Mixed-Effects Association of Single Cells Identifies an Expanded Effector CD4+ T Cell Subset in Rheumatoid Arthritis (original) (raw)
. Author manuscript; available in PMC: 2019 Apr 17.
Published in final edited form as: Sci Transl Med. 2018 Oct 17;10(463):eaaq0305. doi: 10.1126/scitranslmed.aaq0305
Abstract
High dimensional single-cell analyses have improved the ability to resolve complex mixtures of cells from human disease samples; however, identifying disease-associated cell types or cell states in patient samples remains challenging due to technical and inter-individual variation. Here we present Mixed-effects modeling of Associations of Single Cells (MASC), a reverse single cell association strategy for testing whether case-control status influences the membership of single cells in any of multiple cellular subsets while accounting for technical confounders and biological variation. Applying MASC to mass cytometry analyses of CD4+ T cells from the blood of rheumatoid arthritis (RA) patients and controls revealed a significantly expanded population of CD4+ T cells, identified as CD27- HLA-DR+ effector memory cells, in RA patients (OR = 1.7; p = 1.1 × 10−3). The frequency of CD27- HLA-DR+ cells was similarly elevated in blood samples from a second RA patient cohort, and CD27- HLA-DR+ cell frequency decreased in RA patients who responded to immunosuppressive therapy. Mass cytometry and flow cytometry analyses indicated that CD27- HLA-DR+ cells were associated with RA (meta-analysis p = 2.3 × 10−4). Compared to peripheral blood, synovial fluid and synovial tissue samples from RA patients contained ~5-fold higher frequencies of CD27- HLA-DR+ cells, which comprised ~10% of synovial CD4+ T cells. CD27- HLA-DR+ cells expressed a distinctive effector memory transcriptomic program with Th1- and cytotoxicity-associated features, and produced abundant IFN-γ and granzyme A protein upon stimulation. We propose that MASC is a broadly applicable method to identify disease-associated cell populations in high-dimensional single cell data.
One Sentence Summary:
Mixed-effects regression of single-cell data accounts for confounding variation and reveals an expanded CD4+ T cell population in rheumatoid arthritis.
Introduction
The advance of single cell technologies has enabled investigators to resolve cellular heterogeneity with unprecedented resolution. Single cell assays have been particularly useful in the study of the immune system, in which diverse cell populations often consisting of rare and transitional cell states may play an important role (1). Application of single cell transcriptomic and cytometric assays in a case-control study has the potential to reveal expanded pathogenic cell populations in immune-mediated diseases.
Rheumatoid arthritis (RA) is a chronic, systemic disease affecting 0.5–1% of the adult population, making it one of the most common autoimmune disorders worldwide (2). RA is triggered by environmental and genetic risk factors, leading to activation of autoreactive T cells and B cells that mediate an autoimmune response directed at the joints (3, 4). CD4+ T cells have been strongly implicated in RA pathogenesis (5, 6). For one, the strongest genetic association to RA is with the HLA-DRB1 gene within the MHC; these polymorphisms affect the range of antigens that MHCII molecules can bind and present in order to activate CD4+ T cells (3, 7, 8). Furthermore, many RA risk alleles outside of the MHC locus also lie in pathways important for CD4+ T cell activation, differentiation into effector (Teff) and regulatory (Treg) subsets, and maintenance of subset identity (4, 8–12). Defining the precise CD4+ T cell subsets that are expanded or dysregulated in RA patients is critical to deciphering pathogenesis. Such cell populations may be enriched in antigen-specific T cells and may aid in discovery of dominant disease-associated autoantigens. In addition, these populations may directly carry out pathologic effector functions that can be targeted therapeutically (13).
For many autoimmune diseases, directly assaying affected tissues is difficult because samples are only available through invasive procedures. Instead, querying peripheral blood for altered immune cell populations is a rapidly scalable strategy that achieves larger sample sizes and allows for serial monitoring. Flow cytometric studies have identified alterations in specific circulating T cell subsets in RA patients, including an increased frequency of CD28- CD4+ T cells (14–16); however, the expansion of CD28- T cells represents one of the relatively few T cell alterations that has been reproducibly detected by multiple groups. Limited reproducibility may be the consequence of differences in clinical cohorts, small sample sizes, methodologic variability, and use of limited, idiosyncratic combinations of phenotypic markers (17).
The advent of mass cytometry now allows for relatively broad assessment of circulating immune cell populations with >30 markers (18), enabling detailed, multiparametric characterization of lymphocyte subsets. This technology provides the potential to define and quantify lymphocyte subsets at high resolution using multiple markers. While the discovery of novel cellular populations has been enabled by rapid progress in developing sensitive clustering methods (19–23), a key challenge that remains is establishing methods to identify cell populations associated with a disease. In particular, inter-individual variation and technical variation can influence cell population frequencies and need to be accounted for in an association framework.
Single cell association studies, with either mass cytometry or single-cell RNA-seq, require statistical strategies robust to inter-individual donor variability and technical effects that can skew cell subset estimates. For example, mass cytometry studies need to control for variability in machine sensitivity, reagent staining, and sample handling that can lead to batch effects. Inter-individual differences can lead to real shifts in cell population frequencies at baseline, while technical effects can lead to apparent shifts in cell population frequencies.
Here we describe a robust statistical method to test for disease associations with single cell data called MASC (Mixed-effects modeling of Associations of Single Cells), which tests at the single cell level the association between population clusters and disease status. It is a ‘reverse’ association strategy where the case-control status is an independent variable, rather than the dependent variable. This approach enables modeling of technical and inter-individual variance as random effects, allowing robust detection of disease associated cellular populations. We applied MASC to identify T cell subsets associated with RA in a mass cytometry case-control immunophenotyping dataset that we generated focused on CD4+ T cells. This high-dimensional analysis enabled us to identify disease-specific changes in canonical as well as non-canonical CD4+ T cell populations using a panel of 32 markers to reveal cell lineage, activation, and function (24, 25). Using MASC, we identified a population of memory CD4+ T cells, characterized as CD27- HLA-DR+, which was expanded in the circulation of RA patients. Further, we found that CD27- HLA-DR+ T cells were enriched within inflamed RA joints, rapidly produced IFN-γ and cytolytic factors, and contracted with successful treatment of RA.
Results
Statistical and computational strategy
We acquired single cell data from RA case and osteoarthritis (OA) control samples (Figure 1), and then applied MASC after stringent quality controls to remove technical artifacts and poorly stained cells that are typically observed in mass cytometry data (Materials and Methods). First, we objectively defined subsets of cells using an equal number of random cells from each sample so that they contributed equally to the subsequent analyses. We then used DensVM (26) to cluster the mass cytometry data. Finally, we applied MASC to identify differentially abundant cellular populations associated with disease.
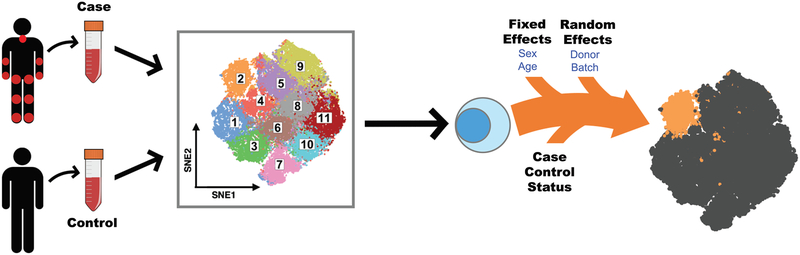
Figure 1: Mixed-effects modeling of Associations of Single Cells (MASC) overview.
Single cell transcriptomics or proteomics are used to assay samples from cases and controls, such as immunoprofiling of peripheral blood. The data is then clustered to define populations of similar cells. Mixed-effects logistic regression is used to predict individual cell membership in previously defined populations. The addition of a case-control term to the regression model allows the user to identify populations for which case-control status is significantly associated.
MASC is a reverse association strategy that uses single cell logistic mixed-effect modeling to test individual cellular populations for association by predicting the subset membership of each cell based upon fixed effects (e.g. sex) and random effects (e.g. batch, donor). It assumes a null model where the subset membership of each single cell is estimated by fixed and random effects without considering the case-control status of the samples. Thus, under this null framework, we assume that variation in cluster frequencies are not associated with case-control status. We then measured the improvement in model fit when a fixed effect term for the case-control status of the sample was included with a likelihood ratio test. This framework allowed us to evaluate the significance and effect size of the case-control association for each subset while controlling for inter-individual and technical variability.
To ensure that MASC was had appropriately calibrated type 1 error, we ran the analysis 10,000 times after permuting case-control labels using the dataset described below to obviate almost any case-control association that might be present. We then recorded the reported p-values for each cluster. Under ideal circumstances, 5% of these 10000 trials would achieve a p-value < 0.05 purely by chance in this random a data set; conversely, an inflated method would have much greater than 5% of trials obtaining a p-value < 0.05. This approach demonstrated that MASC has only a modestly inflated type I error rate for the 19 clusters in the dataset, with 6.5% of trials obtaining p<0.05 (Figure S1A). We found that including both donor and batch random effects was critical; eliminating random effects in the model led to highly inflated p-values with 66.1% of trials obtaining p<0.05 (Figure S1B). As an alternative and frequently used strategy, we also tested a simple binomial test for case-control association. This approach is limited in our view as it fails to model donor specific and technical effects. Unsurprisingly, we found that this commonly used approach produced highly inflated results in comparison to MASC, with 65.7% of trials obtaining p<0.05 (Figure S1C).
Experimental strategy
We applied MASC to mass cytometric analysis of memory CD4+ T cells that were magnetically isolated from peripheral blood mononuclear cells from patients with established RA (cases) and non-inflammatory controls (Table 1). We either (1) rested the cells for 24 hours or (2) stimulated the cells with anti-CD3/anti-CD28 beads for 24 hours before analyzing cells with a 32-marker mass cytometry panel that included 22 markers of lineage, activation, and function (Table S1). This experimental design allowed us to interrogate immune states across cases and controls and also capture stimulation-dependent changes. After stringent quality control measures, we analyzed a total of 26 RA cases and 26 controls. We randomly sampled 1000 cells from each of the 52 samples so that each sample contributed equally to the analysis, preventing samples that happened to have more cells captured by the mass cytometry assay from being overrepresented. We projected resting and stimulated T cell data separately using the Barnes-Hut modification to the t-SNE (_t_-distributed stochastic neighbor embedding) algorithm (60) so that all cells from all samples were projected into the same two dimensions using all markers in the panel with the exception of CD4 and CD45RO, which we used to gate CD4+ memory T cells for analysis.
