETATM 58 H26 UNL 1 -3.217 3.302 -1.969 1.00 0.00 H HETATM 59 H27 UNL 1 -3.610 1.741 1.493 1.00 0.00 H HETATM 60 H28 UNL 1 -4.506 2.602 0.127 1.00 0.00 H HETATM 61 H29 UNL 1 -3.762 3.537 1.484 1.00 0.00 H CONECT 1 2 33 34 35 CONECT 2 3 36 37 CONECT 3 4 38 39 CONECT 4 5 40 41 CONECT 5 6 6 7 CONECT 7 8 26 CONECT 8 9 42 43 CONECT 9 10 10 25 CONECT 10 11 44 CONECT 11 12 12 45 CONECT 12 13 24 CONECT 13 14 14 18 CONECT 14 15 46 CONECT 15 16 16 47 CONECT 16 17 48 CONECT 17 18 18 49 CONECT 18 19 CONECT 19 20 23 23 CONECT 20 21 21 CONECT 21 22 CONECT 22 23 50 CONECT 24 25 25 51 CONECT 25 52 CONECT 26 27 30 53 CONECT 27 28 28 29 CONECT 29 54 CONECT 30 31 32 55 CONECT 31 56 57 58 CONECT 32 59 60 61 END CCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)[C@@H](C(C)C)C(O)=OInChI=1S/C24H29N5O3/c1-4-5-10-21(30)29(22(16(2)3)24(31)32)15-17-11-13-18(14-12-17)19-8-6-7-9-20(19)23-25-27-28-26-23/h6-9,11-14,16,22H,4-5,10,15H2,1-3H3,(H,31,32)(H,25,26,27,28)/t22-/m0/s1 MOL3D MOLSDF3D SDFPDB3D PDBSMILESInChI177 Mrv0541 02231214292D 32 34 0 0 1 0 999 V2000 2.0930 1.2209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -0.0167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 0.8084 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 7.8559 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 9.1909 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.1109 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.9359 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 1.2209 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 3.5220 2.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -2.4917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 16 1 0 0 0 0 2 12 2 0 0 0 0 3 16 2 0 0 0 0 9 4 1 6 0 0 0 4 11 1 0 0 0 0 4 12 1 0 0 0 0 5 7 1 0 0 0 0 5 31 2 0 0 0 0 6 8 2 0 0 0 0 6 31 1 0 0 0 0 7 8 1 0 0 0 0 9 10 1 0 0 0 0 9 16 1 0 0 0 0 10 13 1 0 0 0 0 10 14 1 0 0 0 0 11 17 1 0 0 0 0 12 15 1 0 0 0 0 15 18 1 0 0 0 0 17 20 2 0 0 0 0 17 21 1 0 0 0 0 18 19 1 0 0 0 0 19 25 1 0 0 0 0 20 23 1 0 0 0 0 21 24 2 0 0 0 0 22 23 2 0 0 0 0 22 24 1 0 0 0 0 22 26 1 0 0 0 0 26 27 1 0 0 0 0 26 28 2 0 0 0 0 27 29 2 0 0 0 0 27 31 1 0 0 0 0 28 30 1 0 0 0 0 29 32 1 0 0 0 0 30 32 2 0 0 0 0 M END HMDB0014323 RDKit 3D Valsartan 61 63 0 0 0 0 0 0 0 0999 V2000 -6.2147 -1.9040 -1.0518 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.0355 -2.8131 -0.9456 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.9909 -2.3573 0.0362 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.4229 -1.0028 -0.3172 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.4060 -0.6505 0.7095 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.2138 -1.4268 1.6804 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6351 0.5486 0.6299 N 0 0 0 0 0 0 0 0 0 0 0 0 -0.6676 0.8211 1.6323 C 0 0 0 0 0 0 0 0 0 0 0 0 0.7239 0.4141 1.3796 C 0 0 0 0 0 0 0 0 0 0 0 0 1.1455 -0.8857 1.2415 C 0 0 0 0 0 0 0 0 0 0 0 0 2.4895 -1.2208 1.0227 C 0 0 0 0 0 0 0 0 0 0 0 0 3.4365 -0.2537 0.9384 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8594 -0.4711 0.7246 C 0 0 0 0 0 0 0 0 0 0 0 0 5.7763 -0.1927 1.7553 C 0 0 0 0 0 0 0 0 0 0 0 0 7.1152 -0.3824 1.6223 C 0 0 0 0 0 0 0 0 0 0 0 0 7.6227 -0.8631 0.4484 C 0 0 0 0 0 0 0 0 0 0 0 0 6.7475 -1.1422 -0.5730 C 0 0 0 0 0 0 0 0 0 0 0 0 5.3580 -0.9504 -0.4494 C 0 0 0 0 0 0 0 0 0 0 0 0 4.4759 -1.2505 -1.5492 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8669 -1.7353 -2.7368 N 0 0 0 0 0 0 0 0 0 0 0 0 3.8199 -1.8913 -3.5132 N 0 0 0 0 0 0 0 0 0 0 0 0 2.7481 -1.5156 -2.8517 N 0 0 0 0 0 0 0 0 0 0 0 0 3.1587 -1.1190 -1.6348 N 0 0 0 0 0 0 0 0 0 0 0 0 3.0098 1.0634 1.0781 C 0 0 0 0 0 0 0 0 0 0 0 0 1.6985 1.4189 1.2933 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.9218 1.4452 -0.4675 C 0 0 2 0 0 0 0 0 0 0 0 0 -0.8498 1.5300 -1.4787 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.9414 2.3819 -2.3815 O 0 0 0 0 0 0 0 0 0 0 0 0 0.2273 0.6902 -1.4339 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.3922 2.7957 0.0052 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.6942 3.7419 -1.1081 