Middle East Respiratory Syndrome in 3 Persons, South Korea, 2015 (original) (raw)
Abstract
In May 2015, Middle East respiratory syndrome coronavirus infection was laboratory confirmed in South Korea. Patients were a man who had visited the Middle East, his wife, and a man who shared a hospital room with the index patient. Rapid laboratory confirmation will facilitate subsequent prevention and control for imported cases.
Keywords: Middle East respiratory syndrome, MERS, MERS-CoV, South Korea, viruses, respiratory infections
Middle East respiratory syndrome (MERS) is characterized by mild-to-severe respiratory distress and is caused by a novel coronavirus (MERS-CoV) (1). Since its first identification in 2012, MERS-CoV infection has been reported for 1,413 persons from 26 countries; case-fatality rate is 40.92% (2). We describe an outbreak comprising 3 laboratory-confirmed cases of MERS-CoV infection in South Korea (3–5).
The Study
In accordance with national MERS control guidelines in South Korea (6), specimens are collected from hospitalized patients suspected of having MERS on the basis of epidemiologic history linked to the Middle East; these specimens are then transferred to the Korea National Institute of Health for examination. The index patient, a 68-year-old man engaged in farming-related business, reported that he had traveled to Bahrain on April 24, 2015, the United Arab Emirates on April 29–30, Saudi Arabia on May 1–2, and Qatar on May 2–3 before returning to South Korea on May 4 (Figure 1). While in these countries, he was not exposed to any patients, health care facilities, or animals (including camels and bats) or their excreta. On May 11, the patient experienced chills and a fever (>37°C), and an allopathic medicine was prescribed when he first visited a local clinic. However, his symptoms worsened (temperature >38°C, myalgia, cough, and dyspnea), and after he had visited 4 hospitals (hospitals A–D, in or around Seoul), he was admitted to a general hospital (hospital D, Seoul, South Korea) on May 18. A nasopharyngeal aspiration specimen was collected for MERS-CoV laboratory testing on May 19**.**
Figure 1.
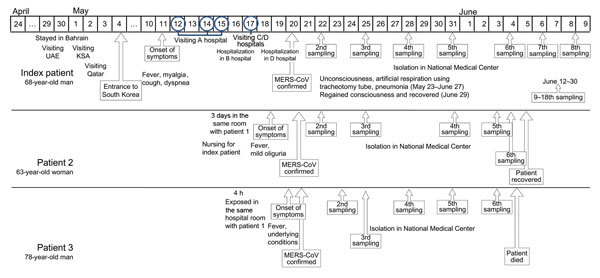
Timeline of events for patients infected with Middle East respiratory syndrome coronavirus (MERS-CoV). The laboratory diagnostic methods used for molecular detection of MERS-CoV RNA were multiplex MERS-CoV real-time reverse transcription PCRs targeting an upstream MERS-CoV envelope protein gene and an open reading frame 1a gene (8,9). KSA, Kingdom of Saudi Arabia; UAE, United Arab Emirates.
Sputum samples from 2 persons who had been in contact with the index patient were also tested for MERS-CoV. Patient 2 was the 63-year-old wife of the index patient; she had had physical contact with him while caring for him during the 3 days of hospitalization. Fever (38°C) and slight oliguria developed in patient 2 on May 19. The other contact, patient 3, was a 78-year-old man who had chronic obstructive pulmonary disease, asthma, and cholangiocarcinoma and who had shared a hospital room with the index patient and had been within 2 meters from him for 4 hours on May 16. Fever (37.8°C) and respiratory symptoms developed in patient 3 on May 20. In the hospital room, the index patient did not undergo any aerosol-generating procedures, but a severe cough developed. The same health care workers cared for the index patient and patient 3.
Laboratory diagnostic methods were performed according to World Health Organization guidelines for molecular detection of MERS-CoV (7–9). To check for contamination derived from the positive control, we designed and synthesized the MERS-CoV real-time reverse transcription PCR (rRT-PCR)–positive transcripts for an upstream MERS-CoV envelope protein gene (upE) and the open reading frame 1a (ORF1a) gene containing 50 bp of a foreign gene (centipede).