Table 1:
Clinical characteristics of patient samples used. Mean ± SD is shown. Parentheses indicate percentages. CRP: C-reactive protein. CDAI: clinical disease activity index. “Other biologics” includes rituximab, tofacitinib, and abatacept.
| Mass cytometry cohort | Flow cytometry cohort | ||||
|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | ||
| Blood Cross-sectional cohorts | Number | 26 | 26 | 27 | 39 |
| Age | 57 ± 15 | 66 ± 9 | 61 ± 14 | 58 ± 14 | |
| Female | 19 (73) | 20 (77) | 18 (64) | 30 (77) | |
| ACPA- or RF-positive | N/A | 22 (85) | N/A | 39 (100) | |
| CRP (mg/L) | N/A | 8.6 ± 16.9 | N/A | 9.8 ± 17.9 | |
| CDAI | N/A | 9.3 ± 4.4 | N/A | 13.7 ± 7.4 | |
| Methotrexate | 0 | 18 (69) | 0 | 18 (46) | |
| Anti-TNF | 0 | 10 (38) | 0 | 16 (41) | |
| Other biologics | 0 | 5 (19) | 0 | 9 (23) |
| Longitudinal cohort | ||
|---|---|---|
| Blood Longitudinal cohort | Number | 18 |
| Age | 49 ± 17 | |
| Female | 17 (94) | |
| ACPA- or RF-positive | 18 (100) | |
| CDAI Before | 17.6 ± 9.3 | |
| CDAI After | 6.3 ± 4.2 | |
| Started methotrexate | 7 | |
| Started anti-TNF | 4 | |
| Started other biologic | 7 |
| Patient | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Synovial Tissue Donors | Age | 57 | 54 | 76 | 46 | 46 | 79 | 62 | 63 | 52 |
| Sex | F | F | F | F | F | F | M | M | F | |
| CRP (mg/L) | 25 | 8 | 8 | 11 | 17 | 19 | 13 | 66 | 76 | |
| CDAI | 14 | 9 | 17 | 15 | 21 | 25 | 5 | 9 | N/A | |
| Methotrexate | No | Yes | No | No | No | No | No | Yes | No | |
| Biologic therapy | Yes | Yes | Yes | Yes | Yes | No | No | No | No |
Clustering Approach
Projecting the data into t-SNE space revealed areas of local density that consisted of predominantly cells from RA or OA samples (Figure S2). We wanted to identify CD4+ memory T cell populations in an unsupervised way. Currently, clustering high dimensional single cell data (such as mass cytometry or single cell RNA-seq data) is an active area of research, and there is no consensus on the best clustering strategy. Hence we objectively considered multiple clustering algorithm options. We evaluated DensVM (26), FlowSOM (27), and Phenograph (23), which were identified as among the best performing in a recent benchmarking comparison of clustering methods for high dimensional cytometry data (28). The DensVM method uses an t-SNE projection of the dataset to first estimate the number of clusters by searching for local densities on the projection with varying bandwidths before classifying cells based on the similarity of expression. Phenograph works by creating a graph representing phenotypic similarities between cells and identifying clusters using Louvain community detection. The FlowSOM algorithm builds a self-organizing map with a minimum spanning tree to detect populations, then classifies cells in a meta-clustering step using consensus hierarchical clustering. We clustered cells with all three methods using the same selection of markers that was used to create t-SNE projections.
After running all three algorithms on the dataset, we identified 19, 19, and 21 clusters for DensVM, FlowSOM, and Phenograph respectively. An ideal clustering algorithm would define clusters that are distinct from each other in terms of marker intensities. However, cluster intensity differences should not be driven by batch effects; that is, clusters should not be disproportionately constituted by cells from any individual batch.
In order to quantify the extent to which clustering approaches defined clusters with distinct marker intensities, we utilized an Marker Informativeness Metric (MIM) (Materials and Methods). All three methods were similar in their ability to generate clusters with distinct marker intensities (Figure S3A). Then, to determine whether those intensity differences were dependent on batch differences, we used Cluster Informativeness Metric (CIM) based metric to assess whether clusters were disproportionately represented by individual batches (Materials and Methods). Here, we observed that Phenograph and FlowSOM were much more sensitive to batch effects than clusters identified by DensVM (p < 2 × 10−3, Wilcoxon rank sum test, Figure S3B). Consistent with this quantitative assessment, we observed that in our data FlowSOM and Phenograph produced clusters that constituted exclusively of or dominated by cells from a specific batch. Thus, we chose to analyze clusters identified by the DensVM algorithm going forward.
Landscape of CD4+ Memory T Cell Subsets
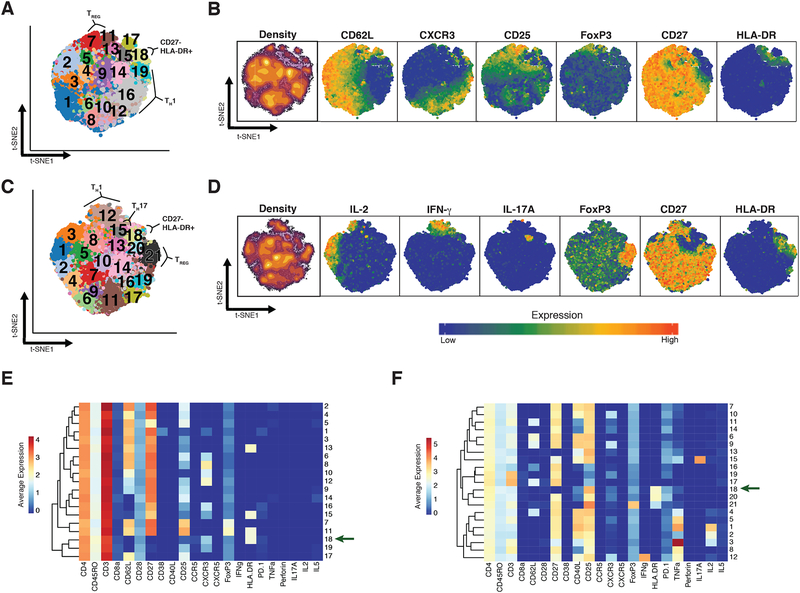
We observed substantial diversity among resting CD4+ memory T cells, consistent with previous reports demonstrating the breadth of phenotypes in CD8+ T cells and CD4+ T cells (29–31). We identified 19 distinct subsets in resting (R) memory CD4+ T cells (Figure 2A, S4A). Central memory T cells (TCM) segregated from effector memory T cells (TEM) by the expression of CD62L (Figure 2B). Five subsets (subsets 1 – 5) of TCM cells, all expressing CD62L, varied in the expression of CD27 and CD38, highlighting the heterogeneity within the TCM compartment (Figure 2E). We identified two Th1 subsets (subsets 8 and 12) as well as two Treg subsets (subsets 7 and 11). Both Treg subsets expressed high levels of CD25 and FoxP3, and subset 11 also expressed HLA-DR, reflecting a known diversity among Treg populations in humans (32).
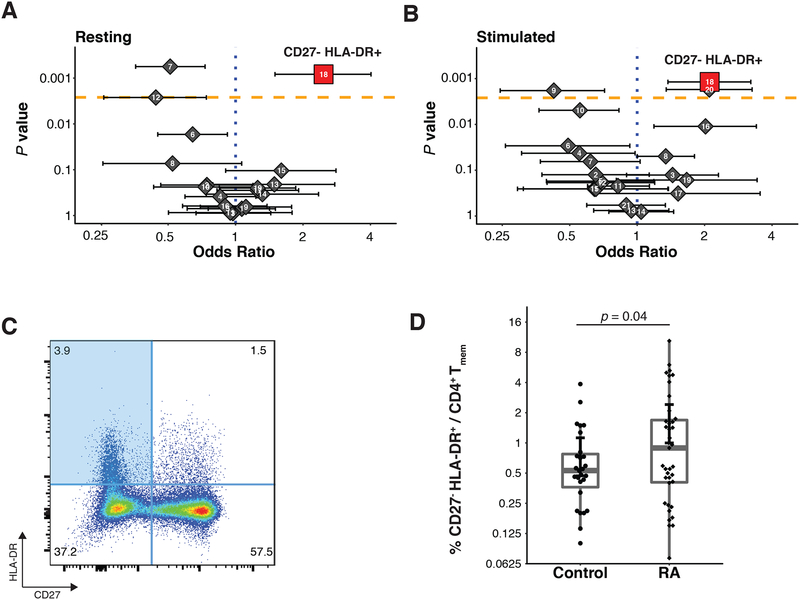
Figure 3: MASC identifies a population that is expanded in RA.
(A, B) Odds ratios and association p-values were calculated by MASC for each population identified the resting (A) and stimulated (B) datasets. The yellow line indicates the significance threshold after applying the Bonferroni correction for multiple testing. (C) Flow cytometry dot plot of gated memory CD4+ T cells from a single RA donor shows the gates used to identify CD27− HLA-DR+ memory CD4+ T cells (blue quadrant). (D) Flow cytometric quantification of the percentage of CD27−, HLA-DR+ cells among blood memory CD4+ T cells in an independent cohort of seropositive RA patients (n = 39) and controls (n = 27). Statistical significance was assessed using a one-tailed _t_-test after assessing normality with a Shapiro-Wilk test (p > 0.52).
When applied to the stimulated CD4+ T cell data in a separate analysis, DensVM identified 21 subsets (Figure 2C, S4B). As expected, certain activation markers, such as CD25 and CD40L, were broadly expressed across most subsets after stimulation (Figure 2D). Mass cytometry robustly detected cytokine production from stimulated CD4+ memory T cells (Figure 2D, 2F). Activated effector cells identified by the production of IL-2 were separated into three groups (subsets 1 – 3) according to relative expression of TNF. Cytokine expression after stimulation also improved the ability to resolve certain CD4+ T effector subsets, such as Th17 cells (subset 15) and Th1 cells (subset 12). However, we were unable to resolve the Treg subtypes that were observed in the resting T cells; after stimulation, all CD25+ FoxP3+ cells were grouped together (subset 21). The set of cells that did not activate after stimulation (subsets 16, 17, and 19) were easily identified by the lack of expression of activation markers such as CD25, CD40L, and cytokines, and the retention of high CD3 expression.
Identifying Populations that are Enriched or Depleted in RA Samples
We next sought to identify subsets that were significantly overrepresented or underrepresented in patient cells. The frequency of RA cells in each subset ranged from 36.7% to 63.6% in the resting state, and 38.9% to 65.7% in the stimulated state (Table S2, Figure S5A). Visualizing the density of the t-SNE projections revealed that related cells clustered into dense groups both at rest and after stimulation (Figure 2B, 2D), and plotting t-SNE projections of cells from cases and controls separately while coloring by density c6learly suggested differential abundance of RA cells among clusters (Figure S2). Accounting for subject-specific and batch-specific random effects with MASC (Materials and Methods), we observed three populations with significantly altered proportions of cells from cases in the resting T cell data. Most notably, we observed enrichment in subset 18 (p = 5.9 × 10−4 Table 2, Figure 3A); this subset consisted 3.1% of total cells from cases compared to 1.7% of cells from controls and achieved an odds ratio (OR) of 1.9 (95% confidence interval [CI] = 1.3 – 2.7). Conversely, subset 7 (p = 8.8 × 10−4, OR = 0.6, 95% CI = 0.5 – 0.8) and subset 12 (p = 2.0 × 10−3, OR = 0.5, 95% CI = 0.4 0.8) were underrepresented for RA cells.
Table 2:
Overview of the subsets found to be significantly expanded in RA. RA proportion reflects the fraction of cells in the subset that were from RA donors. The 95% confidence interval is shown next to the odds ratio.
| Condition | Description | Subset | RA Proportion | P value | Odds Ratio | Test |
|---|---|---|---|---|---|---|
| Resting | HLA-DR+,CD27− | 18 | 0.636 | 5.9 × 10 −4 | 1.9 (1.3 – 2.7) | MASC |
| Stimulated | HLA-DR+, CD27− | 18 | 0.619 | 1.3 × 10 −3 | 1.7 (1.2 – 2.2) | MASC |
| Flow cytometry replication | Gated HLA-DR+, CD27− | NA | NA | 4. 4 × 10 −2 | NA | One-tailed t test |
| Meta-analysis | HLA-DR+, CD27− | NA | NA | 4.8 × 10 −5 | NA | Stouffer’s Z-score method |
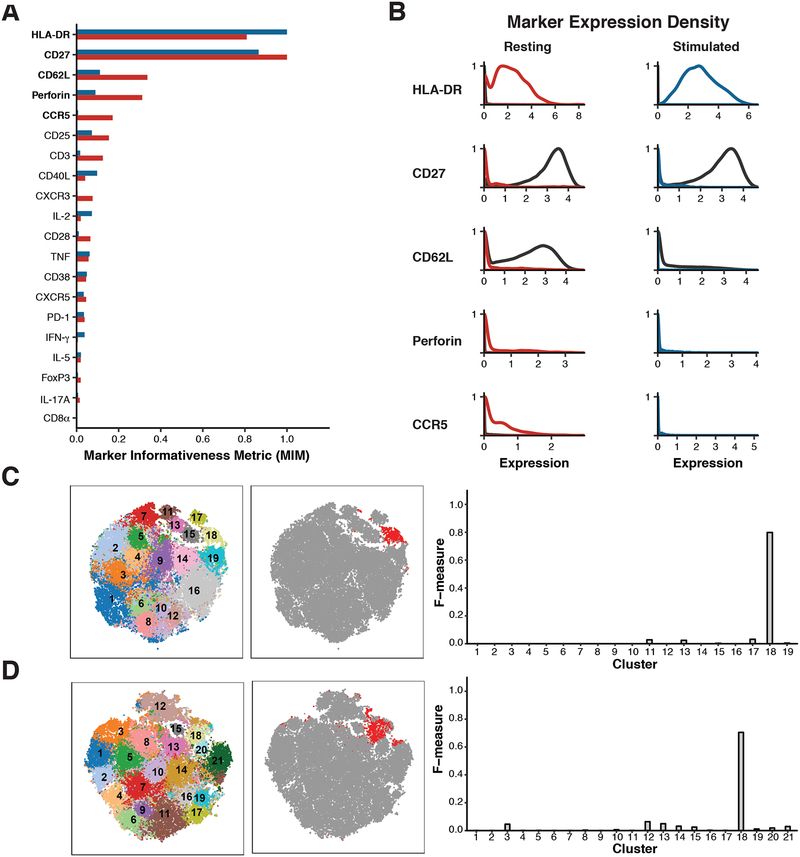
Figure 4: CD27 and HLA-DR expression specifically mark the expanded population.