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.6646 2.6400 0.8265 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.6399 -1.6632 -0.0394 H 0 0 0 0 0 0 0 0 0 0 0 0 -7.0342 -2.4928 -1.5585 H 0 0 0 0 0 0 0 0 0 0 0 0 -6.0590 -1.0185 -1.6701 H 0 0 0 0 0 0 0 0 0 0 0 0 -5.4056 -3.8044 -0.5814 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.6399 -2.9895 -1.9646 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2074 -3.1270 0.0604 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.4626 -2.3597 1.0407 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.9251 -1.0705 -1.3004 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.2246 -0.2413 -0.4069 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.0061 0.3259 2.6037 H 0 0 0 0 0 0 0 0 0 0 0 0 -0.6706 1.9126 1.9457 H 0 0 0 0 0 0 0 0 0 0 0 0 0.4348 -1.6972 1.2996 H 0 0 0 0 0 0 0 0 0 0 0 0 2.7814 -2.2629 0.9184 H 0 0 0 0 0 0 0 0 0 0 0 0 5.4013 0.1909 2.7015 H 0 0 0 0 0 0 0 0 0 0 0 0 7.7801 -0.1454 2.4665 H 0 0 0 0 0 0 0 0 0 0 0 0 8.7020 -1.0122 0.3517 H 0 0 0 0 0 0 0 0 0 0 0 0 7.1111 -1.5263 -1.5219 H 0 0 0 0 0 0 0 0 0 0 0 0 1.7630 -1.5331 -3.2294 H 0 0 0 0 0 0 0 0 0 0 0 0 3.7359 1.8855 1.0178 H 0 0 0 0 0 0 0 0 0 0 0 0 1.3883 2.4594 1.3996 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.7904 1.0165 -1.0741 H 0 0 0 0 0 0 0 0 0 0 0 0 0.6782 0.3828 -2.2955 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.6578 3.2897 0.6760 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.7840 4.3173 -1.3797 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.4028 4.5187 -0.6938 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2174 3.3023 -1.9687 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.6100 1.7410 1.4927 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.5060 2.6018 0.1267 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.7617 3.5370 1.4843 H 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 2 3 1 0 3 4 1 0 4 5 1 0 5 6 2 0 5 7 1 0 7 8 1 0 8 9 1 0 9 10 2 0 10 11 1 0 11 12 2 0 12 13 1 0 13 14 2 0 14 15 1 0 15 16 2 0 16 17 1 0 17 18 2 0 18 19 1 0 19 20 1 0 20 21 2 0 21 22 1 0 22 23 1 0 12 24 1 0 24 25 2 0 7 26 1 0 26 27 1 0 27 28 2 0 27 29 1 0 26 30 1 0 30 31 1 0 30 32 1 0 25 9 1 0 18 13 1 0 23 19 2 0 1 33 1 0 1 34 1 0 1 35 1 0 2 36 1 0 2 37 1 0 3 38 1 0 3 39 1 0 4 40 1 0 4 41 1 0 8 42 1 0 8 43 1 0 10 44 1 0 11 45 1 0 14 46 1 0 15 47 1 0 16 48 1 0 17 49 1 0 22 50 1 0 24 51 1 0 25 52 1 0 26 53 1 6 29 54 1 0 30 55 1 0 31 56 1 0 31 57 1 0 31 58 1 0 32 59 1 0 32 60 1 0 32 61 1 0 M END 177 Mrv0541 02231214292D 32 34 0 0 1 0 999 V2000 2.0930 1.2209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 -0.4292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -0.0167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 0.8084 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 7.8559 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 9.1909 1.2933 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.1109 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 8.9359 2.0779 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 1.2209 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 3.5220 2.0459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.9509 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 4.2365 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 3.5220 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 1.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 5.6655 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.0944 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 6.3799 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 2.8075 -2.4917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -0.0167 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 7.8089 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -0.4292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 -1.6667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 8.5234 0.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 9.2379 -1.