Initially, nasopharyngeal samples from the index patient were positive for MERS-CoV by multiplex rRT-PCR. Sputum samples from patients 2 and 3 were also positive, supporting a diagnosis of MERS-CoV infection. Multiplex rRT-PCR results for upE and ORF1a were positive (Table 1). According to rRT-PCR, the respiratory samples from the 3 patients were negative for 5 other human coronaviruses (SARS-CoV and human CoV-229E, -OC43, -NL63, and -HKU1) and 7 viruses that cause acute respiratory infection (influenza virus A and B; human adenovirus; bocavirus; human parainfluenza virus types 1, 2, and 3; respiratory syncytial virus A and B; human rhinovirus; human metapneumovirus).
Table 1. Real-time reverse transcription PCR results for 2 patients infected with Middle East respiratory syndrome coronavirus, South Korea, 2015*.
| Sample | Index patient (day 9 after illness onset) | Wife of index patient (day 2 after illness onset) | ||
|---|---|---|---|---|
| upE, Ct | ORF1a, Ct | upE, Ct | ORF1a, Ct | |
| Sputum | 18.61 | 19.32 | 26.23 | 26.63 |
| Nasal swab | 29.34 | 29.58 | ND | 36.64 |
| Nasopharyngeal aspirate | 29.35 | 31.45 | NT | NT |
| Serum | 34.81 | 35.50 | ND | ND |
| Urine | ND | ND | 34.34 | ND |
For the index patient, MERS-CoV RNA was detectable in sputum, throat swab, and serum samples but not in a urine sample collected 9 days after symptom onset (Table 1). The viral load, indicated by cycle threshold values, was high in the lower respiratory tract sample but almost undetectable in the throat swab and serum samples. After sequential sampling repeated every 2–5 days, MERS-CoV RNA was detected in sputum until 44 days after symptom onset, although viral RNA was inconsistently detected and patterns of viral load fluctuated (Table 2). Other than initial fever (>37°C), clinical features differed for all 3 patients. The index patient had respiratory symptoms with cough, dyspnea, and myalgia. Patient 2 did not have a relevant medical history and showed mild symptoms. Patient 3 had underlying concurrent conditions and died 16 days after confirmation of MERS-CoV infection (Figure 1).
Table 2. Real-time reverse transcription PCR results for sputum samples serially collected from 3 patients infected with Middle East respiratory syndrome coronavirus, South Korea, 2015*.
| Date | Patient 1 (index patient) | Patient 2† | Patient 3‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Days after illness onset | upE, Ct | ORF1a, Ct | Days after illness onset | upE, Ct | ORF1a, Ct | Days after illness onset | upE, Ct | ORF1a, Ct | |
| May 20 | 9 | 18.61 | 19.32 | 2 | 26.23 | 26.63 | 1 | 28.10 | 28.65 |
| May 22 | 11 | 25.24 | 25.67 | 5 | 35.53 | 35.10 | 3 | 24.67 | 25.04 |
| May 25 | 14 | 26.48 | 27.99 | 11 | 25.20 | 26.10 | 5 | 26.37 | 25.53 |
| May 28 | 17 | 31.23 | 32.05 | 14 | 32.58 | 34.80 | 8 | ND | 35.94 |
| May 31 | 20 | 36.94 | ND | 17 | ND | ND | 11 | 26.30 | 28.01 |
| Jun 4 | 24 | ND | 36.27 | 18 | ND | ND | 14 | ND | ND |
| Jun 6 | 26 | 33.92 | 36.70 | 19 | Discharged | 15 | Died | ||
| Jun 8 | 29 | 36.50 | ND | ||||||
| Jun 9 | 30 | 36.90 | 37.46 | ||||||
| Jun 12 | 33 | ND | ND | ||||||
| Jun15 | 36 | ND | ND | ||||||
| Jun 16 | 37 | 35.46 | ND | ||||||
| Jun 17 | 38 | ND | ND | ||||||
| Jun 22 | 43 | ND | ND | ||||||
| Jun 23 | 44 | 32.97 | 36.31 | ||||||
| Jun 26 | 47 | ND | ND | ||||||
| Jun 29 | 50 | ND | ND | ||||||
| Jun 30 | 51 | ND | ND |
Virus isolation on Vero cells was attempted for each respiratory specimen from the 3 patients. The culture supernatant after inoculation was serially assessed for virus growth by using rRT-PCR and was used for blind passages every 3–7 days after inoculation. After 3 blind passages, cytopathic effect was observed, and we isolated the MERS-CoV strain from South Korea (KOR/KNIH/002_05_2015) from the Vero cells after inoculation by using the sputum from patient 2. We constructed a phylogenetic tree by using the general time reversible plus gamma model of the RAxML version 8.8.0 software (10) and FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree) and by using complete genomes of the MERS-CoV isolate from South Korea (GenBank accession no. KT029139) and 67 reference MERS-CoVs (Figure 2).
