Characterizing cell subsets in heterogeneous tissues using marker enrichment modeling (original) (raw)
. Author manuscript; available in PMC: 2017 Jul 30.
Published in final edited form as: Nat Methods. 2017 Jan 30;14(3):275–278. doi: 10.1038/nmeth.4149
Abstract
Learning cell identity from single-cell data presently relies on human experts. Here, we present Marker Enrichment Modeling (MEM), an algorithm that objectively describes cells by quantifying contextual feature enrichment and reporting a human and machine-readable text label. MEM outperformed traditional metrics in describing immune and cancer cell subsets from fluorescence and mass cytometry. MEM provides a quantitative language to communicate characteristics of new and established cytotypes observed in complex tissues.
Introduction
Quantitative cytometry workflows have developed diverse approaches to grouping cells into populations and visualizing results in graphs that arrange populations based on phenotype1,2. Significant features of populations are typically assumed to be those most highly or differentially expressed. This approach works well when feature variability is low and cells match established types, but computational analysis of single cell data routinely reveals novel cells with non-canonical phenotypes3-5. This is especially common in diseases where abnormal expression profiles and signaling responses distinguish clinically significant cell subsets6-10. Existing statistical approaches can be used to characterize a population's degree of difference from a reference, but may be limited to a normal distribution or may not account for intra- and inter-population variability in a single metric.
The MEM equation (Eq. 1) produces signed value for each population feature by quantifying positive and negative, population-specific, contextual feature enrichment relative to a reference cell population (Supplementary Note 1).
| MEMscore=∣MAGPOP−MAGREF∣+(IQRREFIQRPOP)−1,(MAGPOP−MAGREF)<0→MEM=−MEM | (Eq. 1) |
|---|
In Eq. 1, POP denotes the population of interest, REF denotes the reference population to which POP will be compared, MAG is feature magnitude (here, median protein expression detected by mass or fluorescence flow cytometry), and IQR indicates the interquartile range. A reference population (REF) is chosen based on a biological comparison of interest (Supplementary Note 1, Supplementary Fig. 1). MEM was designed to quantify enrichment, whereas other metrics used in cytometry, such as Kolmogorov-Smirnov (K-S)11, area under the ROC curve (AUC)12, and Earth Mover's Distance (EMD)13, capture other differences between frequency distributions (Supplementary Note 1). In datasets including healthy human blood, bone marrow, and tonsil, murine tissues, and human tumors, MEM identified key proteins used by experts to distinguish rare and novel cell subsets.
Results
Four cytometry studies, Dataset A14, Dataset B15, Dataset C4, and Dataset D, collected as described by Leelatian and Doxie, et al.16, were used to evaluate the ability of MEM to identify biological features of expert and machine identified cell subsets. For datasets A, B, and C, populations had been previously identified by experts and by computational tools including viSNE17 and SPADE18, which are used in mass cytometry for dimensionality reduction and cell clustering1, respectively.
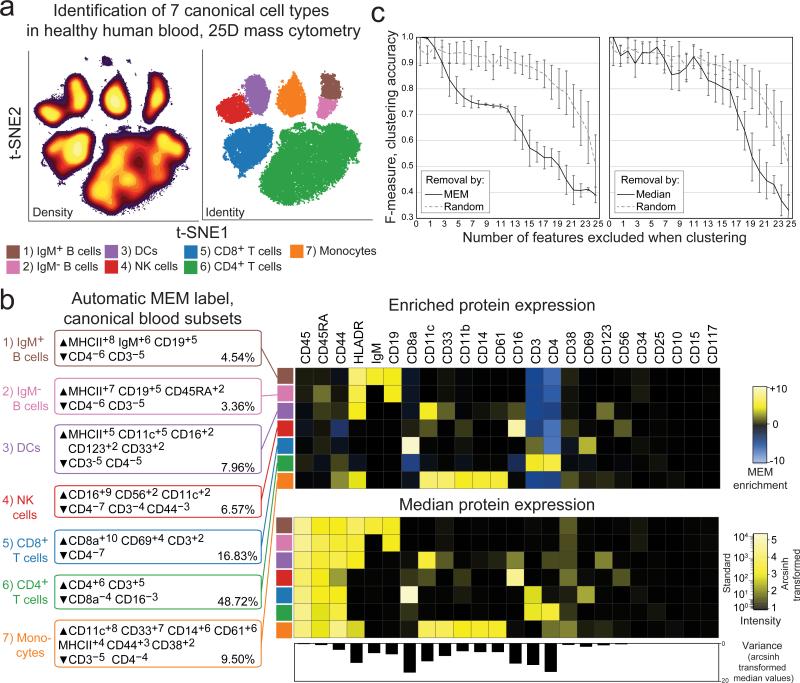
Dataset A was mass cytometry data quantifying expression of 25 proteins on healthy human peripheral blood mononuclear cells (PBMC)14. This dataset was chosen for two reasons: 1) the 7 cell subsets present are well-established, phenotypically distinct populations that served as a gold standard of biological ‘truth’ and 2) the cells in each of the 7 subsets were characterized for 25 proteins that displayed varying homogeneous and heterogeneous expression patterns. Populations were expert gated following viSNE analysis and each population was compared to the other cells in the sample (Fig. 1, Supplementary Table 2). MEM returned labels that matched prior expert analysis14 and correctly assigned high positive enrichment values to canonical protein features of each subset (Fig. 1b), including CD4 on CD4+ T cells (▲CD4+6 CD3+5 ▼CD8a−4 CD16−3), IgM on IgM+ B cells (▲MHC II+8 IgM+6 CD19+5 ▼CD4−6 CD3−5), CD11c and MHC II on monocytes (▲CD11c+8 CD33+7 CD14+6 CD61+6 MHC II+4 CD44+3 ▼CD3−5 CD4−4), and CD16 on NK cells (▲CD16+9 CD56+2 CD11c+2 ▼CD4−7 CD3−4 CD44−3). Proteins that were