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 16 1 0 0 0 0 2 12 2 0 0 0 0 3 16 2 0 0 0 0 9 4 1 6 0 0 0 4 11 1 0 0 0 0 4 12 1 0 0 0 0 5 7 1 0 0 0 0 5 31 2 0 0 0 0 6 8 2 0 0 0 0 6 31 1 0 0 0 0 7 8 1 0 0 0 0 9 10 1 0 0 0 0 9 16 1 0 0 0 0 10 13 1 0 0 0 0 10 14 1 0 0 0 0 11 17 1 0 0 0 0 12 15 1 0 0 0 0 15 18 1 0 0 0 0 17 20 2 0 0 0 0 17 21 1 0 0 0 0 18 19 1 0 0 0 0 19 25 1 0 0 0 0 20 23 1 0 0 0 0 21 24 2 0 0 0 0 22 23 2 0 0 0 0 22 24 1 0 0 0 0 22 26 1 0 0 0 0 26 27 1 0 0 0 0 26 28 2 0 0 0 0 27 29 2 0 0 0 0 27 31 1 0 0 0 0 28 30 1 0 0 0 0 29 32 1 0 0 0 0 30 32 2 0 0 0 0 M END > <DATABASE_ID> HMDB0014323 > <DATABASE_NAME> hmdb > CCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)[C@@H](C(C)C)C(O)=O > <INCHI_IDENTIFIER> InChI=1S/C24H29N5O3/c1-4-5-10-21(30)29(22(16(2)3)24(31)32)15-17-11-13-18(14-12-17)19-8-6-7-9-20(19)23-25-27-28-26-23/h6-9,11-14,16,22H,4-5,10,15H2,1-3H3,(H,31,32)(H,25,26,27,28)/t22-/m0/s1 > <INCHI_KEY> ACWBQPMHZXGDFX-QFIPXVFZSA-N > C24H29N5O3 > <MOLECULAR_WEIGHT> 435.5188 > <EXACT_MASS> 435.227039819 > <JCHEM_ACCEPTOR_COUNT> 6 > <JCHEM_AVERAGE_POLARIZABILITY> 47.27314752925239 > <JCHEM_BIOAVAILABILITY> 1 > <JCHEM_DONOR_COUNT> 2 > <JCHEM_FORMAL_CHARGE> 0 > <JCHEM_GHOSE_FILTER> 0 > <JCHEM_IUPAC> (2S)-3-methyl-2-[N-({4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl}methyl)pentanamido]butanoic acid > <ALOGPS_LOGP> 3.68 > <JCHEM_LOGP> 5.2693789780000015 > <ALOGPS_LOGS> -4.27 > <JCHEM_MDDR_LIKE_RULE> 1 > <JCHEM_NUMBER_OF_RINGS> 3 > <JCHEM_PHYSIOLOGICAL_CHARGE> -1 > <JCHEM_PKA> 7.399818067465363 > <JCHEM_PKA_STRONGEST_ACIDIC> 4.366355827724827 > <JCHEM_PKA_STRONGEST_BASIC> -0.11251135131002898 > <JCHEM_POLAR_SURFACE_AREA> 112.07000000000001 > <JCHEM_REFRACTIVITY> 134.7733 > <JCHEM_ROTATABLE_BOND_COUNT> 10 > <JCHEM_RULE_OF_FIVE> 0 > <ALOGPS_SOLUBILITY> 2.34e-02 g/l > <JCHEM_TRADITIONAL_IUPAC> valsartan > <JCHEM_VEBER_RULE> 0 $$ HMDB0014323 RDKit 3D Valsartan 61 63 0 0 0 0 0 0 0 0999 V2000 -6.2147 -1.9040 -1.0518 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.0355 -2.8131 -0.9456 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.9909 -2.3573 0.0362 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.4229 -1.0028 -0.3172 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.4060 -0.6505 0.7095 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.2138 -1.4268 1.6804 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6351 0.5486 0.6299 N 0 0 0 0 0 0 0 0 0 0 0 0 -0.6676 0.8211 1.6323 C 0 0 0 0 0 0 0 0 0 0 0 0 0.7239 0.4141 1.3796 C 0 0 0 0 0 0 0 0 0 0 0 0 1.1455 -0.8857 1.2415 C 0 0 0 0 0 0 0 0 0 0 0 0 2.4895 -1.2208 1.0227 C 0 0 0 0 0 0 0 0 0 0 0 0 3.4365 -0.2537 0.9384 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8594 -0.4711 0.7246 C 0 0 0 0 0 0 0 0 0 0 0 0 5.7763 -0.1927 1.7553 C 0 0 0 0 0 0 0 0 0 0 0 0 7.1152 -0.3824 1.6223 C 0 0 0 0 0 0 0 0 0 0 0 0 7.6227 -0.8631 0.4484 C 0 0 0 0 0 0 0 0 0 0 0 0 6.7475 -1.1422 -0.5730 C 0 0 0 0 0 0 0 0 0 0 0 0 5.3580 -0.9504 -0.4494 C 0 0 0 0 0 0 0 0 0 0 0 0 4.4759 -1.2505 -1.5492 C 0 0 0 0 0 0 0 0 0 0 0 0 4.8669 -1.7353 -2.7368 N 0 0 0 0 0 0 0 0 0 0 0 0 3.8199 -1.8913 -3.5132 N 0 0 0 0 0 0 0 0 0 0 0 0 2.7481 -1.5156 -2.8517 N 0 0 0 0 0 0 0 0 0 0 0 0 3.1587 -1.1190 -1.6348 N 0 0 0 0 0 0 0 0 0 0 0 0 3.0098 1.0634 1.0781 C 0 0 0 0 0 0 0 0 0 0 0 0 1.6985 1.4189 1.2933 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.9218 1.4452 -0.4675 C 0 0 2 0 0 0 0 0 0 0 0 0 -0.8498 1.5300 -1.4787 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.9414 2.3819 -2.3815 O 0 0 0 0 0 0 0 0 0 0 0 0 0.2273 0.6902 -1.4339 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.3922 2.7957 0.0052 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.6942 3.7419 -1.1081 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.6646 2.6400 0.8265 C 0 0 0 0 0 0 0 0 0 0 0 0 -6.6399 -1.6632 -0.0394 H 0 0 0 0 0 0 0 0 0 0 0 0 -7.0342 -2.4928 -1.5585 H 0 0 0 0 0 0 0 0 0 0 0 0 -6.0590 -1.0185 -1.6701 H 0 0 0 0 0 0 0 0 0 0 0 0 -5.4056 -3.8044 -0.5814 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.6399 -2.9895 -1.9646 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2074 -3.1270 