Figure 2.
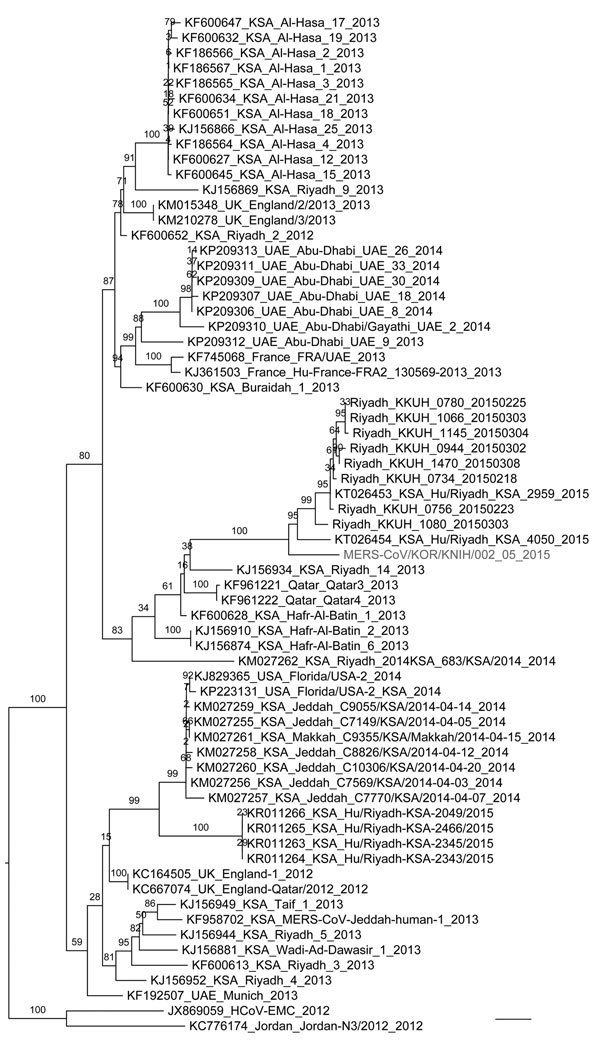
Phylogenetic tree comparing complete genome nucleotide sequences of Middle East respiratory syndrome coronavirus (MERS-CoV) isolate from South Korea (KOR/KNIH/002_05_2015) with those of 67 reference MERS-CoVs (GenBank database). The tree was constructed by using the general time reversible plus gamma model of RAxML version 8.8.0 software (10) and visualized by using FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). RAxML bootstrap values (1,000 replicates) are shown above the branches. Bootstrap values >75 are shown on the branches. The MERS-CoV strain from South Korea is shown in gray. National Center for Biotechnology Information accession numbers are shown before each taxon name. The unit of branch length is the number of substitutions per site. Scale bar indicates 4 × 10–4 nucleotide substitutions per site.
Conclusions
Because, to our knowledge, cases of MERS-CoV infection in South Korea have not been reported, we had to establish laboratory testing protocols to overcome vulnerabilities in the absence of appropriate epidemiologic support (i.e., generate positive controls to check for contamination and repeat testing). Positive controls containing foreign genes have been generated to check for laboratory contamination. For patients with an unclear exposure history (such as the index patient) and for patients with short exposure durations and unusual clinical symptoms (such as patients 2 and 3), it would be useful if the positive results of rRT-PCR could be confirmed through agarose gel electrophoresis to exclude contamination from the positive control (3,4).
The index patient had no history of potential exposure to camels, bats, or their excreta; to symptomatic persons; or to health care workers during his trip to the Middle East, including Saudi Arabia. Although the source of infection for the index patient is unclear, phylogenetic analysis of the whole viral genome showed that the isolate from South Korea was closely related to the MERS-CoV strains isolated in Saudi Arabia in 2015.