(A) Plot of the Kullback-Liebler divergence for each marker comparing cluster 18 to all other cells (grey) in both the resting dataset (red) and the stimulated dataset (blue). (B) Density plots showing expression of the five markers most different between cluster 18 cells (resting = red, stimulated = blue) and all other cells in the same dataset (black line). (C) Left: t-SNE projection of clusters identified in resting dataset; Middle: Same t-SNE projection, with cells gated as CD27- HLA-DR+ colored in red; Right: F-measure scores were calculated for the overlap between gated cells and each cluster in the resting dataset. (D) Left: t-SNE projection of clusters identified in stimulated dataset; Middle: Same t-SNE projection, with cells gated as CD27- HLA-DR+ colored in red; Right: F-measure scores were calculated for the overlap between gated cells and each cluster in the stimulated dataset.
To confirm the robustness of this analytical model, we conducted a stringent permutation test that was robust to potential batch effects. To control for batch, we reassigned case-control labels randomly to each sample, retaining the same number of cases and controls in each batch. For each T cell subset we recorded the number of case cells after randomization to define a null distribution. In the real data, we observed that 753 out of the 1184 cells in subset 18 were from case samples. In contrast, when we permuted the data we observed a mean of 550 cells and a standard deviation of 63 cells from case samples. In only 93 out of 100,000 permutations did we observe more than 753 cells from case samples in the randomized data (permutation p = 9.4 × 10−4, Table S2). We applied permutation testing to every subset and found that the _p_-values produced by permutation were similar to _p_-values derived from the mixed-effects model framework (Spearman’s r = 0.86, Figure S5B, S5C). When we considered the subsets identified as significant by MASC, both subsets 18 (permutation p = 9.4 × 10−4) and 7 (permutation p = 1.8 × 10−4) retained significance whereas subset 12 demonstrated nominal evidence of case-control association (permutation p = 1.3 × 10−2).
Next, for each resting T cell subset, we identified corresponding subsets in the stimulated T cell dataset using a cluster centroid alignment strategy to calculate the distance between subsets across datasets (Materials and Methods). Subset 18 in the resting dataset was most similar to subset 18 in the stimulated dataset, while subset 7 in the resting dataset was most similar to subset 21 in the stimulated dataset (Figure S5D, S5E). Applying MASC, we observed case-control association for subset 18 in the stimulated data (p = 1.2 × 10−3, OR = 1.7, 95% CI = 1.2 – 2.2), while subset 21 (p = 0.55) was not significant in the stimulated data (Table 2, Figure 3B). We identified one additional population subset that was significant in the stimulated dataset: subset 20 (p = 1.7 × 10−3, OR = 1.7, 95% CI = 1.2 − 2.3) (Table S3).
We wanted to ensure that the results we observed were not an artifact of using DensVM to cluster the cytometry data. When we used Phenograph (23) and FlowSOM (27) to cluster the same mass cytometry dataset, we observed a CD27-, HLA-DR+ cluster with either method (Figure S6A, S6C) with association to RA by MASC (Figure S6B, S6D). We also assessed how analysis by MASC compared to Citrus (43), an algorithm that uses hierarchical clustering to define cellular subsets and then builds a set of models using the clusters to stratify cases and controls. When applied to the same case-control resting dataset, Citrus was unable to produce models with an acceptable cross-validation error rate, regardless of the method used (Figure S7).
CD27- HLA-DR+ CD4+ Effector Memory T Cell Expansion in RA
After noting that subset 18 demonstrated robust association with RA, we interrogated the key features of this population. The lack of CD62L expression iin subset 18 ndicates that this subset is an effector memory T cell population. To define the markers that best differentiated this subset from other cells, we calculated the MIM for the expression of each marker in the subset for both the resting and stimulated datasets (Materials and Methods, Figure 4A). The expression of HLA-DR and perforin were notably increased in subset 18 compared to all other cells, while the expression of CD27 was decreased in this subset (Figure 4B). We observed that gating on CD27 and HLA-DR largely recapitulated subset 18 in both the resting cells and the stimulated cells (Figure 4C and 4D), with an F measure of 0.8 and 0.7 respectively (Materials and Methods). Although HLA-DR is a known to be expressed on T cells in response to activation, it takes several days to induce strong HLA-DR expression (33). Thus, it is likely that cells in subset 18 expressed HLA-DR prior to stimulation, such that analyses of both resting and stimulated cells identified the same HLA-DR+ CD27- effector memory T cell population.
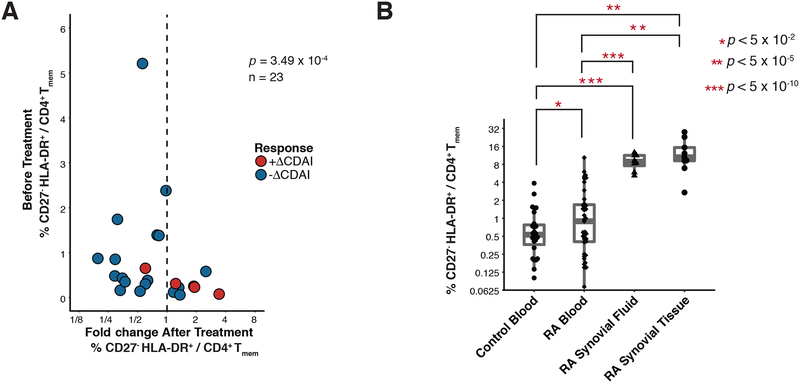
Figure 5: CD27− HLA-DR+ memory CD4+ T cells are expanded in the blood and joints of patients with active RA.
(A) Flow cytometric quantification of the frequency of CD27- HLA-DR+ memory CD4+ T cells in 18 RA patients prior to starting a new medication, plotted against change in cell frequency after 3 months of new therapy. Treatment significantly reduced CD27- HLA-DR+ cell frequency as determined by a Wilcoxon signed-rank test. (B) Flow cytometric quantification of the percentage of memory CD4+ T cells with a CD27- HLA-DR+ phenotype in cells from seropositive RA synovial fluid (n=8) and synovial tissue (n=9), compared to blood samples from RA patients and controls. Blood sample data are the same as shown in panel 3d. Significance was assessed using one-tailed _t_-test after determining normality with a Shapiro-Wilk test (p > 0.52) and applying a Bonferroni correction for multiple testing.
In order to confirm the expansion of the CD27- HLA-DR+ T cell population in RA patients that we observed by mass cytometry, we evaluated the frequency of CD27- HLA-DR+ T cells in an independent cohort of 39 seropositive RA patients and 27 non-inflammatory controls using conventional flow cytometry (Table 1). We determined the percentage of memory CD4+ T cells with a CD27- HLA-DR+ phenotype by gating individual samples from each group (Figure 3C, Figure S8A, Figure S9). Consistent with the mass cytometry analysis, CD27- HLA-DR+ cells were significantly expanded in the RA patient samples (p = 0.044, one-tailed t test, Shapiro Wilk normality test p > 0.52, Figure 3D). The frequency of this subset was 0.8% in controls and 1.7% in RA samples, which is similar to the two-fold enrichment we observed in the mass cytometry data. We then considered the mass cytometry and flow cytometry association results together in a meta-analysis, confirming that CD27- HLA-DR+ cells significantly associated with RA (p = 2.3 × 10−4, Stouffers Z-score method, Table 2).
To assess the effect of RA treatment on CD27- HLA-DR+ cell frequency, we quantified CD27- HLA-DR+ cell frequencies in 23 RA patients before and 3 months after initiation of a new medication for RA. We dichotomized patients as those who experienced a clinical response, defined as a reduction in CDAI (Clinical Disease Activity Index) (61) scores (ΔCDAI-), versus those that did not, defined as an increase in CDAI scores (ΔCDAI+). We observed that changes in CD27- HLA-DR+ cell frequency tracked with clinical response to treatment initiation (p = 3.49×10−4, Wilcoxon signed-rank test, Figure 5A). Specifically, in the 18 ΔCDAI- patients, the frequency of CD27- HLA-DR+ cells significantly reduced by 0.7 fold; in contrast, the 5 ΔCDAI+ patients in this same trial had an 1.8-fold increase in CD27- HLA-DR+ cells (p = 0.006, Wilcoxon signed-rank test, Figure S10A). We observed a significant difference in the CD27- HLA-DR+ frequency fold-change between patients experiencing a CDAI reduction versus not (p = 0.02, Wilcoxon rank sum test, Figure S10B). The decrease in frequency of CD27- HLA-DR+ cells in patients responding to treatment escalation was accompanied by a slight increase in the frequency of CD27+ HLA-DR- cells: CD27+ HLA-DR- cells represented an average of 86.2% of CD4+ memory T cells before treatment, and an average of 87.9% of CD4+ memory T cells afterwards (p = 0.009, Wilcoxon signed-rank test, Figure S10C). These results confirm that CD27- HLA-DR+ CD4+ T cells were expanded in the circulation of RA patients and decreased with effective disease treatment.
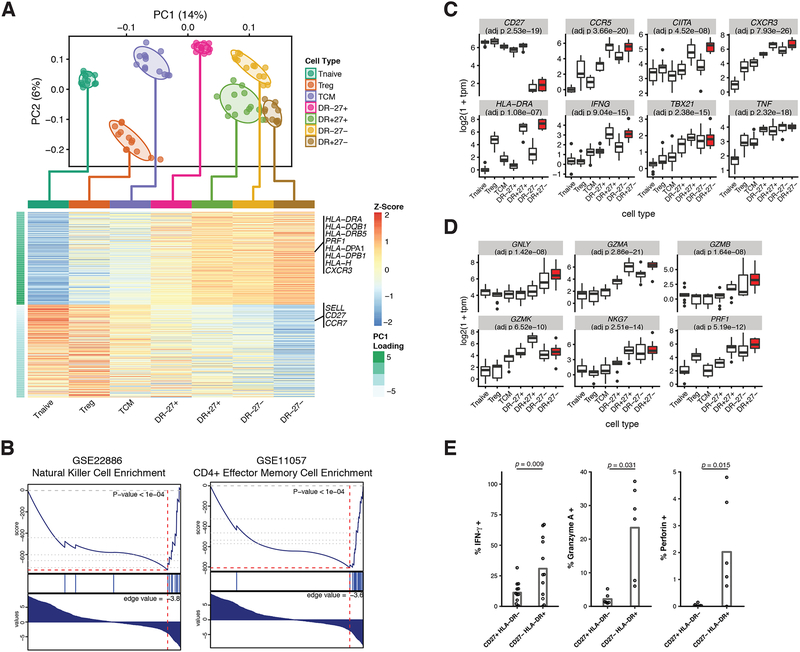
Figure 6: Transcriptomic characterization of CD27- HLA-DR+ memory CD4+ T cells identified a Th1-skewed cytotoxic phenotype.