0.0604 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.4626 -2.3597 1.0407 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.9251 -1.0705 -1.3004 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.2246 -0.2413 -0.4069 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.0061 0.3259 2.6037 H 0 0 0 0 0 0 0 0 0 0 0 0 -0.6706 1.9126 1.9457 H 0 0 0 0 0 0 0 0 0 0 0 0 0.4348 -1.6972 1.2996 H 0 0 0 0 0 0 0 0 0 0 0 0 2.7814 -2.2629 0.9184 H 0 0 0 0 0 0 0 0 0 0 0 0 5.4013 0.1909 2.7015 H 0 0 0 0 0 0 0 0 0 0 0 0 7.7801 -0.1454 2.4665 H 0 0 0 0 0 0 0 0 0 0 0 0 8.7020 -1.0122 0.3517 H 0 0 0 0 0 0 0 0 0 0 0 0 7.1111 -1.5263 -1.5219 H 0 0 0 0 0 0 0 0 0 0 0 0 1.7630 -1.5331 -3.2294 H 0 0 0 0 0 0 0 0 0 0 0 0 3.7359 1.8855 1.0178 H 0 0 0 0 0 0 0 0 0 0 0 0 1.3883 2.4594 1.3996 H 0 0 0 0 0 0 0 0 0 0 0 0 -2.7904 1.0165 -1.0741 H 0 0 0 0 0 0 0 0 0 0 0 0 0.6782 0.3828 -2.2955 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.6578 3.2897 0.6760 H 0 0 0 0 0 0 0 0 0 0 0 0 -1.7840 4.3173 -1.3797 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.4028 4.5187 -0.6938 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.2174 3.3023 -1.9687 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.6100 1.7410 1.4927 H 0 0 0 0 0 0 0 0 0 0 0 0 -4.5060 2.6018 0.1267 H 0 0 0 0 0 0 0 0 0 0 0 0 -3.7617 3.5370 1.4843 H 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 2 3 1 0 3 4 1 0 4 5 1 0 5 6 2 0 5 7 1 0 7 8 1 0 8 9 1 0 9 10 2 0 10 11 1 0 11 12 2 0 12 13 1 0 13 14 2 0 14 15 1 0 15 16 2 0 16 17 1 0 17 18 2 0 18 19 1 0 19 20 1 0 20 21 2 0 21 22 1 0 22 23 1 0 12 24 1 0 24 25 2 0 7 26 1 0 26 27 1 0 27 28 2 0 27 29 1 0 26 30 1 0 30 31 1 0 30 32 1 0 25 9 1 0 18 13 1 0 23 19 2 0 1 33 1 0 1 34 1 0 1 35 1 0 2 36 1 0 2 37 1 0 3 38 1 0 3 39 1 0 4 40 1 0 4 41 1 0 8 42 1 0 8 43 1 0 10 44 1 0 11 45 1 0 14 46 1 0 15 47 1 0 16 48 1 0 17 49 1 0 22 50 1 0 24 51 1 0 25 52 1 0 26 53 1 6 29 54 1 0 30 55 1 0 31 56 1 0 31 57 1 0 31 58 1 0 32 59 1 0 32 60 1 0 32 61 1 0 M END HEADER PROTEIN 23-FEB-12 NONE TITLE NULL COMPND MOLECULE: 177 SOURCE NULL KEYWDS NULL EXPDTA NULL AUTHOR Marvin REVDAT 1 23-FEB-12 0 HETATM 1 O UNK 0 3.907 2.279 0.000 0.00 0.00 O+0 HETATM 2 O UNK 0 9.242 -0.801 0.000 0.00 0.00 O+0 HETATM 3 O UNK 0 5.241 -0.031 0.000 0.00 0.00 O+0 HETATM 4 N UNK 0 7.908 1.509 0.000 0.00 0.00 N+0 HETATM 5 N UNK 0 14.664 2.414 0.000 0.00 0.00 N+0 HETATM 6 N UNK 0 17.156 2.414 0.000 0.00 0.00 N+0 HETATM 7 N UNK 0 15.140 3.879 0.000 0.00 0.00 N+0 HETATM 8 N UNK 0 16.680 3.879 0.000 0.00 0.00 N+0 HETATM 9 C UNK 0 6.574 2.279 0.000 0.00 0.00 C+0 HETATM 10 C UNK 0 6.574 3.819 0.000 0.00 0.00 C+0 HETATM 11 C UNK 0 9.242 2.279 0.000 0.00 0.00 C+0 HETATM 12 C UNK 0 7.908 -0.031 0.000 0.00 0.00 C+0 HETATM 13 C UNK 0 5.241 4.589 0.000 0.00 0.00 C+0 HETATM 14 C UNK 0 7.908 4.589 0.000 0.00 0.00 C+0 HETATM 15 C UNK 0 6.574 -0.801 0.000 0.00 0.00 C+0 HETATM 16 C UNK 0 5.241 1.509 0.000 0.00 0.00 C+0 HETATM 17 C UNK 0 10.576 1.509 0.000 0.00 0.00 C+0 HETATM 18 C UNK 0 6.574 -2.341 0.000 0.00 0.00 C+0 HETATM 19 C UNK 0 5.241 -3.111 0.000 0.00 0.00 C+0 HETATM 20 C UNK 0 11.909 2.279 0.000 0.00 0.00 C+0 HETATM 21 C UNK 0 10.576 -0.031 0.000 0.00 0.00 C+0 HETATM 22 C UNK 0 13.243 -0.031 0.000 0.00 0.00 C+0 HETATM 23 C UNK 0 13.243 1.509 0.000 0.00 0.00 C+0 HETATM 24 C UNK 0 11.909 -0.801 0.000 0.00 0.00 C+0 HETATM 25 C UNK 0 5.241 -4.651 0.000 0.00 0.00 C+0 HETATM 26 C UNK 0 14.577 -0.801 0.000 0.00 0.00 C+0 HETATM 27 C UNK 0 15.910 -0.031 0.000 0.00 0.00 C+0 HETATM 28 C UNK 0 14.577 -2.341 0.000 0.00 0.00 C+0 HETATM 29 C UNK 0 17.244 -0.801 0.000 0.00 0.00 C+0 HETATM 30 C UNK 0 15.910 -3.111 0.000 0.00 0.00 C+0 HETATM 31 C UNK 0 15.910 1.509 0.000 0.00 0.00 C+0 HETATM 32 C UNK 0 17.244 -2.341 0.000 0.00 0.00 C+0 CONECT 1 16 CONECT 2 12 CONECT 3 16 CONECT 4 9 11 12 CONECT 5 7 31 CONECT 6 8 31 CONECT 7 5 8 CONECT 8 6 7 CONECT 9 4 10 16 CONECT 10 9 13 14 CONECT 11 4 17 CONECT 12 2 4 15 CONECT 13 10 CONECT 14 10 CONECT 15 12 18 CONECT 16 1 3 9 CONECT 17 11 20 21 CONECT 18 15 19 CONECT 19 18 25 CONECT 20 17 23 CONECT 21 17 