Because the index patient initially concealed his travel history to Saudi Arabia, United Arab Emirates and Qatar, MERS-CoV infection was not considered and the patient was not isolated until MERS-CoV infection was suspected 7 days after symptom onset. Meanwhile, other patients and health care workers had multiple opportunities for exposure to the index patient (3–5). The 2 contacts reported here had each been exposed to the index patient. Patient 3 was probably infected via droplet transmission in the hospital room. The hospital room, originally built for 6 persons, had been divided into 2 rooms and lacked ventilation. Furthermore, an air conditioning unit cycled the air in the room with the door and window closed. Thus, poor ventilation might have played a major role in droplet transmission. Detection of MERS-CoV RNA in the respiratory tract varies up to day 33 (11–13). In this study, virus was detected in the respiratory tract, inconsistently, for up to 44 days.
Development of effective preventive measures for the MERS-CoV prevention will require systemic and prospective studies associated with viral shedding and use of specimens in addition to those obtained from the respiratory tract to define the kinetics of MERS-CoV. Rapid detection of MERS-CoV, using multiplex rRT-PCR to detect upE and ORF1a genes, would be helpful for countries outside the Arabian Peninsula.
Acknowledgments
We thank the division of epidemic intelligence service and National Medical Center for assistance with identifying patients, abstracting medical records, and collecting specimens.
This study was supported by grants from the Intramural Research Fund no.2014-N47001-00 of the Korea National Institute of Health and the Intramural Fund no.4834-300-210-13 of the Korea Centers for Disease Control and Prevention.
Biography
Dr. Yang is a staff scientist in Korea National Institute of Health, Korea Centers for Disease Control and Prevention. Her research encompasses the diagnosis, immune response, viral pathogenesis, epidemiology of respiratory viruses with a particular interest in emerging virus identification.
Footnotes
Suggested citation for this article: Yang JS, Park S, Kim YJ, Kang HJ, Kim H, Han YW, et al. Middle East respiratory syndrome in 3 persons, South Korea, 2015. Emerg Infect Dis. 2015 Nov [_date cited_]. http://dx.doi.org/10.3201/eid2111.151016
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Frouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20 . 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Emergencies preparedness, response: Middle East respiratory syndrome coronavirus (MERS-CoV) maps and epicurves. 10–16 August 2015. [cited 2015 Aug 20]. http://www.who.int/csr/disease/coronavirus_infections/maps-epicurves/en
- 3.Korea Society of Infectious Diseases, Korean Society for Healthcare-associated Infection Control and Prevention. An unexpected outbreak of Middle East respiratory syndrome coronavirus infection in the Republic of Korea, 2015. Infect Chemother. 2015;47:120–2.http:// [DOI] [PMC free article] [PubMed]
- 4.Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, Wu JT. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Euro Surveill. 2015;20:21163 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SS, Wong NS. Probable transmission chains of Middle East respiratory syndrome coronavirus and the multiple generations of secondary infection in South Korea. Int J Infect Dis. 2015;38:65–7 . 10.1016/j.ijid.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korea Centers for Disease Control and Prevention. MERS countermeasure guideline (revised December 2014) [in Korean]. Cheongju (South Korea): The Centers; 2014.
- 7.World Health Organization. Laboratory testing for Middle East respiratory syndrome coronavirus. Interim recommendations (revised). September 2014. Geneva: The Organization; 2014. [Google Scholar]
- 8.Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285 . [DOI] [PubMed] [Google Scholar]
- 9.Corman VM, Muller MA, Costabel U, Timm J, Binger T, Meyer B, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:pii20334 . [DOI] [PubMed] [Google Scholar]
- 10.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spanakis N, Tsiodras S, Haagmans BL, Raj VS, Pontikis K, Koutsoukou A, et al. Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int J Antimicrob Agents. 2014;44:528–32. 10.1016/j.ijantimicag.2014.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drosten C, Seilmaier M, Corman VM, Hartmann W, Scheible G, Sack S, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–51. 10.1016/S1473-3099(13)70154-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poissy J, Goffard A, Parmentier-Decrucq E, Favory R, Kauv M, Kipnis E, et al. Kinetics and pattern of viral excretion in biological specimens of two MERS-CoV cases. J Clin Virol. 2014;61:275–8. 10.1016/j.jcv.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]