not significantly enriched on any of the 7 subsets of mature human blood mononuclear cells were correctly assigned near-zero MEM scores (e.g. CD34 and CD117 proteins expressed on hematopoietic stem cells, Fig. 1b). Similarly, proteins with little variability across cell subsets were assigned low, near-zero MEM scores, even for highly expressed proteins (e.g. CD45 on all subsets, CD45RA on non-T cells, Fig. 1b). Incorporating information about feature variability allowed MEM to capture negative enrichment that was not reflected in magnitude difference (MAGDIFF, Supplementary Note 2). Highly enriched proteins were more important to accurate population identification than proteins characterized by high median expression alone (Fig. 1c; Supplementary Fig. 2; Supplementary Fig. 3).
Figure 1. Marker enrichment modeling (MEM) automatically labels human blood cell populations in Dataset A.
a) Cells from normal human blood were previously grouped into 7 canonical populations using viSNE analysis and expert review of 25D mass cytometry data14. b) MEM labels were computationally generated for each canonical cell subset using the other six populations as reference. The population labeled by immunologists as “CD4+ T cells” was labeled by MEM as ▲CD4+6 CD3+5 ▼CD8a−4 CD16−3 and comprised 48.72% of PBMC in this sample. In contrast, the MEM label ▲CD16+9 CD56+2 CD11c+2▼CD4−7 CD3−4 CD44−3 was generated for the population gated as “NK cells”. Heatmaps show protein enrichment values used to generate MEM labels and the median protein expression values for each protein on each cell subset. Variability in protein expression across the 7 canonical cell populations is shown below to highlight proteins that were expressed homogeneously (low variability, e.g. CD45) and those that were expressed heterogeneously (high variability, e.g. CD8a, CD4). c) Graphs show decreasing f-measure (clustering accuracy) as markers were excluded from k-means cluster analysis based on high to low absolute MEM or median values, compared to random exclusion.
To test the hypothesis that features with high MEM scores would be important for computational cluster formation, the 25 proteins measured in Dataset A (Figure 1b) were sorted in six ways: 1) high to low MEM score, 2) high to low median value, 3) high to low MAGDIFF, 4) high to low z-score, 5) high to low K-S statistic, and 6) randomly (Supplementary Table 3). Z-score and K-S statistic values are shown in Supplementary Table 5. The proteins were then sequentially, cumulatively excluded from use in k-means clustering and f-measure was calculated to measure clustering accuracy (Fig. 1c and Supplementary Fig. 2). The order in which markers were excluded is shown in Supplementary Table 3. Random exclusion was performed 15 times and the average result is shown (Fig. 1c). Clustering accuracy was most impacted by excluding proteins based on MEM score. F-measure dropped to 0.75 after removing the proteins with the top 6 MEM scores, whereas a comparable F-measure decrease was only observed after removing the 14 highest markers based on MAGDIFF, the 13 highest markers based on z-score, and the 12 highest markers based on K-S statistic values (Supplementary Fig. 2). Removing markers based on median was not significantly different from removing markers randomly until the 15 markers with the highest median signal intensity were excluded (Supplementary Fig. 2). The same analysis was performed with viSNE in place of k-means clustering to visualize loss of population resolution (Supplementary Fig. 3c). In this case, loss of accuracy was reflected in the viSNE map as a loss of separation between “islands” of cells. These results indicated that MEM enrichment scores captured markers that were important to cell identity better than traditional comparisons based solely on median protein expression.
Dataset B was mass cytometry data quantifying expression of 31 proteins on healthy human bone marrow15. Computational and expert analysis had previously identified 23 populations of cells that were analyzed here by MEM (Supplementary Note 3). For example, the cell subset labeled as HSCs was highly enriched for CD34 (CD34+6) and negatively enriched for CD45 (CD45−5). Dataset B also illustrated the general rule that MEM scores will approach median values as feature variability within populations decreases (Supplementary Fig. 4). MEM captured feature enrichment and heterogeneity better than median in diverse populations, as in Fig. 1c.