24 CONECT 22 23 24 26 CONECT 23 20 22 CONECT 24 21 22 CONECT 25 19 CONECT 26 22 27 28 CONECT 27 26 29 31 CONECT 28 26 30 CONECT 29 27 32 CONECT 30 28 32 CONECT 31 5 6 27 CONECT 32 29 30 MASTER 0 0 0 0 0 0 0 0 32 0 68 0 END COMPND HMDB0014323 HETATM 1 C1 UNL 1 -6.215 -1.904 -1.052 1.00 0.00 C HETATM 2 C2 UNL 1 -5.036 -2.813 -0.946 1.00 0.00 C HETATM 3 C3 UNL 1 -3.991 -2.357 0.036 1.00 0.00 C HETATM 4 C4 UNL 1 -3.423 -1.003 -0.317 1.00 0.00 C HETATM 5 C5 UNL 1 -2.406 -0.651 0.710 1.00 0.00 C HETATM 6 O1 UNL 1 -2.214 -1.427 1.680 1.00 0.00 O HETATM 7 N1 UNL 1 -1.635 0.549 0.630 1.00 0.00 N HETATM 8 C6 UNL 1 -0.668 0.821 1.632 1.00 0.00 C HETATM 9 C7 UNL 1 0.724 0.414 1.380 1.00 0.00 C HETATM 10 C8 UNL 1 1.145 -0.886 1.241 1.00 0.00 C HETATM 11 C9 UNL 1 2.489 -1.221 1.023 1.00 0.00 C HETATM 12 C10 UNL 1 3.436 -0.254 0.938 1.00 0.00 C HETATM 13 C11 UNL 1 4.859 -0.471 0.725 1.00 0.00 C HETATM 14 C12 UNL 1 5.776 -0.193 1.755 1.00 0.00 C HETATM 15 C13 UNL 1 7.115 -0.382 1.622 1.00 0.00 C HETATM 16 C14 UNL 1 7.623 -0.863 0.448 1.00 0.00 C HETATM 17 C15 UNL 1 6.748 -1.142 -0.573 1.00 0.00 C HETATM 18 C16 UNL 1 5.358 -0.950 -0.449 1.00 0.00 C HETATM 19 C17 UNL 1 4.476 -1.250 -1.549 1.00 0.00 C HETATM 20 N2 UNL 1 4.867 -1.735 -2.737 1.00 0.00 N HETATM 21 N3 UNL 1 3.820 -1.891 -3.513 1.00 0.00 N HETATM 22 N4 UNL 1 2.748 -1.516 -2.852 1.00 0.00 N HETATM 23 N5 UNL 1 3.159 -1.119 -1.635 1.00 0.00 N HETATM 24 C18 UNL 1 3.010 1.063 1.078 1.00 0.00 C HETATM 25 C19 UNL 1 1.699 1.419 1.293 1.00 0.00 C HETATM 26 C20 UNL 1 -1.922 1.445 -0.467 1.00 0.00 C HETATM 27 C21 UNL 1 -0.850 1.530 -1.479 1.00 0.00 C HETATM 28 O2 UNL 1 -0.941 2.382 -2.382 1.00 0.00 O HETATM 29 O3 UNL 1 0.227 0.690 -1.434 1.00 0.00 O HETATM 30 C22 UNL 1 -2.392 2.796 0.005 1.00 0.00 C HETATM 31 C23 UNL 1 -2.694 3.742 -1.108 1.00 0.00 C HETATM 32 C24 UNL 1 -3.665 2.640 0.826 1.00 0.00 C HETATM 33 H1 UNL 1 -6.640 -1.663 -0.039 1.00 0.00 H HETATM 34 H2 UNL 1 -7.034 -2.493 -1.559 1.00 0.00 H HETATM 35 H3 UNL 1 -6.059 -1.018 -1.670 1.00 0.00 H HETATM 36 H4 UNL 1 -5.406 -3.804 -0.581 1.00 0.00 H HETATM 37 H5 UNL 1 -4.640 -2.990 -1.965 1.00 0.00 H HETATM 38 H6 UNL 1 -3.207 -3.127 0.060 1.00 0.00 H HETATM 39 H7 UNL 1 -4.463 -2.360 1.041 1.00 0.00 H HETATM 40 H8 UNL 1 -2.925 -1.070 -1.300 1.00 0.00 H HETATM 41 H9 UNL 1 -4.225 -0.241 -0.407 1.00 0.00 H HETATM 42 H10 UNL 1 -1.006 0.326 2.604 1.00 0.00 H HETATM 43 H11 UNL 1 -0.671 1.913 1.946 1.00 0.00 H HETATM 44 H12 UNL 1 0.435 -1.697 1.300 1.00 0.00 H HETATM 45 H13 UNL 1 2.781 -2.263 0.918 1.00 0.00 H HETATM 46 H14 UNL 1 5.401 0.191 2.701 1.00 0.00 H HETATM 47 H15 UNL 1 7.780 -0.145 2.466 1.00 0.00 H HETATM 48 H16 UNL 1 8.702 -1.012 0.352 1.00 0.00 H HETATM 49 H17 UNL 1 7.111 -1.526 -1.522 1.00 0.00 H HETATM 50 H18 UNL 1 1.763 -1.533 -3.229 1.00 0.00 H HETATM 51 H19 UNL 1 3.736 1.885 1.018 1.00 0.00 H HETATM 52 H20 UNL 1 1.388 2.459 1.400 1.00 0.00 H HETATM 53 H21 UNL 1 -2.790 1.017 -1.074 1.00 0.00 H HETATM 54 H22 UNL 1 0.678 0.383 -2.296 1.00 0.00 H HETATM 55 H23 UNL 1 -1.658 3.290 0.676 1.00 0.00 H HETATM 56 H24 UNL 1 -1.784 4.317 -1.380 1.00 0.00 H HETATM 57 H25 UNL 1 -3.403 4.519 -0.694 1.00 0.00 H HETATM 58 H26 UNL 1 -3.217 3.302 -1.969 1.00 0.00 H HETATM 59 H27 UNL 1 -3.610 1.741 1.493 1.00 0.00 H HETATM 60 H28 UNL 1 -4.506 2.602 0.127 1.00 0.00 H HETATM 61 H29 UNL 1 -3.762 3.537 1.484 1.00 0.00 H CONECT 1 2 33 34 35 CONECT 2 3 36 37 CONECT 3 4 38 39 CONECT 4 5 40 41 CONECT 5 6 6 7 CONECT 7 8 26 CONECT 8 9 42 43 CONECT 9 10 10 25 CONECT 10 11 44 CONECT 11 12 12 45 CONECT 12 13 24 CONECT 13 14 14 18 CONECT 14 15 46 CONECT 15 16 16 47 CONECT 16 17 48 CONECT 17 18 18 49 CONECT 18 19 CONECT 19 20 23 23 CONECT 20 21 21 CONECT 21 22 CONECT 22 23 50 CONECT 24 25 25 51 CONECT 25 52 CONECT 26 27 30 53 CONECT 27 28 28 29 CONECT 29 54 CONECT 30 31 32 55 CONECT 31 56 57 58 CONECT 32 59 60 61 END CCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)[C@@H](C(C)C)C(O)=OInChI=1S/C24H29N5O3/c1-4-5-10-21(30)29(22(16(2)3)24(31)32)15-17-11-13-18(14-12-17)19-8-6-7-9-20(19)23-25-27-28-26-23/h6-9,11-14,16,22H,4-5,10,15H2,1-3H3,(H,31,32)(H,25,26,27,28)/t22-/m0/s1 View in JSmolView Stereo Labels View in JSmolView Stereo Labels |