RNA-seq characterization of CD627- HLA-DR+ (DR+27-) cells and 6 related CD4+ T cell populations: naive T cells (Tnaive), regulatory T cells (Treg), central memory t cells (TCM), and three populations of effector memory T cells, CD27+ HLA-DR- (DR-27+), CD27+ HLA-DR+ (DR+27+), and CD27- HLA-DR- (DR-27-). (A) PCA plot (top) and PC1 gene loadings (bottom) of 90 samples from the 7 CD4+ T cell populations. Cells were colored on the PCA plot according to known cell type. Normal confidence ellipses at 1 standard deviation were plotted for each cell type. The 300 most positive and 300 most negative PC1 gene loadings for each cell type were averaged and plotted in the heatmap. Genes relevant to the CD27- HLA-DR+ population were labeled. (B) Gene set enrichment analysis was performed on all genes, ranked on their PC1 loadings. Two significantly enriched gene sets: NK signature (GSE22886 NAIVE CD4 T CELL VS NK CELL DN) and effector memory t cell signature (GSE11057 NAIVE VS EFF MEMORY CD4 T CELL) are shown. (C) Distribution of log-scaled expression of six canonical Th1 genes: CCR5, CIITA, CXCR3, IFNG, TBX21 (Tbet), and TNF. Populations are ordered by PC1 loading values, with CD27- HLA-DR+ population highlighted in red. (D) Distribution of log-scaled gene expression of six canonical cytotoxic genes: GNLY, GZMA, GZMB, GMZK, NKG7, and PRF1. Populations are ordered by PC1 loading values, with the CD27- HLA-DR+ population highlighted in red. Reported p-values in (C) and (D) correspond to a linear model of gene expression against ordered cell type (as an ordinal variable), with p-values adjusted for multiple testing by the Benjamini Hochberg procedure. (E) Cytokine expression determined by intracellular cytokine staining of peripheral effector memory CD4+ T cells after in vitro stimulation with PMA/ionomycin. The percentage of cells positive for each stain is plotted for CD27+ HLA-DR- and CD27- HLA-DR+ subsets. Each dot represents a separate donor (n = 12; 6 RA patients and 6 controls, except for the quantification of Granyzme A and perforin where n = 6; 3 RA patients and 3 controls). Statistical significance was assessed using a Wilcoxon signed-rank test.
To determine whether CD27- HLA-DR+ T cells were further enriched at the sites of inflammation in seropositive RA patients, we evaluated T cells in inflamed synovial tissue samples obtained at the time of arthroplasty (Table 1) and in inflammatory synovial fluid samples from RA patients. In a set of 9 synovial tissue samples with lymphocytic infiltrates observed by histology, the frequency of CD27- HLA-DR+ cells was significantly increased 5-fold (p < 0.01, Wilcoxon rank-sum, median 10.5% of memory CD4+ T cells) compared to blood (Figure 5B). Notably, in 2 of the tissue samples, >20% of the memory CD4+ T cells displayed this phenotype. CD27- HLA-DR+ cells were similarly expanded in synovial fluid samples from seropositive RA patients (p < 0.001, Wilcoxon rank-sum, median 8.9% of memory CD4+ T cells, n=8) Thus, CD27- HLA-DR+ T cells are enriched at the primary sites of inflammation in RA patients.
Functional and Transcriptional Features of CD27- HLA-DR+ CD4+ Effector Memory T Cells
To evaluate the potential function of the CD27- HLA-DR+ cell subset, we performed RNA-seq on CD27- HLA-DR+ CD4+ T cells with other effector populations to identify transcriptomic signatures for this subset (Figure S8B–D). We sorted and sequenced the following CD4+ T cell populations: naïve CD4+ T cells (TN), central memory CD4+ T cells (TCM), regulatory CD4+ T cells (TReg), and all four CD27−/+ HLA-DR−/+ subsets – defined as DR+27+ Effector T cells (DR+27+), DR+27- Effector T Cells (DR+27-), DR-27+ Effector T Cells (DR-27+), and DR-27- Effector T Cells (DR-27-) – from peripheral blood mononuclear cells (PBMCs) from 7 RA cases and 6 OA controls. In total we generated 1.1 billion reads for 90 samples. We aligned reads with Kallisto (34) and applied stringent quality controls to remove genes that lacked sufficient expression (Materials and Methods), ultimately resulting in a set of 15,234 genes for analysis.
We performed principal components analysis (PCA) on the expression data (Figure 6A). Strikingly, the first PC, capturing 4% of the total variation, separated cell types along a naïve to effector axis, with the CD27- HLA-DR+ subset representing the extreme case with the highest PC1 values, and naïve T cells with the lowest PC1 values. We examined the genes with the highest PC1 loadings, and found that PC1 was associated negatively with CCR7 and positively with CXCR3 and CCR5. This finding was consistent with the elevated expression of CXCR3 and CCR5 we observed in the mass cytometry analyses of CD27- HLA-DR+ cells. We note that CXCR3 and CCR5 are both chemokine receptors associated with a Th1 phenotype. In addition, PRF1 (perforin), a cytotoxic factor, was also strongly associated with PC1. The extreme position of CD27- HLA-DR+ cells along the continuum of CD4+ T cells suggested a possible late or terminal effector memory phenotype. To further explore this naïve to effector gradient, we used the gene loadings along PC1 to perform gene set enrichment analysis (GSEA) and identify the pathways that were most associated. Intriguingly, the naïve to CD27- HLA-DR+ axis was strongly correlated with naïve vs effector and natural killer (NK) vs CD4+ T cell gene signatures (Figure 6B).
The association of CXCR3 and CCR5 with PC1 prompted us to examine the expression of Th1-associated genes in these cells. The effector memory cell populations showed increased expression of multiple Th1-associated genes, including IFNG (IFN-γ) and TBX21 (Tbet), compared to naïve, Treg, and central memory cells. In addition, expression of these Th1-associated genes was higher in CD27- HLADR+ compared to CD27+ HLADR- effector memory cells, which constituted the majority of the effector memory T cell pool (Figure 6C). In contrast, cytokines characteristic of other polarized Th subsets (IL17A, IL4, IL21, TGFB1) and transcription factors associated with other Th subsets (RORC, GATA3, BCL6, FOXP3) were not elevated in CD27- HLA-DR+ cells compared to other effector populations (Figure S11A). We noted that the transcription factor CIITA was also increased in CD27- HLA-DR+ cells (Figure 6C). CIITA is well-known for its role as a regulator of MHC class II; indeed, we observed that targets of CIITA such as HLA genes were significantly overexpressed in the CD27- HLA-DR+ population compared to other populations (Figure S11B).
As the expression of PRF1 was positively associated with PC1, we wanted to assess the relationship between the expression of cytotoxic gene programs and the naïve to effector PC1 gradient. We used gene set enrichment analysis (GSEA) to query whether NK cell associated genes were upregulated in the CD27-HLA-DR+ population. Specifically, we ranked genes by their degree of differential expression, comparing expression in CD27-HLA-DR+ versus that in all other cell types. For NK cell associated genes, we used the previously defined geneset from MSigDB that defines genes upregulated in NK cells versus CD4+ T cells. Indeed, genes that were highly expressed in NK cells similarly showed high expression in CD27- HLA-DR+ cells, while genes with low expression in NK cells showed similarly low expression in CD27- HLADR+ cells (Table S4). Consistent with the enrichment results, a set of genes characteristically associated with cytolytic cells, including PRF1, GZMB, GZMA, GNLY, and NKG7 were all increased in expression in CD27- HLADR+ cells compared to most other T cell populations analyzed (Figure 6D). These results indicate that CD27- HLA-DR+ cells express a transcriptomic signature characteristic of cytotoxic cells.
We also evaluated the production and expression of cytokines and cytolytic factors at the protein level by intracellular flow cytometry. We assessed production of effector molecules by cells after in vitro stimulation with PMA + ionomycin, a stimulation method that readily reveals T cell capacity for cytokine production. Consistent with RNA-seq analyses, CD27- HLA-DR+ cells also more frequently expressed perforin (p = 0.031, one-tailed Wilcoxon signed-rank test) and granzyme A (p = 0.015, one-tailed Wilcoxon signed-rank test), than did CD27+ HLA-DR- cells, which constitute the majority of memory CD4+ T cells. In addition, CD27- HLA-DR+ cells produced IFN-γ at a much higher frequency (p = 0.009, one-tailed Wilcoxon signed-rank test, Figure 6E). Percent positivity for each marker was determined by selecting an expression level threshold to determine positive staining (Figure S12). Taken together, broad analyses of gene expression and targeted measures of effector molecule production at the protein level indicated that CD27 HLA-DR+ T cells are a Th1-skewed T cell population capable of producing cytotoxic molecules.
Discussion
Mass cytometry has been successfully applied to decipher the heterogeneity of human T cells in multiple settings, including for the identification of disease-specific changes to circulating immune cell populations and mapping of developmental pathways (29, 35–38). It and other emerging single cell strategies offer a promising avenue to characterize the T cell and other immunological features of a wide-range of diseases in humans. However, successful application of mass cytometry on a larger scale (>20 samples) to conduct association studies on clinical phenotypes in humans has been limited thus far (39–42), in part due to the limited availability of effective association testing strategies.
Although there are many approaches to cluster single cell data (19–23, 26, 27), extension of these methods to perform case-control association testing is not straightforward. The simplest strategy would be to define subsets of interest by clustering the cells and then comparing frequencies of these subsets in cases and controls with a univariate test such as a t-test or a non-parametric Mann-Whitney test. While commonly used in flow cytometry analysis, this approach is dramatically underpowered, as it relies upon reducing single-cell data to potentially inaccurate per-sample subset frequencies.
In contrast, our methodology takes full advantage of single cell measurements by using a “reverse-association” framework to test whether case-control status influences the membership of a given cell in a population - that is, each single cell was treated as a single event. However, these cells are not actually entirely statistically independent. Failing to account for dependencies, for example by using a binomial test or not accounting for random effects, can result in considerably inflated statistics and irreproducible associations (Figure S1). Using a mixed-effects logistic regression model allowed us to account for covariance in single cell data induced by technical and biological factors that could confound association signals (Figure 1, Figure S1B), without inflated association tests. MASC also allows users to utilize technical covariates that might be relevant to a single cell measurement (e.g. read depth per cell for single cell RNA-seq or signal quality for mass cytometry) that might influence cluster membership for a single cell.
We note that Citrus, an association strategy to automatically highlight statistical differences between experimental groups and identify predictive populations in mass cytometry data (43), was unable to identify the expanded CD27- HLA-DR+ population. We believe that our methodology compared favorably to Citrus because it incorporated both technical covariates (e.g. batch) and clinical covariates when modeling associations, a key feature when analyzing high-dimensional datasets of large disease cohorts. Additionally, the agglomerative hierarchical clustering framework in Citrus substantially increased the testing burden and limited power.
Although we found that clustering with DensVM outperformed FlowSOM and Phenograph on our data (Figure S3), we want to emphasize that many clustering methods have emerged to analyze both cytometry and single cell RNA-seq data; clustering single cell data remains an active field of research (44, 45). There continues to be controversy as to the best strategies, most appropriate metrics, and the optimal parameter choices (44, 46–48). Available clustering approaches utilize a variety of methods, such as graph-based community detection, self-organizing maps (27), and density-based clustering (20, 23, 49). As neural networks and as a subclass, autoencoders, have been proven to be powerful general nonlinear models, we implemented an autoencoder-based clustering approach that we tested on our datasets. We found this clustering method also had potential, and should be further investigated in the future (Figure S13). As different clustering strategies may be more appropriate for different datasets, we have implemented MASC specifically to allow for different clustering options based on user preference.
The CD27- HLA-DR+ T cells identified by MASC in blood samples from RA patients were further enriched in both synovial tissue and synovial fluid of RA patients. The accumulation of these cells in chronically inflamed RA joints, combined with lack of CD27, suggests that these cells have been chronically activated. Loss of CD27 is characteristic of T cells that have been repeatedly activated, for example T cells that recognize common restimulation antigens (50–52). Importantly, the broader CD27- CD4+ T cell population did not itself differ in frequency between RA patients and controls, in contrast to the expanded population of CD27- HLA-DR+ cells identified by MASC. We hypothesize the CD27- HLA-DR+ T cell population in RA patients may be enriched in RA antigen-specific T cells, offering a potential tool to identify relevant antigens in RA.