Dataset C was mass cytometry data quantifying expression of 38 proteins on murine cells from eight tissues4 (Supplementary Note 4). In this dataset, “cluster 28” was a novel population identified as CD11bint NK cells. The MEM label for cluster 28 within ILCs was ▲CD11b+5 CD62L+3 ▼CD4−7 CD103−4 Terr119−3 (Supplementary Note 4 and Supplementary Fig. 5). This MEM label captured the key feature of this novel innate lymphoid cell subset (CD11bint) and highlighted additional features that can be used to match this subset to cells identified by others (i.e., to cytotype the population). These results indicate that MEM labels complement unbiased population discovery and effectively characterize cyto incognito19 by providing unbiased descriptions that correctly capture key features of novel cell types.
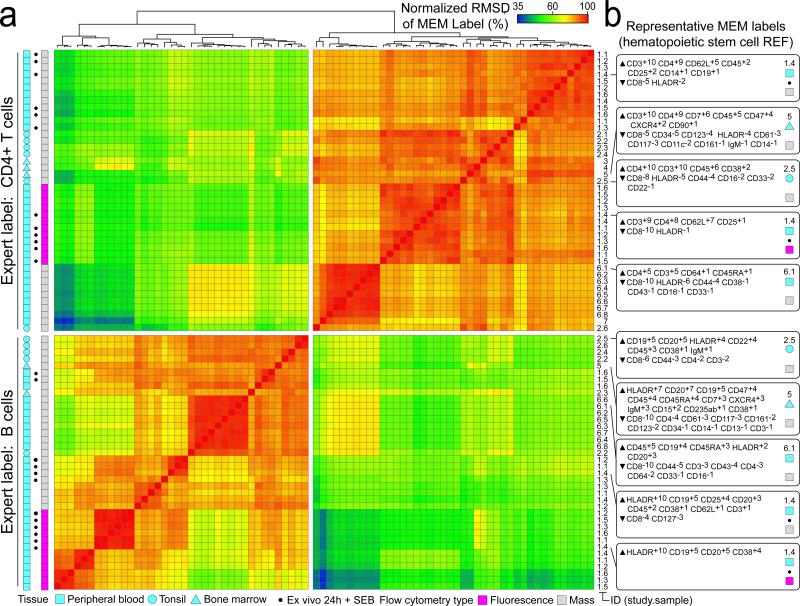
An important aspect of MEM is generation of machine-readable quantitative labels that can be used to register population identities across samples and studies. A MEM label for a newly discovered population can be compared quantitatively against a reference set of established MEM labels or a MEM label reported in a paper. To illustrate this idea, the pairwise, normalized root-mean-squared distance (RMSD) of MEM scores was calculated as a measure of similarity between 80 populations of cells from 7 different studies including healthy CD4+ T cell and B cell (Fig. 2). Cells had highly similar MEM scores within each major cell type, regardless of platform (mass or fluorescence flow cytometry), study, or tissue source. For example, T cells run on mass cytometry from different blood donors were 97% ± 1.3 similar to each other, 85% ± 1.9 similar to T cells from blood run on fluorescence flow cytometry, and 87% ± 2.1 similar to T cells from tonsil run on mass cytometry (Fig. 2). However, these cells were 66.9% ± 13 similar to any B cell population. This indicates that MEM scores provide a way to communicate cell identity and to quantify similarities of cell types from the text label alone.
Figure 2. Hierarchical clustering based solely on MEM label groups T cells and B cells measured in diverse studies using different cytometry platforms.
A) MEM label values were compared for each of 80 populations (CD4+ T cells and B cells) from 3 human tissues representing 6 mass cytometry studies and 1 fluorescence flow cytometry study. The normalized RMSD (i.e. similarity) for two populations was 100% when MEM label exponents were identical for all of the shared proteins. Populations are shown clustered according to MEM label percent similarity. Tissue type, source study (numbered 1-7 and referenced in online methods), and individual sample IDs are indicated to the right. *indicates samples stimulated by bacterial superantigen Staphlococcus enterotoxin B (SEB). B) Representative MEM labels for CD4+ T cells (top) and B cells (bottom) from SEB-stimulated normal human blood (1.4, top, mass cytometry), normal human bone marrow (5, mass cytometry), normal human tonsil (2.5, mass cytometry), SEB-stimulated normal human blood (1.4, bottom, fluorescence flow cytometry), and normal human blood (6.1, mass cytometry).