| Synonyms |
ValueSource(S)-N-Valeryl-N-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]-methyl}-valineChEBIDiovanChEBIN-(p-(O-1H-Tetrazol-5-ylphenyl)benzyl)-N-valeryl-L-valineChEBIN-Pentanoyl-N-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl}-L-valineChEBIValsHMDBN-Valeryl-N-((2'-(1H-tetrazol-5-yl)biphenyl-4-yl)methyl)valineHMDBTaregHMDBKalpressHMDBMitenHMDBProvasHMDBNisisHMDB |
| Chemical Formula |
C24H29N5O3 |
| Average Molecular Weight |
435.5188 |
| Monoisotopic Molecular Weight |
435.227039819 |
| IUPAC Name |
(2S)-3-methyl-2-[N-({4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl}methyl)pentanamido]butanoic acid |
| Traditional Name |
valsartan |
| CAS Registry Number |
137862-53-4 |
| SMILES |
CCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)[C@@H](C(C)C)C(O)=O |
| InChI Identifier |
InChI=1S/C24H29N5O3/c1-4-5-10-21(30)29(22(16(2)3)24(31)32)15-17-11-13-18(14-12-17)19-8-6-7-9-20(19)23-25-27-28-26-23/h6-9,11-14,16,22H,4-5,10,15H2,1-3H3,(H,31,32)(H,25,26,27,28)/t22-/m0/s1 |
| InChI Key |
ACWBQPMHZXGDFX-QFIPXVFZSA-N |
| Chemical Taxonomy |
|
| Description |
Belongs to the class of organic compounds known as valine and derivatives. Valine and derivatives are compounds containing valine or a derivative thereof resulting from reaction of valine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
| Kingdom |
Organic compounds |
| Super Class |
Organic acids and derivatives |
| Class |
Carboxylic acids and derivatives |
| Sub Class |
Amino acids, peptides, and analogues |
| Direct Parent |
Valine and derivatives |
| Alternative Parents |
N-acyl-L-alpha-amino acids Biphenyls and derivatives Phenyltetrazoles and derivatives N-acyl amines Tertiary carboxylic acid amides Heteroaromatic compounds Monocarboxylic acids and derivatives Carboxylic acids Azacyclic compounds Organopnictogen compounds Organonitrogen compounds Organic oxides Hydrocarbon derivatives Carbonyl compounds |
| Substituents |
N-acyl-alpha-amino acid Valine or derivatives N-acyl-alpha amino acid or derivatives N-acyl-l-alpha-amino acid Biphenyl Phenyltetrazole Monocyclic benzene moiety N-acyl-amine Benzenoid Azole Heteroaromatic compound Tertiary carboxylic acid amide Tetrazole Carboxamide group Monocarboxylic acid or derivatives Carboxylic acid Azacycle Organoheterocyclic compound Organooxygen compound Organonitrogen compound Hydrocarbon derivative Organic oxide Organopnictogen compound Carbonyl group Organic nitrogen compound Organic oxygen compound Aromatic heteromonocyclic compound |
| Molecular Framework |
Aromatic heteromonocyclic compounds |
| External Descriptors |
monocarboxylic acid (CHEBI:9927 )monocarboxylic acid amide (CHEBI:9927 )biphenylyltetrazole (CHEBI:9927 ) |
| Ontology |
|
| Physiological effect |
Not Available |
| Disposition |
Biological locationExcretaBiofluid or ExcretaUrine (PMID: 21059682)Non-excretory biofluidBiofluid or ExcretaBlood (PMID: 21059682)Cellular substructureExtracellular (HMDB: HMDB0014323)Membrane (HMDB: HMDB0014323) |
| Process |
Naturally occurring processBiological processBiochemical pathwayDrug action pathwayValsartan Action Pathway (PathBank: SMP0000165) |
| Role |
Not Available |
| Physical Properties |
|
| State |
Solid |
| Experimental Molecular Properties |
PropertyValueReferenceMelting Point116 - 117 °CNot AvailableBoiling PointNot AvailableNot AvailableWater Solubility0.023 g/LNot AvailableLogP5.8Not Available |
| Experimental Chromatographic Properties |
Experimental Collision Cross SectionsAdduct TypeData SourceCCS Value (Å2)Reference[M+H]+Not Available202.007http://allccs.zhulab.cn/database/detail?ID=AllCCS00001280 |
| Predicted Molecular Properties |