Expression of HLA-DR suggests recent or continued activation of these cells in vivo. The well-known HLA class II association to RA may also suggest an important role in disease susceptibility for this subset. Despite the suspected chronic activation of these cells, CD27- HLA-DR+ cells did not appear functionally exhausted. CD27- HLA-DR+ cells rapidly produced multiple effector cytokines upon stimulation in vitro, with an increased predisposition to IFN-γ production. These cells also showed increased expression of cytolytic molecules such as granzyme A and perforin on both the transcriptomic and proteomic level.
CD4+ cytotoxic T cells expressing granzyme A and perforin, also with a CD27- phenotype, are reported to be expanded in patients with chronic viral infections (53). Of note, CD4+ cells that are perforin+ and granzyme A+ have been observed in RA synovial samples (54, 55) and other chronic inflammatory conditions (56, 57). Interestingly, cytolytic CD4+ T cells lack the capacity to provide B cell help, suggesting that this is a distinct population from the expanded T peripheral helper cell population in seropositive RA (13, 55). These findings nominate CD27- HLA-DR+ T cells as a potential pathogenic T cell population that may participate in the chronic autoimmune response in RA.
Although we were able to detect significant associations between the frequency of CD27- HLA-DR+ cells and clinical response (Figure 5A), we recognize the need for a larger study to detect other subtle subphenotypic associations, such as association with specific therapeutics or disease activity (Figure S14). To this end, larger prospective cohorts with deep immunophenotyping of CD4+ T cells in blood and tissue will be critical.
We note that this study has certain limitations. First, the degree of expansion of CD27- HLA-DR+ T cells in the periphery is modest and varies between individuals. While we were able to recover the majority of cells identified as expanded by MASC in our mass cytometry experiments using CD27 and HLA-DR as markers; we note that the expression of cells has some degree of heterogeneity (Figure S15). More fine-grained immunophenotyping may help to refine phenotypes that even more specifically pinpoint the disease-associated T cell population. While we have characterized these cells as effector memory CD4+ T cells that produce molecules associated with cytotoxicity and Th1 phenotype, further characterization is needed in future studies. For example, we assigned an effector memory phenotype to the CD27- HLADR+ cell subset based on upon mass cytometry data; however, the mass cytometry panel did not specifically measure the expression of CD45RA. While this would appear to leave open the possibility that these cells might belong to a TEMRA (CD45RO+ CD45RA+) subset, we note that our sorting strategy for cells prior to assay by mass cytometry specifically excluded CD45RA+ cells and consider this possibility unlikely. In addition, our study focused exclusively on CD45RO+ memory CD4+ T cells, thus we have not assessed other CD4+ T cells populations that may also have cytotoxic features, including T effector memory CD45RA+ (TEMRA) cells (62). A broader cytometric assessment of T cell and other immune cell phenotypes in RA will be of interest in subsequent studies. Also, we did observe that the mRNA and protein expression of specific markers was not always concordant. Applying more recently developed single cell techniques that ascertain protein and mRNA expression simultaneously would better describe the dynamics of CD27- HLA-DR+ T cells in response to stimulation (63, 64). We anticipate that MASC will be applied test for case-control associated differential abundance across multiple cell types in the future.
In summary, the MASC single-cell association modeling framework identified a Th1-skewed cytotoxic effector memory CD4+ T cell population expanded in RA using a case-control mass cytometry dataset. The MASC method is adaptable to any case-control experiment in which single cell data are available, including flow cytometry, mass cytometry, and single cell RNA-seq datasets. Although current single cell RNA-seq studies are not yet large scale, ongoing projects may benefit from using the MASC framework in case-control testing, or testing for other clinical subphenotypes such as specific treatment response or disease progression.
Materials and Methods
Study Design
The objective of this research study was to profile immune subsets in peripheral blood samples from RA patients and osteoarthriris (OA) controls to identify disease-related alterations or changes in frequency among CD4+ T cells.
Human subject research was performed in accordance with the Institutional Review Boards at Partners HealthCare and Hospital for Special Surgery via approved protocols (Partners HealthCare Protocol 2014P00255) with appropriate informed consent as required. Patients with RA fulfilled the ACR 2010 Rheumatoid Arthritis classification criteria, and electronic medical records were used to ascertain patients’ rheumatoid factor and anti-CCP antibody status, C-reactive protein level, and medication use. Synovial tissue samples for mass and flow cytometry were collected from seropositive RA patients undergoing arthroplasty at the Hospital for Special Surgery, New York or at Brigham and Women’s Hospital, Boston. Samples with lymphocytic infiltrates on histology were selected for analysis. Sample inclusion criteria were established prospectively, with the exception of samples that were excluded from the study due to poor acquisition by the mass cytometer. The study sample size was a result of including all samples that passed quality control and not set prospectively.
Synovial fluid samples were obtained as excess material from a separate cohort of patients undergoing diagnostic or therapeutic arthrocentesis of an inflammatory knee effusion as directed by the treating rheumatologist. These samples were de-identified; therefore, additional clinical information was not available.
Blood samples for clinical phenotyping were obtained from consented patients seen at Brigham and Women’s Hospital. We performed medical record review for ACPA (anti-citrullinated protein antibody) positivity according to CCP2 (cyclic citrullinated protein) assays, C-reactive protein (CRP), and RA-specific medications including methotrexate and biologic DMARDS. For blood cell analyses in the cross-sectional cohort, the treating physician measured the clinical disease activity index (CDAI) on the day of sample acquisition. For RA patients followed longitudinally, a new disease-modifying antirheumatic drug (DMARD) was initiated at the discretion of the treating rheumatologist, and CDAIs were determined at each visit by trained research study staff. Blood samples were acquired before initiation of a new biologic DMARD or within 1 week of starting methotrexate and 3 months after initiating DMARD therapy(58). Concurrent prednisone at doses ≤10mg/day were permitted. All synovial fluid and blood samples were subjected to density centrifugation using Ficoll-Hypaque to isolate mononuclear cells, which were cryopreserved for batched analyses.
Sample Preparation for Mass Cytometry
We rapidly thawed cryopreserved PBMCs and isolated total CD4+ Memory T cells by negative selection using MACS magnetic bead separation technology (Miltenyi). Subsequently, we rested the CD4+ Memory T cells for 24 hours in complete RPMI (Gibco) sterile-filtered and supplemented with 15% FBS, 1% Pen/Strep (Gibco), 0.5% Essential and Non-Essential Amino Acids (Gibco), 1%Sodium Pyruvate (Gibco), 1% HEPES (Gibco), and 55μM 2-mercaptoethanol (Gibco). We activated the cells using Human T-Activator CD3/CD28 Dynabeads (ThermoFisher) at a density of 1 bead:2 cells. At 6 hours prior to harvesting (t=18 hours of stimulation), we added Monensin and Brefeldin A 1:1000 (BD GolgiPlug and BD GolgiStop). After 24 hours of stimulation, we incubated the cells with a rhodium metallointercalator (Fluidigm) in culture at a final dilution of 1:500 for 15 minutes as a viability measure. We then harvested cells into FACS tubes and washed with CyTOF Staining Buffer (CSB) composed of PBS with 0.5% BSA (Sigma-Aldrich), 0.02% sodium azide (Sigma-Aldrich), and 2μM EDTA (Ambion). We spun the cells at 500xg for 7 minutes at room temperature. We incubated the resulting cell pellets in 10ul Fc Receptor Binding Inhibitor Polyclonal Antibody (eBioscience) and 40ul of CSB for 10 minutes at 4C. The samples were then incubated for 30 minutes at 4C on a shaker rack with 1ul of the following eighteen CyTOF surface antibodies in a cocktail brought to a volume of 50ul/sample in CSB: Anti-Human CD49D (9F10)-141Pr (Fluidigm), Anti-Human CCR5 (CD195)(–P-6G4) - 144Nd (Fluidigm), Anti-Human CD4 (RPA-T4) −145Nd (Fluidigm), Anti-Human CD8a (RPA-T8) −146Nd (Fluidigm), Anti-Human CD45RO-147Sm (Brigham and Women’s Hospital CyTOF Core), Anti-Human CD28–148Nd (BWH CyTOF Core), Anti-Human CD25 (IL-2R- (2A3) - 149Sm (Fluidigm), Anti-Human PD1–151Eu (BWH CyTOF Core), Anti-Human CD62L (DREG-56)-153Eu (Fluidigm), Anti-Human CD3 (UCHT1) −154Sm (Fluidigm), Anti-Human CD27 (L128) −155Gd (Fluidigm), Anti-Human CD183[CXCR3](G025H7)-156Gd (Fluidigm), Anti-Human CCR7–170Er (BWH CyTOF Core), Anti-Human ICOS-160Gd (BWH CyTOF Core), Anti-Human CD38 (HIT2)-167Er (Fluidigm), Anti-Human CD154 (CD40L) (24–31)-168Er (Fluidigm), Anti-Human CXCR5[CD185](51505)-171Yb (Fluidigm), and Anti-Human HLA-DR (L243) −174Yb (Fluidigm). We washed the cells with 1 ml of CSB and spun at 700xg for 5 minutes at RT. Post spin, we aspirated the buffer from pellet and added 1 ml 1:4 ratio of concentrate to diluent of a Foxp3 / Transcription Factor Staining Buffer Set (eBioscience) supplemented with formaldehyde solution (Sigma-Aldrich #F1268) to a final concentration of 1.6%. We incubated the cells at room temperature on a gentle shaker in the dark for 45 minutes. We washed the cells with two ml of CSB + 0.3% saponin (CSB-S), and spun at 800xg for 5 minutes. We incubated the cell pellet with 1ul of the following fourteen intracellular antibodies and 10ul of a solution of Iridium (1:25) in CSB-S brought to a total volume of 100ul for 35 minutes at r.t. on a gentle shaker: Anti-Human IL-4 (MP4–25D2)-142 (Fluidigm), Anti-Mouse/Human IL-5 (TRFK5) −143Nd (Fluidigm), Anti-Human IL-22 (22URTI) −150Nd (Fluidigm), Anti-Human TNFα (Mab11) −152Sm (Fluidigm), Anti-Human IL-2 (MQ1–17H12)-158Gd (Fluidigm), Anti-Human IL21–159Tb (BWH CyTOF Core), Anti-Human IFNg (B27) −165Ho (Fluidigm), Anti-Human GATA3–166Er (BWH CyTOF Core), Anti-Human IL9–172Yb (BWH CyTOF Core), Anti-Human Perforin (B-D48)-175Lu (Fluidigm), Anti-Human IL10–176Yb (BWH CyTOF Core), Anti-Human IL17A-169Tm (BWH CyTOF Core), Anti-Human Foxp3 (PCH101)-162Dy (Fluidigm), and Anti-Human Tbet-164Dy (BWH CyTOF Core). Post-incubation, we washed the cells in PBS and spun them at 800xg for 5 minutes. We resuspended the pellet in 1 ml of 4% formaldehyde prepared in CSB and incubated the cells for 10 minutes at r.t. on a gentle shaker, followed by another PBS wash and spin at 800xg for 5 minutes. We washed the pellet in 1ml of MilliQ deionized water, spun at 800xg for 6 minutes, and subsequently resuspended the resulting pellet in deionized water at a concentration of 700,000 cells per ml for analysis via the CyTOF 2. We transferred the suspensions to new FACS tubes through a 70μm cell strainer and added MaxPar EQ Four Element Calibration Beads (Fluidigm #201078) at a ratio of 1:10 by volume prior to acquisition.
Mass Cytometry Panel Design
We designed an antibody panel for mass cytometry with the goal of both accurately identifying CD4+ effector memory T cell populations and measuring cellular heterogeneity within these populations. We chose markers that fell into one of five categories to generate a broadly informative panel: chemokine receptors, transcription factors, lineage markers, effector molecules, and markers of cellular activation and exhaustion (Table S1).