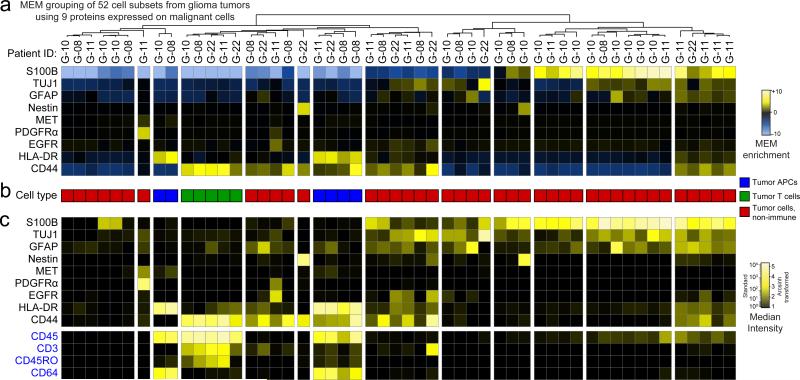
Dataset D included 52 populations of tumor infiltrating APCs, tumor infiltrating T cells, and non-immune malignant tumor cells identified in human glioma tumors16. To obtain these populations, each tumor was analyzed by viSNE and cell subsets were expert gated solely on t-SNE cluster density (Supplementary Fig. 6). To determine whether MEM could distinguish immune cell subsets from other tumor cell types with limited information, MEM scores were calculated using only 9 markers that were expected to be expressed on cancer cells (S100B, TUJ1, GFAP, Nestin, MET, PDGFRα, EGFR, HLA-DR, and CD44, Fig. 3a). The 52 populations were grouped into 13 major cell types based on MEM enrichment of 9 analyzed proteins, and these groups were interpreted as tumor infiltrating APCs (Fig. 3b, blue), tumor infiltrating T cells (Fig. 3b, green), or non-immune tumor cells (Fig. 3b, red). To confirm cell identity, four protein features that had been excluded from MEM analysis were assessed (Fig. 3c, CD45, CD3, CD45RO, and CD64). CD45 and CD3 were used to confirm T cell identity and CD45 and CD64 were used to confirm APC identity. MEM correctly identified both immune cell subsets from all tumor types without using key immune lineage markers and without using healthy populations (e.g. APCs from blood or tonsil) to guide the clustering. Thus, MEM labels distinguished populations of cells based on non-traditional features and in a disease context.
Figure 3. MEM correctly grouped immune and cancer cell populations from glioma tumors using nine proteins expressed on cancer cells in Dataset D.
(A) A heatmap of MEM enrichment scores is shown for 52 populations of cells identified in tumors from 4 glioblastoma patients (G-08, G-10, G-11, G22) in an unsupervised manner using viSNE. MEM scores were then calculated based only on the nine measured proteins expected to be expressed on cancer cells (S100B, TJF1, GFAP, Nestin, MET, PGFRα, HLA-DR, and CD44). (B) Each population was annotated for a cell type based on review of the MEM label and classified as tumor infiltrating APCs (blue), tumor infiltrating T cells (green), or non-immune tumor cells (red). (C) A heatmap of median intensity values is shown for the 13 measured proteins from each of the 52 tumor cell populations. Expression of CD45, CD3, and CD64 was used to assess the respective identity of leukocytes, T cells, and antigen presenting cells.
Discussion
MEM labels provided a quantitative language to objectively communicate characteristics of new and established cell types observed in complex tissue microenvironments. Algorithmic comparison of MEM labels correctly identified 80 cell populations from 7 studies of 3 human tissues measured using different instrumentation and distinguished tumor-infiltrating immune cell subsets and malignant cell populations from human glioma tumors. Following additional validation in other cell types, tissues, and instrumentation platforms, it may be possible for machines and humans to use MEM labels to learn and clearly communicate cell identity (cytotype). Given widespread adoption and reporting, MEM labels could be used to communicate cytotypes in a manner analogous to cluster of differentiation (CD) naming of antigen targets of antibodies20. MEM can compare populations against a common reference (Supplementary Note 5) and guide feature selection for computational and experimental analysis. MEM can also be used to monitor changes in tissues over time during treatment. Deviation from a stable MEM score for peripheral blood cell subsets would be expected in the case of emerging malignant cells9, and lack of change towards a healthy set of MEM scores for blood or bone marrow cell subsets might indicate a lack of response to chemotherapy for a leukemia patient. MEM is expected to assist in machine learning applications by providing quantitative text descriptions of cytotype that can be algorithmically parsed and used to classify newly identified cell subpopulations.
Data Availability Statement
The normal human PBMC dataset (Figure 1) were generated by CyTOF analysis as described by Leelatian, et al.14 and is available as an FCS file in Flow Repository (https://flowrepository.org/experiments/1043).