PropertyValueSourceWater Solubility0.023 g/LALOGPSlogP3.68ALOGPSlogP5.27ChemAxonlogS-4.3ALOGPSpKa (Strongest Acidic)4.37ChemAxonpKa (Strongest Basic)-0.11ChemAxonPhysiological Charge-1ChemAxonHydrogen Acceptor Count6ChemAxonHydrogen Donor Count2ChemAxonPolar Surface Area112.07 ŲChemAxonRotatable Bond Count10ChemAxonRefractivity134.77 m³·mol⁻¹ChemAxonPolarizability47.27 ųChemAxonNumber of Rings3ChemAxonBioavailabilityYesChemAxonRule of FiveNoChemAxonGhose FilterNoChemAxonVeber's RuleNoChemAxonMDDR-like RuleYesChemAxon |
| Predicted Chromatographic Properties |
Predicted Collision Cross SectionsPredictorAdduct TypeCCS Value (Å2)ReferenceDarkChem[M+H]+202.5431661259 DarkChem[M-H]-202.04231661259 DeepCCS[M+H]+204.74230932474 DeepCCS[M-H]-202.34630932474 DeepCCS[M-2H]-235.22930932474 DeepCCS[M+Na]+211.3230932474 AllCCS[M+H]+207.832859911 AllCCS[M+H-H2O]+205.632859911 AllCCS[M+NH4]+209.832859911 AllCCS[M+Na]+210.432859911 AllCCS[M-H]-203.932859911 AllCCS[M+Na-2H]-204.832859911 AllCCS[M+HCOO]-206.032859911 Predicted Kovats Retention IndicesUnderivatizedMetaboliteSMILESKovats RI ValueColumn TypeReferenceValsartanCCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)C@@HC(O)=O4361.3Standard polar33892256 ValsartanCCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)C@@HC(O)=O3237.6Standard non polar33892256 ValsartanCCCCC(=O)N(CC1=CC=C(C=C1)C1=CC=CC=C1C1=NNN=N1)C@@HC(O)=O3559.8Semi standard non polar33892256 DerivatizedDerivative Name / StructureSMILESKovats RI ValueColumn TypeReferenceValsartan,1TMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=N[NH]N=N2)C=C1)C@HC(C)C3416.4Semi standard non polar33892256 Valsartan,1TMS,isomer #2CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C)N=N2)C=C1)C@HC(C)C3626.8Semi standard non polar33892256 Valsartan,2TMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C)N=N2)C=C1)C@HC(C)C3585.9Semi standard non polar33892256 Valsartan,2TMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C)N=N2)C=C1)C@HC(C)C3530.6Standard non polar33892256 Valsartan,2TMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C)N=N2)C=C1)C@HC(C)C4470.1Standard polar33892256 Valsartan,1TBDMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=N[NH]N=N2)C=C1)C@HC(C)C3613.3Semi standard non polar33892256 Valsartan,1TBDMS,isomer #2CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C(C)(C)C)N=N2)C=C1)C@HC(C)C3743.1Semi standard non polar33892256 Valsartan,2TBDMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C(C)(C)C)N=N2)C=C1)C@HC(C)C3890.1Semi standard non polar33892256 Valsartan,2TBDMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C(C)(C)C)N=N2)C=C1)C@HC(C)C3989.9Standard non polar33892256 Valsartan,2TBDMS,isomer #1CCCCC(=O)N(CC1=CC=C(C2=CC=CC=C2C2=NN(Si(C)C(C)(C)C)N=N2)C=C1)C@HC(C)C4493.5Standard polar33892256 |
| Spectra |
|
| GC-MS SpectraSpectrum TypeDescriptionSplash KeyDeposition DateSourceViewPredicted GC-MSPredicted GC-MS Spectrum - Valsartan GC-MS (Non-derivatized) - 70eV, Positivesplash10-000i-3094000000-1a2b93c2e5e760118ffd2017-09-01Wishart LabView SpectrumPredicted GC-MSPredicted GC-MS Spectrum - Valsartan GC-MS (1 TMS) - 70eV, Positivesplash10-052u-9236700000-2f92b4642db0f6e1db762017-10-06Wishart LabView SpectrumPredicted GC-MSPredicted GC-MS Spectrum - Valsartan GC-MS (Non-derivatized) - 70eV, PositiveNot Available2021-10-12Wishart LabView SpectrumMS/MS SpectraSpectrum TypeDescriptionSplash KeyDeposition DateSourceViewExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-qTof , Positive-QTOF splash10-0a4i-0122900000-e2d5ae1b4433254a8ebe 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-qTof , Positive-QTOF splash10-0a4u-1492000000-bc19c44b1bd382b3180f 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-QTOF , negative-QTOF splash10-001i-0000900000-45397dbc5403f1111d85 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-QTOF , negative-QTOF splash10-003r-0303900000-1056988fef13dc9becc8 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-QTOF , negative-QTOF splash10-004i-0900000000-65d8dfbf9280fcea77fb 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-QTOF , negative-QTOF splash10-004i-0900000000-77a9588a3e936c9d2e28 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-0udi-0009000000-160f82ad0952fc9b3350 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-001i-0000900000-d7bf9d8976f47e44ab4a 