Mass Cytometry Data Acquisition
We analyzed samples at a concentration of 700,000 cells/ml on a Fluidigm-DVS CyTOF 2 mass cytometer. We added Max Par 4-Element EQ calibration beads to every sample that was run on the CyTOF 2, which allowed us to normalize variability in detector sensitivity for samples run in different batches using previously described methods(59). We used staining for iridium and rhodium metallointercalators to identify viable singlet events. We excluded samples where acquisition failed or only yielded a fraction of the input cells (< 5%). The criteria for sample exclusion were not set prospectively but were maintained during all data collection runs.
As samples were processed and analyzed on different dates, we ran equal numbers of cases and controls each time to guard against batch effects. However, as CyTOF data are very sensitive to day-to-day variability, we took extra steps to pre-process and normalize data across the entire study.
Flow Cytometry Sample Preparation
For flow cytometry analysis of the validation and longitudinal blood cohorts and synovial samples, cryopreserved cells were thawed into warm RPMI/10% FBS, washed once in cold PBS, and stained in PBS/1% BSA with the following antibodies for 45 minutes: anti-CD27-FITC (TB01), anti-CXCR3-PE (CEW33D), anti-CD4-PE-Cy7 (RPA-T4), anti-ICOS-PerCP-Cy5.5 (ISA-3), anti-CXCR5-BV421 (J252D4), anti-CD45RA-BV510 (HI100), anti-HLA-DR-BV605 (G46–6), anti-CD49d-BV711 (9F10), anti-PD-1-APC (EH12.2H7), anti-CD3-AlexaFluor700 (HIT3A), anti-CD29-APC-Cy7 (TS2/16), propidium iodide. Cells were washed in cold PBS, passed through a 70-micron filter, and data acquired on a BD FACSAria Fusion or BD Fortessa using FACSDiva software. Samples were analyzed in uniformly processed batches containing both cases and controls.
Flow Cytometry Intracellular Cytokine Staining
Effector memory CD4+ T cells were purified from cryopreserved PBMCs by magnetic negative selection (Miltenyi) and rested overnight in RPMI/10%FBS media. The following day, cells were stimulated with PMA (50ng/mL) and ionomycin (1μg/mL) for 6 hours. Brefeldin A and monensin (both 1:1000, eBioscience) were added for the last 5 hours. Cells were washed twice in cold PBS, incubated for 30 minutes with Fixable Viability Dye eFluor 780 (eBioscience), washed in PBS/1%BSA, and stained with anti-CD4-BV650 (RPA-T4), anti-CD27-BV510 (TB01), anti-HLADR-BV605 (G46–6), anti-CD20-APC-Cy7 (2H7), and anti-CD14-APC-Cy7 (M5E2). Cells were then washed and fixed and permeabilized using the eBioscience Transcription Factor Fix/Perm Buffer. Cells were then washed in PBS/1%BSA/0.3% saponin and incubated with anti-IFN-γ-FITC (B27), anti-TNF-PerCp/Cy5.5 (mAb11), anti-IL-10-PE (JES3–9D7) and anti-IL-2-PE/Cy7 (MQ1–17H12) or anti-granzyme A-AF647 (CB9) and anti-perforin-PE/Cy7 (B-D48) for 30 minutes, washed once, filtered, and data acquired on a BD Fortessa analyzer. Gates were drawn to identify singlet T cells by FSC/SSC characteristics, and dead cells and any contaminating monocytes and B cells were excluded by gating out eFluor 780-positive, CD20+, and CD14+ events.
Synovial Tissue Processing
Synovial samples were acquired after removal as part of standard of care during arthroplasty surgery. Synovial tissue was isolated by careful dissection, minced, and digested with 100μg/mL LiberaseTL and 100μg/mL DNaseI (both Roche) in RPMI (Life Technologies) for 15 minutes, inverting every 5 minutes. Cells were passed through a 70μm cell strainer, washed, subjected to red blood cell lysis, and cryopreserved in Cryostor CS10 (BioLife Solutions) for batched analyses.
RNA Library Preparation and Sequencing
Seven RA case samples and six OA control samples were flow sorted into the following seven subsets for low-input RNA-Sequencing: Central Memory T cells (TCM), Naïve T cells (TN), Regulatory T cells (TReg), DR+27+ Effector T cells (DR+27+), DR+27- Effector T Cells (DR+27-), DR-27+ Effector T Cells (DR-27+), and DR-27- Effector T Cells (DR-27-). We rapidly thawed the case and control samples of cryopreserved peripheral blood mononuclear cells, and MACS enriched the samples for CD4+ T cells (Miltenyi). We rested the samples overnight in complete RPMI/10% FBS. Following the rest, we prepared the samples for fluorescence activated cell sorting (FACS). We washed the cells once in cold PBS, and incubated them with eBioscience human FC Receptor Binding Inhibitor (Thermo). Subsequently, we stained the samples in PBS/5% FBS for 45 minutes with the following antibodies: FITC CD27 (Biolegend), PE CD25 (Biolegend), Pe/Cy7 CD127(IL-7Ra) (Biolegend), Brilliant Violet 510 HLA-DR (Biolegend), Brilliant Violet 605 CD45RA (Biolegend), APC CD62L (Biolegend), Alexa Fluor 700 CD4 (Biolegend), APC/Cy7 CD14 (Biolegend), and APC/Cy7 CD19 (Biolegend). Post-stain, we washed the cells in cold PBS, passed them through a 70-micron filter, and acquired the samples on a BD FACSAria Fusion cytometer. 1000 cells from each subset were sorted and collected into 5ul of TCL Lysis Buffer (Qiagen) with 1% b-me. We processed and collected the samples uniformly in batches containing both cases and controls, and randomized the samples within the plate. We prepared sequencing libraries using the Smart-Seq2 protocol. Sequenced libraries were pooled and sequenced with the Illumina HiSeq 2500 using 25bp paired-end reads. We removed one outlier with low read depth. The remaining libraries were sequenced to a depth of 6–19M reads.
Flow Cytometry Data Analysis
Flow cytometry data were analyzed using FlowJo 10.0.7 (TreeStar Inc.), with serial gates drawn to identify singlet lymphocytes by FSC/SSC characteristics. Viable memory CD4+ T cells were identified as propidium iodide-negative CD3+ CD4+ CD45RA- cells. We then calculated the frequency of various populations from the pool of memory CD4+ T cells.
Gene Expression Quantification
We quantified cDNAs on canonical chromosomes (autosomal, X, Y, and Mitochondrial) in Ensembl release 83 with Kallisto v0.43.1 in transcripts per million (TPM). This analysis quantified inferred counts and length-normalized expression (TPM) of transcripts. To quantify gene expression, we collapsed transcripts mapping to the same HGNC genes symbol by summing the TPM values over. We filtered transcripts that were not sufficiently well expressed, omitting those that did not have at least 5 counts in at least 10 samples. This resulted in 15,234 well expressed genes, from 27,717 total. For differential expression analysis and principal components analysis (PCA), we used log (base 2) transformed TPM values.
Principal Component Analysis
Using the gene level log2 TPM expression described above, we selected the top 500 most variably expressed genes for PCA, excluding genes with lower than 1 mean or 0.75 standard deviation expression. We used the removeBatchEffect function in the R Limma package, with default parameters, to regress out donor-specific contributions to gene expression. To prevent the absolute range of a strongly expressed gene from dominating the signal in the PCA, we scaled gene expression using the base R scale function on the rows of the expression matrix. This function centers and scales a vector by subtracting the mean and dividing by standard deviation. We then re-normalized the samples by centering and scaling the columns, with the same R function. Finally, we used prcomp in R to perform PCA on the resulting gene expression matrix.
Correlation Analysis
For individual genes, we computed association of their expression with the proposed ordering of cell types, along the naïve to effector gradient. In this association, we modeled expression as a linear function of cell type, encoded as an ordinal variable, and donor, one-hot encoded as a categorical variable. The statistical significance of the association was estimated with the lm function in R, followed by adjustment using the Benjamini-Hochberg procedure.
Gene Set Enrichment Analysis
We performed gene set enrichment analysis using the R package gage directly on orderings defined by previous analyses. To enrich pathways from the PCA results, we used gene loadings for each principle component. For differential expression, we used the estimated t statistics. In order to produce barcode plots for select pathways, we reanalyzed these pathways using the R liger package.
Statistical Analysis
Mixed-effects modeling of Associations of Single Cells (MASC)
Here, we present a flexible method of finding significant associations between subset abundance and case-control status that we have named MASC (Mixed-effects modeling of Associations of Single Cells). The MASC framework has three steps: (1) stringent quality control, (2) definition of population clusters, and (3) association testing. Here we assume that we have single cell assays each quantifying M possible markers where markers can be genes (RNA-seq) or proteins (cytometry).
Quality control.
To mitigate the influence of batch effects and spurious clusters, we first removed poorly recorded events and low-quality markers before further analysis. We removed those markers (1) that have little expression, as these markers are not informative, and (2) with significant batch variability. First, we concatenated samples by batch and measured the fraction of cells negative and positive for each marker. We then calculated the ratio of between-batch variance to total variance for each marker’s negative and positive populations, allowing us to rank and retain 20 markers that were the least variable between batches. We also removed markers that were either uniformly negative or positive across batches, as this indicated that the antibody for that marker was not binding specifically to its target. For single-cell transcriptomic data, an analogous step would involve removing genes with low numbers of supporting reads or genes whose expression varies widely between batches.
Once low-quality markers were identified and removed, we removed events that were likely to be artifacts. We first removed events that had extremely high signal for a single marker: events that have recorded expression values at or above the 99.9th percentile for that marker are removed. These events were considered unlikely to be intact, viable cells. Next, a composite “information content” score (eq. 1) for eacI event i was created in the following manner: the expression x for each marker M is rescaled from 0 to 1 across the entire dataset to create normalized expression values y i for each event i. The sum of these normalized expression values was used to create the event’s information content score.
The information content score reflects that events with little to no expression in every channel are less informative than events that have more recorded expression. Events with low scores (INFOi < 0.05) were considered unlikely to be informative in downstream analysis and were removed. In addition, events that derived more than half of their information content score from expression in a single channel were also removed (eq. 2):
| INFOi∗0.5<maxm∈M(yi,m) | 2) |
|---|
Potential explanations for these events include poorly stained cells or artifacts caused by the clumping of antibodies with DNA fragments. A final filtering step retained events that were recorded as having detectable expression in at least M min markers, where M min may vary from experiment to experiment based on the panel design and expected level of co-expression between channels. The quality control steps described here are specific for mass cytometry analysis and will need to be optimized for use with transcriptomic data.
Clustering.
After applying quality control measures to each sample, we combined data from cases and controls into a single dataset. It was critical to ensure that each sample contributed equal numbers of cells to this dataset, as otherwise the largest samples would dominate the analysis and confound association testing. After sampling an equal number of cells from each sample, we partitioned these cells into populations using DensVM (26), which performs unsupervised clustering based on marker expression. We note that partitioning the data can be accomplished with different clustering approaches – such as SPADE or PhenoGraph for mass cytometry data – or even by using traditional bivariate gating, as MASC is not dependent on any particular method of clustering (Figure S5)
Association testing.
Once all cells were assigned to a given cluster, the relationship between single cells and clusters was modeled using mixed-effects logistic regression to account for donor or technical variation (eq. 3). We modeled the age and sex of sample k as fixed effect covariates, whereas the donor and batch that cell i belongs to were modeled as random effects. The random effects variance-covariance matrix treated each sample and batch as independent gaussians. Each cluster was individually modeled. Note that this baseline model did not explicitly include any single cell expression measures.
| log[Yi,j1−Yi,j]=θj+βclinicalXi,k+(ϕi|k)+(κi|m) | 3) | | -------------------------------------------------- | -- |
where Y i,j is the odds of cell i belonging to cluster j, θ j is the intercept for cluster j, β clinical is a vector of clinical covariates for the kth sample, (ϕ i |k) is the random effect for cell i from kth sample, (κ i |m) is the random effect for cell i from batch m.