The normal human bone marrow data set from Bendall and Simonds, et al15 (Dataset B, Supplementary Note 3) was downloaded from Cytobank24 as FCS files that included the cell population IDs defined by Bendall and Simonds, et al.15 (https://reports.cytobank.org/1/v1). MEM enrichment scores from Dataset B were compared to the authors’ analysis and prior studies of proteins marking stem cells, progenitor cells, and mature cells25,26
The murine myeloid CyTOF dataset from Becher, et al4 (Dataset C, Supplementary Note 4) was downloaded from Cytobank as FCS files that contained gated cell events and cluster IDs as designated by automated analysis conducted by Becher et al4. MEM enrichment scores from Dataset C were compared to the authors’ analysis and prior studies of neutrophils27,28.
Datasets for Figure 2 were generated in 7 separate fluorescence and mass cytometry studies by 1) Nicholas et al. 23, 2) Polikowsky et al.22, 3) Ferrell et al. 21, 4) Amir et al.17, 5) Bendall and Simonds et al.15, 6) Greenplate et al., previously unpublished data, and 7) Leelatian et al14.
The phospho-flow AML data set generated by Irish et al.6 (Supplementary Note 5-Fig.2) was downloaded from Cytobank as FCS files.
The human GBM mass cytometry dataset (Fig. 3) was generated and analyzed as described by Leelatian and Doxie et al.16 and are available on Flow Repository as text files (https://flowrepository.org/experiments/1044/).
Online Methods
CyTOF data pre-processing and analysis
Data analysis was performed using the online analysis platform Cytobank24 and the statistical programming environment R. Raw median intensity (MI) values were transformed to a hyperbolic arcsine scale. A cofactor of 15 was used for the PBMC dataset (Fig. 1), and 5 was used for the normal human bone marrow data set and for the murine myeloid data set. Single, intact cells were gated based on cell length (30-60) and nucleic acid intercalator (iridium). Major PBMC subsets were gated based on CD45 expression (leukocytes) and on canonical lineage marker expression to identify major blood cell subsets.
FCS files were exported from Cytobank as FCS or tab-delimited text files that were parsed for expression intensity information using the R package flowCore 29. MEM was calculated using the arcsinh transformed MI values, as described above. Heatmaps were generated using the heatmap.2 function in the gplots R package30.
Fluorescence Phospho-Flow AML Data Analysis
Data were downloaded from Cytobank as FCS files and processed in R as described above. MFI values were transformed to a log normal scale. For each AML patient, a median value and an IQR value was calculated for each marker in the unstimulated condition and for the stimulated conditions. The unstimulated median values were subtracted from the stimulated median values, and likewise for the IQR values. MEM was then calculated by comparing each patient's subtracted median and IQR values to those of the other patients. This enabled a comparison of fold change signaling values rather than raw values.
Marker Enrichment Modeling (MEM)
MEM analysis begins after populations have been identified and aims to provide a simple way to compare findings from experts working with different platforms or performing analysis using different computational tools for population discovery 18,31-34 and graphical visualization 6,8,15,35,36. These tools have differing strengths that depend greatly on the structure of the datasets and controls, the biological goals of the study, and the quality of the existing knowledge in the field 1,2,37.
MEM equation
The MEM equation is implemented as an R package (Supplementary Software). Currently, MEM uses medians as the magnitude value; however, depending on the data type, mean may be a more appropriate magnitude statistic and mean could be substituted for median in the equation. Similarly, other statistics, such as variance, might be substituted for IQR. The MEM equation was developed with the intention of capturing and quantifying population-specific feature enrichment in a simple equation that avoids over-fitting or unnecessary computation. The primary goal of this equation is to scale magnitude differences depending on distribution spread. While other distribution features such as skew or shape could be informative, incorporating only two pieces of information – magnitude and spread – into the equation captured enough information to be useful in quantifying both positive and negative population-specific feature enrichment.
MEM output and score scaling
The MEM R script outputs a heatmap of MEM values with a text label summary of feature enrichment as the population (row) names. The + or − value provided along with the marker name is converted to a −10 to +10 scale and rounded to the nearest integer. As implemented here, the maximum of the scale was set using the highest absolute value MEM score observed across all markers and populations. All values in the matrix are divided by this maximum value and multiplied by 10 to achieve the −10 to +10 scaling. After scaling, the original sign value is reapplied to each MEM score. Scaling the output this way is intended to generate MEM values and labels that are intuitive to human readers and to facilitate comparison of feature enrichment across experiments, samples, batches, time points, and data types.
IQR Threshold
Because MEM uses a ratio of IQR values, near zero values in the denominator, IQRPOP, will greatly increase MEM scores. For each measurement type, it is important to identify a minimum significant IQR value so that small IQR values below the platform's ability to distinguish signal from noise do not inappropriately increase MEM scores. To automatically determine a minimum threshold for IQRPOP, the algorithm here calculated the average of the IQR values that were associated with the lowest quartile of population and reference medians. For the mass and fluorescence cytometry datasets used, the automatically calculated IQR threshold was on average 0.5 ± X and so the IQR threshold for all studies here was set to 0.5. The default IQR threshold in the algorithm is also set to 0.5. To have the IQR threshold re-calculated, investigators should specify the “auto” option for the IQR.thresh argument in the MEM function. It is recommended that investigators applying MEM to datasets from different instruments or who are testing MEM for the first time determine whether a change in the IQR threshold is needed.