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-0fb9-0915000000-1f5a4e134cd8450810e7 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-004i-0900000000-19095db3835430e9fe16 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-004i-0900000000-00e9b9924b37c4db7a8d 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-004i-0900000000-fb8741bb91ee87d8245e 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-001i-0000900000-be5ab1c8cde8851235e3 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-0fb9-0915000000-08501c181e1066f2aa0a 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-004i-0900000000-6cabcfa5948837eddb2e 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-004i-0900000000-d0aedd0bab06b374571a 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-004i-0901000000-a4572421f0a393f299a1 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-ITFT , negative-QTOF splash10-0udi-0009000000-0f743f9e78dce83f2414 2017-09-14HMDB team, MONAView SpectrumExperimental LC-MS/MS LC-MS/MS Spectrum - Valsartan LC-ESI-QFT , negative-QTOF splash10-003r-0912600000-feeef5a05d8ef569460d 2017-09-14HMDB team, MONAView SpectrumPredicted LC-MS/MSPredicted LC-MS/MS Spectrum - Valsartan 10V, Positive-QTOFsplash10-000i-1055900000-71633e9ccff4a2e3fddf2017-07-26Wishart LabView SpectrumPredicted LC-MS/MSPredicted LC-MS/MS Spectrum - Valsartan 20V, Positive-QTOFsplash10-000i-3089200000-21a5c13b314fa930073e2017-07-26Wishart LabView SpectrumPredicted LC-MS/MSPredicted LC-MS/MS Spectrum - Valsartan 40V, Positive-QTOFsplash10-052r-4293000000-5b414a2cac94b4c0bad52017-07-26Wishart LabView SpectrumPredicted LC-MS/MSPredicted LC-MS/MS Spectrum - Valsartan 10V, Negative-QTOFsplash10-000x-0009600000-4157b99813121a9c94ea2017-07-26Wishart LabView SpectrumPredicted LC-MS/MSPredicted LC-MS/MS Spectrum - Valsartan 20V, Negative-QTOFsplash10-0f8c-1119200000-f3f0fc40c02029f258822017-07-26Wishart LabView SpectrumPredicted LC-MS/MSPredicted LC-MS/MS Spectrum - Valsartan 40V, Negative-QTOFsplash10-00xu-5906000000-9ea0192d8fb20dac6ee72017-07-26Wishart LabView Spectrum |
|
| Biological Properties |
|
| Cellular Locations |
Extracellular Membrane |
| Biospecimen Locations |
Blood Urine |
| Tissue Locations |
Not Available |
| Pathways |
NameSMPDB/PathBankKEGGValsartan Action Pathway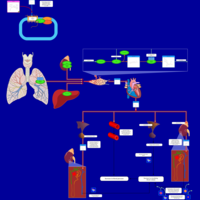 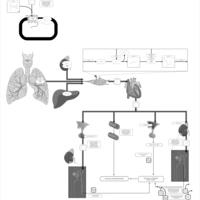  Not Available Not Available |
| Normal Concentrations |
|
| BiospecimenStatusValueAgeSexConditionReferenceDetailsBloodExpected but not QuantifiedNot QuantifiedNot AvailableNot AvailableTaking drug identified by DrugBank entry DB0017721059682 detailsUrineExpected but not QuantifiedNot QuantifiedNot AvailableNot AvailableTaking drug identified by DrugBank entry DB0017721059682 details |
|
| Abnormal Concentrations |
|
| Not Available |
|
| Associated Disorders and Diseases |
|
| Disease References |
None |
| Associated OMIM IDs |
None |
| External Links |
|
| DrugBank ID |
DB00177 |
| Phenol Explorer Compound ID |
Not Available |
| FooDB ID |
Not Available |
| KNApSAcK ID |
Not Available |
| Chemspider ID |
54833 |
| KEGG Compound ID |
Not Available |
| BioCyc ID |
Not Available |
| BiGG ID |
Not Available |
| Wikipedia Link |
Valsartan |
| METLIN ID |
Not Available |
| PubChem Compound |
60846 |
| PDB ID |
Not Available |
| ChEBI ID |
9927 |
| Food Biomarker Ontology |
Not Available |
| VMH ID |
Not Available |
| MarkerDB ID |
Not Available |
| Good Scents ID |
Not Available |
| References |
|
| Synthesis Reference |
Not Available |
| Material Safety Data Sheet (MSDS) |
Not Available |
| General References |
(). Bader, M. (2004). Renin-angiotensin-aldosterone system. In S. Offermanns, & W. Rosenthal (Eds.). Encyclopedic reference of molecular pharmacology_ (pp. 810-814). Berlin, Germany: Springer. . . (). Diovan. (2009). [Electronic version]. e-CPS. Retrieved December 28, 2009.. . (). Stanfield, C.L., & Germann, W.J. (2008). Principles of human physiology (3rd ed.). San Francisco, CA: Pearson Education, Inc.. . |