To determine if any clusters were associated with case-control status, we included an additional covariate that indicated whether the kth sample is a case or control (eq. 4)
| log[Yi,j1−Yi,j]=θj+βclinicalXi,k+(ϕi|k)+(κi|m)+βcaseXi,k | 4) | | ------------------------------------------------------------ | -- |
Here Y i,j is the odds of cell i belonging to cluster j, θ j is the intercept for cluster j, β clinical is a vector of clinical covariates for the kth sample, (ϕ i |k) is the random effect for cell i from kth sample, (κ i |m) is the random effect for cell i from batch m, β case indicates the effect of kth sample’s case-control status.
| D=−2∗ln (likelihood for null modellikelihood for full model) | 5) |
|---|
| p=1−(t(v−2)/2e−t/22v/2Γ(v2)) | 6) |
|---|
We compared the two models using a likelihood ratio test (eq. 5) to find the test statistic D, which is the ratio of the likelihoods for the baseline and full models. The term D is distributed under the null by a χ2 distribution with 1 degree of freedom, as there is only one additional parameter in the full model compared to the null (case-control status). We derived a _p_-value by comparing test statistic D of the likelihood ratio test to the value of the χ2 distribution with 1 degree of freedom (eq. 6), allowing us to find clusters in which case-control status significantly improves model fit. A significant result (p < 0.05 after multiple testing correction) indicated that cluster membership for a single cell is influenced by case-status after accounting for technical and clinical covariates. The effect size of the case-control association can be estimated by calculating the odds ratio from β case. If a dataset includes multiple groups, then we can test for association between g groups using _g_-1 indicator variables. This approach allowed us to capture inter-individual differences between donors, as well as model the influence of technical and clinical covariates that might influence a cell to be included as a member of one cluster versus another.
Mass Cytometry Data Analysis
We analyzed 50 magnetically sorted peripheral blood samples (26 cases, 24 controls) in the resting condition and 52 samples (26 cases, 26 controls) in the stimulated condition by CyTOF. Here, we stimulated samples by incubating them with Human T-Activator CD3/CD28 Dynabeads (ThermoFisher) at a density of 1 bead:2 cells for 24 hours. Two samples were only analyzed after stimulation due to low numbers of available PBMCs. We ran aliquots of a standard PBMC sample alongside cases and controls with each CyTOF run to allow us to measure batch variability directly, as these aliquots should not be biologically dissimilar. We then used these data to find markers that had stained poorly or varied significantly between batches and removed them analysis. After acquisition, each sample was gated to a CD4+, CD45RO+ population using FlowJo 10.1 (TreeStar, USA) and combined into a single dataset before analyzing the data using MASC as previously described. We performed the data filtration steps requiring cells to demonstrate measurable expression (arcsinh-transformed expression > 0) in at least 5 markers (M min = 5). This removed 3.0–6.0% of all events captured in each sample. We first removed the initial noise factor applied to all zero expression values in mass cytometry by subtracting 1 from expression values and setting any negative values to 0, then applied the inverse hyperbolic sine transform with a cofactor of 5 to the raw expression data, using the following equation: y=sinh−1max(x−1,0)5
To partition the data, we first randomly selected 1000 cells from each sample and applied the t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm (Barnes-Hut implementation(60)) to the reduced dataset with the following parameters: perplexity = 30 and theta = 0.5. We did not include channels for CD4 or CD45RO in the t-SNE clustering as these markers were only used in gating samples to confirm the purity of CD4 memory T cell selection. We performed separate t-SNE projections for resting and stimulated cells. To identify high-dimensional populations, we used a modified version of DensVM(26). DensVM performs kernel density estimation across the dimensionally reduced t-SNE map to build a training set, then assigns cells to clusters by their expression of all markers using an SVM classifier. We modified the DensVM code to increase the range of potential bandwidths searched during the density estimation step and to return the SVM model generated from the t-SNE projection.
To create elbow plots, we ran DensVM using 25 bandwidth settings evenly spaced along the interval [0.61, 5] for the resting data and [0.66, 5] for the stimulated data. We normalized marker expression to mean 0 variance 1 in each dataset before calculating the fraction of total variance explained by between-cluster variance.
Association testing for each cluster was performed using mixed-effects logistic regression according to the MASC method. Donor and batch were included as random-effect covariates, donor sex and age was included as a fixed-effect covariate, and donors were labeled as either cases (RA) or controls (OA). To confirm the associations found by MASC, we conducted exact permutation testing in which we permuted the association between case-control status and samples within batches 10,000 times, measuring the fraction of cells from RA samples that contributed to each cluster in each permutation. This allowed us to build an empirical null for the case-control skew of each cluster, and we could then determine for each cluster how often a skew equal or greater to the observed skew occurred. We adjusted the p-values for the number of tests we performed (the number of clusters analyzed in each condition) using the Bonferroni correction.
Cluster Alignment
We aligned subsets between experiments using the following strategy: In each experiment, expression data was first scaled to mean zero, variance one to account for differences in sensitivity. We used the mean expression value for marker column to define a centroid for each cluster in both datasets. For a given cluster in the first dataset (query dataset), we calculated the Euclidean distance (eq. 7) between that cluster and cluster centroids in the second dataset (target dataset) across all shared markers. The cluster that is most similar to the cluster in query dataset is the cluster with the lowest distance in the target dataset, relative to all other clusters in that dataset.
| dist(q,t)=∑k=1K(qk−tk)2 | 7) |
|---|
Here, q refers to the query cluster and t to the target cluster, while q k and t k indicate the normalized mean expression of marker k (out of K total markers) in clusters q and t respectively.
Marker Informativeness Metric (MIM)
We wanted to determine which markers best separated a given population from the rest of the data in a quantitative manner, as finding a set of population-specific markers is crucial for isolating the population in vivo. In order to do this, we examined the distribution of expression for each marker individually in the entire dataset (Q) and the population of interest (P). We then grouped expression values for P and Q into 100 bins, normalized the binned vector to 1, and calculated the Kullback-Leibler divergence from Q to P for that marker with the following equation:
DKL(P∥Q)=∑i=1i=100PilogPiQi
The divergence score can be interpreted as a measure of how much the distribution of expression for a given marker in the entire dataset resembles the distribution of expression for that marker in the population of interest. Higher scores represent lower similarity of the marker’s expression distributions for P and Q, indicating that the expression profile of that marker is more specific for that population. By calculating this score for every marker, we can rank and identify markers that best differentiate the population of interest from the dataset.
Biaxial Gating and Cluster Overlap
To determine the concordance between biaxial gating of CD27- HLA-DR+ cells and cluster 18, we first selected cells that had normalized expression values of CD27 < 1 and HLA-DR >= 1. We then calculated an F-measure statistic between the cells selected using the CD27 and HLA-DR gates and each cluster identified in the resting and stimulated datasets (eq. 8). Here, precision is defined as the number of cells in each cluster tested that fall into the CD27- HLA-DR+ gate, while recall is defined as the number of cells gated as CD27-HLA-DR+ that are in each cluster.
| Fmeasure=2×precision×recallprecision+recall | 8) |
|---|
Clustering Informativeness Metric (CIM)
We compared clustering sets that contained either 19 (FlowSOM and DensVM) or 21 (Phenograph) clusters. We independently clustered the resting dataset with Phenograph and FlowSOM using the same cells and markers used to cluster the data with DensVM. We set k to 19 for FlowSOM clustering to match the number of clusters found by DensVM; for Phenograph, we used the default setting of k = 30.
To evaluate quantify the ability of different clustering algorithms to define clusters that was explaining marker fluctuations, we defined an information theory based metric to evaluate the relative information content captured by each set of clusters in terms of marker intensity. We selected this approach since it is separate from the objective functions that the clustering algorithms were attempting to optimize.
First, for each cell, we normalized marker intensities so that they summed to one. Then we defined a null Q i representing the average normalized intenIity for marker i across all cells. We also defined P i,j which is the mean intInsity of marker i of cells from cluster j. Then for each cluster j we calculate their KL divergence for each of the M markers (eq. 9).
| DKL,j(Pj∥Q)=∑iMPi,jlnPi,jQi | 9) |
|---|
A cluster with low divergence from the average expression of markers across the entire dataset will capture less marker intensity information than one with a high divergence, as biologically valid clusters will have unique marker profiles that differ greatly from one another and from the average marker expression profile.
We defined a similar metric to quantify the extent to which individual batches were accounting for differences in cluster composition. In this instance we calculated P i,j which is the proportion of cells from cluIter j that batch i contributed. We also calculate Q i which is the proportion oI cells that batch i contributes overall to the dataset. With this definition we calculate the KL divergence for each of the M batches (eq. 10).
| DKL,j(P.j∥Q)=∑iMPi,jlnPi,jQi | 10) |
|---|
A cluster that contains cells with low divergence from the null distribution of cells across batches is affected less by batch effects than one with a high divergence score, and a cluster completely free of batch effects should have a K-L divergence of zero.
Autoencoder Clustering
We performed clustering analysis using a deep autoencoder and a Gaussian Mixture Model (GMM). The autoencoder was designed with an architecture of 3 hidden layers, with depths of 8, 2, and 8 nodes, respectively. We trained the model with no regularization and 300 epochs. The two modes from the middle hidden layer were then used as features to learn a GMM. The number of clusters was chosen a priori to match the number discovered in the DensVM analysis. All parameters not specific in this section were set to default values.
Meta-Analysis
We used Stouffer’s Z-score method to define a meta-analysis p-value for the significant expansion of CD27- HLA-DR+ cells in RA. We converted p-values from the resting mass cytometry and flow cytometry analyses to Z-scores, and found a meta-analysis Z-score by taking the sum of these scores divided by the square-root of the number of scores – which was two, in our case. We then derived a meta-analysis p-value from the Z-score using the standard normal distribution.
All analyses were performed using custom scripts for R 3.4.0. We used the following packages: flowCore (65) to read and process FCS files for further analysis, lme4 (66) to apply mixed-effects logistic regression, ggplot2 (67), and pheatmap (68) for data visualization, and cytofkit (69) for the implementation of FlowSOM and Phenograph clustering algorithms. RNA-seq analyses were conducted using Kallisto (70) to align reads and gage (71) and liger (72) to perform gene set enrichment analysis. The h2o (73) package and mclust (74) packages were used to implement the autoencoder clustering method.
Supplementary Material
Supplement
Figure S1. MASC Type 1 Error.
Figure S2. t-SNE Projection Density.
Figure S3. Cluster Informativeness Metric Analysis of Clustering Approaches.
Figure S4. DensVM Clustering Elbow Plots.
Figure S5. Association Permutation Testing and Cluster Alignment.
Figure S6. Phenograph and FlowSOM Clustering.
Figure S7. Association Testing with Citrus.
Figure S8. Flow Cytometry and RNA-seq Gating Strategies.
Figure S9. CD27 and HLA-DR Expression in Flow Cytometry Cohort.
Figure S10. CD4+ T Cell Populations in a Clinical Response Cohort.
Figure S11. RNA-seq Analysis of CD4+ T Cell Subsets.
Figure S12. Flow Cytometry Expression Quantification.
Figure S13. Auto-encoder Clustering Method.
Figure S14. CD27- HLA-DR+ Frequency and Clinical Characteristics.
Table S1. Panel design for mass cytometry experiments.
Table S2. MASC analysis of the 19 clusters identified in the resting dataset.
Table S3. MASC analysis of the 21 clusters identified in the stimulated dataset.
Table S4. Gene set enrichment analysis of genes differentially expressed in CD27- HLA-DR+ cells.
Supplemental Figure s15
Figure S15. Marker Expression Distribution Plots for DensVM Clusters.
Figure 2: Diversity of CD4+ memory T cells before and after stimulation.
(A) t-SNE projection of 50,000 resting CD4+ memory T cells sampled equally from RA patients (n=24) and controls (n=26). DensVM identified 19 populations in this dataset. (B) Same t-SNE projections as in (A) colored by the density of cells on the SNE plot or the expression of the markers labeled above each panel. (C) t-SNE projection of 52,000 CD4+ stimulated memory T cells sampled equally from RA patients (n=26) and controls (n=26). Cells were stimulated for 24 hours with anti-CD3/anti-CD28 beads. (D) Same t-SNE projections as in (C) colored by the density of cells on the SNE plot or the expression of the markers labeled above each panel. (E) Heatmap showing mean expression of indicated markers across the 19 populations found in resting cells. (F) Heatmap showing mean expression of indicated markers across the 21 populations found after stimulation. Protein expression data are shown after arcsinh transformation. All markers but CD4 and CD45RO were used to create t-SNE projections and perform clustering.