Reference population selection
MEM scores are contextual; a population's MEM score depends on the reference population(s) to which it is compared. Selection of a reference population should be made deliberately depending on the biological question being addressed. When populations in a MEM analysis arise from different experimental sources, it may be necessary in some cases to normalize measurements prior to MEM analysis to avoid artifacts from experimental variation.
PBMC processing and mass cytometry
PBMC were isolated and cryopreserved as described by Greenplate, et al9. PBMC were stained with metal conjugated antibodies and prepared for the mass cytometry as previously described9. The following antibodies were used in the staining panel: CD19-142, CCR5-144, CD4-145, CD64-146, CD20-147, CCR4-149, CD43-150, CD14-151, TCRγδ-152, CD45RA-153, CD45-154, CXCR3-156, CD33-158, CCR7-159, CD28-169, CD29-162, CD45RO-164, CD16-165, CD44-166, CD27-167, CD8-168, CD25-169, CD3-170, CD57-172, PD-L1-175, and CD56-176 (Fluidigm Sciences). In addition, the following purified antibodies from Biolegend were labeled using MaxPar DN3 kits (Fludigm Sciences), stored at 4°C in antibody stabilization buffer (Candor Bioscience GmbH) and used in the same panel: ICOS-141, TIM-143, CD38-148, CD32-161, HLA-DR-163, CXCR5-171, and PD-1-174.
Cell subpopulation MEM Score Similarity Calculations
Comparison of CD4+ T cells to B cells in Figure 2
In order to assess the robustness of MEM across tissue sample types, donors, experimental runs, and flow cytometry platforms (fluorescence and mass cytometry), MEM scores were calculated for cell subsets from 7 different experiments that included 3 healthy human bone marrow samples15,17,21, 9 healthy human PBMC samples14,23, and 6 healthy human tonsil samples22. MEM scores were calculated for each population using as the reference population a combination of hematopoietic stem cells gated as CD34+ CD38lo/− from two studies of healthy human bone marrow15,21. Population similarity was calculated using root mean squared distance (RMSD) calculated on all population MEM scores in a pairwise fashion. MEM scores were calculated using all markers in common between each dataset and the HSC reference (Supplementary Table 4).
RMSD was calculated here as the square root of the average in squared distance between all MEM values in common for each pair of populations (Supplementary Table 4) and then converted into percent maximum possible RMSD. Given the −10 to 10 MEM scale, an RMSD of 20 was the maximum possible difference and corresponded to 0% similarity, whereas an RMSD of 0 between MEM labels indicated 100% similarity. This approach emphasized differences in marker expression when comparing populations. Calculated statistics for CD4+ T cell comparisons included average MEM value +/− standard deviation and p-value calculated using an unpaired, two-tailed Student's t-test.
Human Glioma and Normal Immune Cell MEM Analysis
Glioblastoma data (G-08, G-10, G-11, and G-22) were collected following a published protocol16. Cells were stained with isotope-tagged antibodies to detect surface and intracellular targets following established protocols16,38. MEM analysis of glioblastoma patient samples was performed with 9 markers (S100B, TUJ1, GFAP, Nestin, MET, PDGFRα, EGFR, HLA-DR, and CD44), using arcsinh transformation of original median intensity values with a cofactor of 5. Each cell subset was the POP, and the remaining cell subsets were the REF in the analysis.
Z-score and K-S statistic calculations
Z-score was calculated between POP and REF as (MEANpop-MEANref)/STDEVref for each marker.
The K-S statistic11,39 was calculated comparing the distribution for each marker on POP and REF using the function ks.test() in R.
F-measure Analysis
PBMC populations were defined by expert human gating on canonical markers. For f-measure analysis (Fig. 1c and Supplementary Fig. 2), the 25 measured markers from the CyTOF analysis of healthy PBMC were sorted based on absolute MEM scores, median values, median difference, z-score, and K-S statistic (shown in Supplementary Fig. 2), or randomly across all PBMC populations and the 25 measured proteins. The 5×25 matrix was converted into an ordered vector (length 25×5) and then sorted by absolute value. The first occurrence of each marker in the list was kept and subsequent occurrences of that marker in the list (i.e. that marker's scores on other populations) were discarded. The order of markers excluded by MEM, median, median difference, z-score, and K-S statistic are shown in Supplementary Table 3. Markers were then sequentially, cumulatively excluded from k-means clustering of cells from high to low absolute for each statistic or score. F-measure was calculated as:
- Sensitivity = True Positives/ (True Positives + False Negatives)
- Specificity = True Negatives/ (True Negatives + False Positives)
- F-measure = 2*(sensitivity*specificity)/ (sensitivity + specificity)
An F-measure was calculated for each round of clustering, where truth was the cell cluster ID resulting from clustering on all 25 markers. The moving average of f-measure with an interval of 3 was calculated in Microsoft Excel. The F-measures for random marker exclusion are the average at each point of 15 different rounds of random marker exclusion from clustering.
Supplementary Material
1
2
3
04
Acknowledgements
This study was supported by R25 CA136440-04 (K.E.D.), F31 CA199993 (A.R.G.), R00 CA143231-03 (J.M.I.), the Vanderbilt-Ingram Cancer Center (VICC, P30 CA68485), VICC Ambassadors, a VICC Hematology Helping Hands award (J.M.I. and K.E.D.), and the Vanderbilt International Scholars Program (N.L.). Thanks to Mikael Roussel for helpful discussions of myeloid cell identity markers, to Deon Doxie for helpful discussions of MEM analysis of tumor and immune cell subsets, and to Lola Chambless and Rebecca Ihrie for use of glioma tumor data generated by N.L.
Footnotes
Author contribution
All authors designed experiments, discussed data visualization and contributed intellectually to the manuscript, and approved the final manuscript. J.M.I. and K.E.D. performed computational analyses, developed analytical tools and protocols, conceived and designed the study, and wrote the manuscript. A.R.G. contributed to Fig. 2 and Fig. 3 and assisted with manuscript revisions. N.L. contributed to Fig. 3 and manuscript revisions. C.E.W. contributed to R code implementation and manuscript revisions.
Competing Financial Interests: J.M.I. is co-founder and board member and Cytobank Inc. and received research support from Incyte Corp.
References
- 1.Diggins KE, Ferrell PB, Jr., Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. doi:10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nature reviews. Immunology. 2016;16:449–462. doi: 10.1038/nri.2016.56. doi:10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 3.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. doi:10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher B, et al. High-dimensional analysis of the murine myeloid cell system. Nature immunology. 2014;15:1181–1189. doi: 10.1038/ni.3006. doi:10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 5.Greenplate AR, Johnson DB, Ferrell PB, Irish JM. Systems immune monitoring in cancer therapy. European journal of cancer. 2016;61:77–84. doi: 10.1016/j.ejca.2016.03.085. doi:10.1016/j.ejca.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. doi:10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Irish JM, et al. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12747–12754. doi: 10.1073/pnas.1002057107. doi:10.1073/pnas.1002057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. doi:10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenplate AR, et al. Myelodysplastic Syndrome Revealed by Systems Immunology in a Melanoma Patient Undergoing Anti-PD-1 Therapy. Cancer Immunol Res. 2016 doi: 10.1158/2326-6066.CIR-15-0213. doi:10.1158/2326-6066.CIR-15-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudilliere B, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Science translational medicine. 2014;6:255ra131. doi: 10.1126/scitranslmed.3009701. doi:10.1126/scitranslmed.3009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young IT. Proof without prejudice: use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1977;25:935–941. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, Donnenberg VS, Wilson JW, Donnenberg AD. The use of simultaneous confidence bands for comparison of single parameter fluorescent intensity data. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2016;89:89–97. doi: 10.1002/cyto.a.22733. doi:10.1002/cyto.a.22733. [DOI] [PubMed] [Google Scholar]
- 13.Orlova DY, et al. Earth Mover's Distance (EMD): A True Metric for Comparing Biomarker Expression Levels in Cell Populations. PloS one. 2016;11:e0151859. doi: 10.1371/journal.pone.0151859. doi:10.1371/journal.pone.0151859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leelatian N, Diggins KE, Irish JM. Characterizing Phenotypes and Signaling Networks of Si ngle Human Cells by Mass Cytometry. Methods in molecular biology. 2015;1346:99–113. doi: 10.1007/978-1-4939-2987-0_8. doi:10.1007/978-1-4939-2987-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. doi:10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leelatian N, et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry. Part B, Clinical cytometry. 2016 doi: 10.1002/cyto.b.21481. doi:10.1002/cyto.b.21481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amir el AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature biotechnology. 2013;31:545–552. doi: 10.1038/nbt.2594. doi:10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nature biotechnology. 2011;29:886–891. doi: 10.1038/nbt.1991. doi:10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irish JM. Beyond the age of cellular discovery. Nature immunology. 2014;15:1095–1097. doi: 10.1038/ni.3034. doi:10.1038/ni.3034. [DOI] [PubMed] [Google Scholar]
- 20.Nomenclature for clusters of differentiation (CD) of antigens defined on human leukocyte populations. IUIS-WHO Nomenclature Subcommittee. Bulletin of the World Health Organization. 1984;62:809–815. [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 21.Ferrell PB, Jr., et al. High-Dimensional Analysis of Acute Myeloid Leukemia Reveals Phenotypic Changes in Persistent Cells during Induction Therapy. PloS one. 2016;11:e0153207. doi: 10.1371/journal.pone.0153207. doi:10.1371/journal.pone.0153207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polikowsky HG, Wogsland CE, Diggins KE, Huse K, Irish JM. Cutting Edge: Redox Signaling Hypersensitivity Distinguishes Human Germinal Center B Cells. Journal of immunology. 2015;195:1364–1367. doi: 10.4049/jimmunol.1500904. doi:10.4049/jimmunol.1500904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas KJ, et al. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: A comparison of mass and fluorescence cytometry. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2016;89:271–280. doi: 10.1002/cyto.a.22799. doi:10.1002/cyto.a.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotecha N, Krutzik PO, Irish JM. Paul Robinson J, editor. Web-based analysis and publication of flow cytometry experiments. Current protocols in cytometry / editorial board. 2010 doi: 10.1002/0471142956.cy1017s53. Chapter 10, Unit10 17, doi:10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Civin CI, et al. Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. Journal of immunology. 1984;133:157–165. [PubMed] [Google Scholar]
- 26.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. doi:10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Basit A, et al. ICAM-1 and LFA-1 play critical roles in LPS-induced neutrophil recruitment into the alveolar space. Am J Physiol Lung Cell Mol Physiol. 2006;291:L200–207. doi: 10.1152/ajplung.00346.2005. doi:10.1152/ajplung.00346.2005. [DOI] [PubMed] [Google Scholar]
- 28.Furze RC, Rankin SM. Neutrophil mobilization and clearance in the bone marrow. Immunology. 2008;125:281–288. doi: 10.1111/j.1365-2567.2008.02950.x. doi:10.1111/j.1365-2567.2008.02950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahne F, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. doi:10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory R, Warnes BB, Bonebakker Lodewijk, Gentleman Robert, Huber Andy Liaw Wolfgang, Lumley Thomas, Maechler Martin, Magnusson Arni, Moeller Steffen, Schwartz Marc, Venables Bill. gplots: Various R Programming Tools for Plotting Data. 2015 [Google Scholar]
- 31.Lo K,HF, Brinkman R, Gottardo R. FlowClust: a Bioconductor package for automated gating of flow cytometry data. BMC bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosmann TR, et al. SWIFT-scalable clustering for automated identification of rare cell populations in large, high-dimensional flow cytometry datasets, Part 2: Biological evaluation. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2014 doi: 10.1002/cyto.a.22445. doi:10.1002/cyto.a.22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2770–2777. doi: 10.1073/pnas.1408792111. doi:10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE). Proceedings of the National Academy of Sciences of the United States of America. 2014;111:202–207. doi: 10.1073/pnas.1321405111. doi:10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. doi:10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitzer MH, et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science. 2015;349:1259425. doi: 10.1126/science.1259425. doi:10.1126/science.1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nature immunology. 2014;15:128–135. doi: 10.1038/ni.2796. doi:10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leelatian N, Diggins KE, Irish JM. Methods in Molecular Biology. 2015:99–-113. doi: 10.1007/978-1-4939-2987-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox C, Reeder JE, Robinson RD, Suppes SB, Wheeless LL. Comparison of frequency distributions in flow cytometry. Cytometry. 1988;9:291–298. doi: 10.1002/cyto.990090404. doi:10.1002/cyto.990090404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
04
Data Availability Statement
The normal human PBMC dataset (Figure 1) were generated by CyTOF analysis as described by Leelatian, et al.14 and is available as an FCS file in Flow Repository (https://flowrepository.org/experiments/1043).
The normal human bone marrow data set from Bendall and Simonds, et al15 (Dataset B, Supplementary Note 3) was downloaded from Cytobank24 as FCS files that included the cell population IDs defined by Bendall and Simonds, et al.15 (https://reports.cytobank.org/1/v1). MEM enrichment scores from Dataset B were compared to the authors’ analysis and prior studies of proteins marking stem cells, progenitor cells, and mature cells25,26
The murine myeloid CyTOF dataset from Becher, et al4 (Dataset C, Supplementary Note 4) was downloaded from Cytobank as FCS files that contained gated cell events and cluster IDs as designated by automated analysis conducted by Becher et al4. MEM enrichment scores from Dataset C were compared to the authors’ analysis and prior studies of neutrophils27,28.
Datasets for Figure 2 were generated in 7 separate fluorescence and mass cytometry studies by 1) Nicholas et al. 23, 2) Polikowsky et al.22, 3) Ferrell et al. 21, 4) Amir et al.17, 5) Bendall and Simonds et al.15, 6) Greenplate et al., previously unpublished data, and 7) Leelatian et al14.
The phospho-flow AML data set generated by Irish et al.6 (Supplementary Note 5-Fig.2) was downloaded from Cytobank as FCS files.
The human GBM mass cytometry dataset (Fig. 3) was generated and analyzed as described by Leelatian and Doxie et al.16 and are available on Flow Repository as text files (https://flowrepository.org/experiments/1044/).