Acknowledgements:
This work was supported in part by funding from the National Institutes of Health: UH2AR067677 (S.R.), U19AI111224 (S.R.), and 1R01AR063759 (S.R.), R01 AR064850–03 (Y.C.L.), the Doris Duke Charitable Foundation Grant #2013097 (S.R.), T32 AR007530–31 (M.B.B., D. A. R.), the William Docken Inflammatory Autoimmune Disease Fund (M.B.B., S. R.), the Ruth L. Kirschstein National Research Service Award F31-AR070582 (K. S.), and the Rheumatology Research Foundation Tobe and Stephen Malawista, MD Endowment in Academic Rheumatology (D.A.R).
Footnotes
Competing interests
All authors declare that they have no competing financial interests. I.K. has been a paid bioinformatics consultant for Outlier Bio LLC since November 2017.
Data and materials Availability
All data associated with this study can be found in the paper or supplementary materials. The data that support the findings of this study has been deposited in the database GEO (GSE118209). RNA sequencing reads have been deposited in the SRA database (SRP156530). MASC and all other custom scripts used in this analysis are available at https://www.github.com/immunogenomics.
References and notes
- 1.Neu KE, Tang Q, Wilson PC, Khan AA, Single-Cell Genomics: Approaches and Utility in Immunology, Trends Immunol. 38, 140–149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross M, Smith E, Hoy D, Carmona L, Wolfe F, Vos T, Williams B, Gabriel S, Lassere M, Johns N, Buchbinder R, Woolf A, March L, The global burden of rheumatoid arthritis: estimates from the global burden of disease 2010 study, Annals of the Rheumatic Diseases 73, 1316–1322 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Viatte S, Plant D, Raychaudhuri S, Genetics and epigenetics of rheumatoid arthritis, Nat Rev Rheumatol 9, 141–153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurkó J, Besenyei T, Laki J, Glant TT, Mikecz K, Szekanecz Z, Genetics of Rheumatoid Arthritis — A Comprehensive Review, Clinic Rev Allerg Immunol 45, 170–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gizinski AM, Fox DA, T cell subsets and their role in the pathogenesis of rheumatic disease, Current Opinion in Rheumatology 26, 204–210 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Shao S, Jiao Z, Guo M, Xu H, Wang S, The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis, Rheumatol. Int 32, 887–893 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee H-S, Jia X, Alfredsson L, Padyukov L, Klareskog L, Worthington J, Siminovitch KA, Bae S-C, Plenge RM, Gregersen PK, de Bakker PIW, Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis, Nature Publishing Group 44, 291–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perricone C, Ceccarelli F, Valesini G, An overview on the genetic of rheumatoid arthritis: A never-ending story, Autoimmunity Reviews 10, 599–608 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Diogo D, Okada Y, Plenge RM, Genome-wide association studies to advance our understanding of critical cell types and pathways in rheumatoid arthritis: recent findings and challenges, Current Opinion in Rheumatology 26, 85–92 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, Zhernakova A, Stahl E, Viatte S, McAllister K, Amos CI, Padyukov L, Toes REM, Huizinga TWJ, Wijmenga C, Trynka G, Franke L, Westra H-J, Alfredsson L, Hu X, Sandor C, de Bakker PIW, Davila S, Khor CC, Heng KK, Andrews R, Edkins S, Hunt SE, Langford C, Symmons D, Concannon P, Onengut-Gumuscu S, Rich SS, Deloukas P, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Ärlsetig L, Martin J, Rantapää-Dahlqvist S, Plenge RM, Raychaudhuri S, Klareskog L, Gregersen PK, Worthington J, High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis, Nat Genet 44, 1336–1340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, Graham RR, Manoharan A, Ortmann W, Bhangale T, Denny JC, Carroll RJ, Eyler AE, Greenberg JD, Kremer JM, Pappas DA, Jiang L, Yin J, Ye L, Su D-F, Yang J, Xie G, Keystone E, Westra H-J, Esko T, Metspalu A, Zhou X, Gupta N, Mirel D, Stahl EA, Diogo D, Cui J, Liao K, Guo MH, Myouzen K, Kawaguchi T, Coenen MJH, van Riel PLCM, van de Laar MAFJ, Guchelaar H-J, Huizinga TWJ, Dieudé P, Mariette X, Bridges SL, Zhernakova A, Toes REM, Tak PP, Miceli-Richard C, Bang S-Y, Lee H-S, Martin J, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Rantapää-Dahlqvist S, Ärlestig L, Choi HK, Kamatani Y, Galan P, Lathrop M, RACI consortium, GARNET consortium, Eyre S Bowes J, Barton A, de Vries N, Moreland LW, Criswell LA, Karlson EW, Taniguchi A, Yamada R, Kubo M, Liu JS, Bae S-C, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Raychaudhuri S, Stranger BE, De Jager PL, Franke L, Visscher PM, Brown MA, Yamanaka H, Mimori T, Takahashi A, Xu H, Behrens TW, Siminovitch KA, Momohara S, Matsuda F, Yamamoto K, Plenge RM, Genetics of rheumatoid arthritis contributes to biology and drug discovery, Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Kim H, Stahl E, Plenge R, Daly M, Raychaudhuri S, Integrating Autoimmune Risk Loci with Gene-Expression Data Identifies Specific Pathogenic Immune Cell Subsets, The American Journal of Human Genetics 89, 496–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, Teslovich NC, Weinblatt ME, Massarotti EM, Coblyn JS, Helfgott SM, Lee YC, Todd DJ, Bykerk VP, Goodman SM, Pernis AB, Ivashkiv LB, Karlson EW, Nigrovic PA, Filer A, Buckley CD, Lederer JA, Raychaudhuri S, Brenner MB, Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis, Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens PB, Goronzy JJ, Schaid D, Weyand CM, Expansion of unusual CD4+ T cells in severe rheumatoid arthritis, Arthritis Rheum 40, 1106–1114 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Scarsi M, Ziglioli T, Airo P, Decreased circulating CD28-negative T cells in patients with rheumatoid arthritis treated with abatacept are correlated with clinical response, J. Rheumatol 37, 911–916 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Warrington KJ, Takemura S, Goronzy JJ, Weyand CM, CD4+,CD28- T cells in rheumatoid arthritis patients combine features of the innate and adaptive immune systems, Arthritis Rheum 44, 13–20 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Fonseka CY, Rao DA, Raychaudhuri S, Leveraging blood and tissue CD4+ T cell heterogeneity at the single cell level to identify mechanisms of disease in rheumatoid arthritis, Current Opinion in Immunology 49, 27–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendall SC, Simonds EF, Qiu P, Amir E-AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP, Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum, Science 332, 687–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sörensen T, Baumgart S, Durek P, Grützkau A, Häupl T, immunoClust-An automated analysis pipeline for the identification of immunophenotypic signatures in high-dimensional cytometric datasets, Cytometry, n/a–n/a (2015). [DOI] [PubMed] [Google Scholar]
- 20.Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP, Automated mapping of phenotype space with single-cell data, Nat Meth 13, 493–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin P, Troup M, Ho JWK, CIDR: Ultrafast and accurate clustering through imputation for single-cell RNA-seq data, Genome Biol. 18, 59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets, Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe’er D, Nolan GP, Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis, Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S, Highly multiparametric analysis by mass cytometry, Journal of Immunological Methods 361, 1–20 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Ermann J, Rao DA, Teslovich NC, Brenner MB, Raychaudhuri S, Immune cell profiling to guide therapeutic decisions in rheumatic diseases, Nat Rev Rheumatol 11, 541–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KWW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, Greter M, Ginhoux F, Newell EW, High-dimensional analysis of the murine myeloid cell system, Nat. Immunol 15, 1181–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y, FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data, Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Weber LM, Robinson MD, Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data, Cytometry A 89, 1084–1096 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Wong MT, Ong DEH, Lim FSH, Teng KWW, McGovern N, Narayanan S, Ho WQ, Cerny D, Tan HKK, Anicete R, Tan BK, Lim TKH, Chan CY, Cheow PC, Lee SY, Takano A, Tan E-H, Tam JKC, Tan EY, Chan JKY, Fink K, Bertoletti A, Ginhoux F, Curotto de Lafaille MA, Newell EW, A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures, Immunity 45, 442–456 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Wong MT, Van Der Maaten L, Newell EW, Categorical Analysis of Human T Cell Heterogeneity with One-Dimensional Soli-Expression by Nonlinear Stochastic Embedding, J. Immunol 196, 924–932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong MT, Chen J, Narayanan S, Lin W, Anicete R, Kiaang HTK, De Lafaille MAC, Poidinger M, Newell EW, Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis, Cell Rep 11, 1822–1833 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Baecher-Allan C, Wolf E, Hafler DA, MHC class II expression identifies functionally distinct human regulatory T cells, The Journal of Immunology 176, 4622–4631 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Oshima S, Eckels DD, Selective expression of class II MHC isotypes by MLC-activated human T lymphocytes, Hum. Immunol 27, 208–219 (1990). [DOI] [PubMed] [Google Scholar]
- 34.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification, Nat Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Mason GM, Lowe K, Melchiotti R, Ellis R, de Rinaldis E, Peakman M, Heck S, Lombardi G, Tree TIM, Phenotypic Complexity of the Human Regulatory T Cell Compartment Revealed by Mass Cytometry, J. Immunol 195, 2030–2037 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D, Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development, Cell 157, 714–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrell PB, Diggins KE, Polikowsky HG, Mohan SR, Seegmiller AC, Irish JM, High-Dimensional Analysis of Acute Myeloid Leukemia Reveals Phenotypic Changes in Persistent Cells during Induction Therapy, PLoS ONE 11 (2016), doi: 10.1371/journal.pone.0153207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Wishbone identifies bifurcating developmental trajectories from single-cell data, Nature (2016), doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mingueneau M, Boudaoud S, Haskett S, Reynolds TL, Nocturne G, Norton E, Zhang X, Constant M, Park D, Wang W, Lazure T, Le Pajolec C, Ergun A, Mariette X, Cytometry by time-of-flight immunophenotyping identifies a blood Sjögren’s signature correlating with disease activity and glandular inflammation, J. Allergy Clin. Immunol (2016), doi: 10.1016/j.jaci.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Hansmann L, Blum L, Ju C-H, Liedtke M, Robinson WH, Davis MM, Mass cytometry analysis shows that a novel memory phenotype B cell is expanded in multiple myeloma, Cancer Immunol Res 3, 650–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, Kinuthia J, Montgomery RR, John-Stewart G, Holmes S, Blish CA, Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility, Sci Transl Med 7, 297ra115–297ra115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaudillière B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, Silva J, Ganio EA, Yeh CG, Maloney WJ, Huddleston JI, Goodman SB, Davis MM, Bendall SC, Fantl WJ, Angst MS, Nolan GP, Clinical recovery from surgery correlates with single-cell immune signatures, Sci Transl Med 6, 255ra131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP, Automated identification of stratifying signatures in cellular subpopulations, Proc. Natl. Acad. Sci. U.S.A 111, E2770–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saeys Y, Gassen SV, Lambrecht BN, Computational flow cytometry: helping to make sense of high-dimensional immunology data, Nature Reviews Immunology 16, 449–462 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Wagner A, Regev A, Yosef N, Revealing the vectors of cellular identity with single-cell genomics, Nat Biotechnol 34, 1145–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chester C, Maecker HT, Algorithmic Tools for Mining High-Dimensional Cytometry Data, J. Immunol 195, 773–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orlova DY, Herzenberg LA, Walther G, Science not art: statistically sound methods for identifying subsets in multi-dimensional flow and mass cytometry data sets, Nature Reviews Immunology 18, 77–77 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Saeys Y, Van Gassen S, Lambrecht B, “Response to Orlova et al. Science not art: statistically sound methods for identifying subsets in multi-dimensional flow and mass cytometry data sets”, Nature Reviews Immunology 18, 78–78 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Qiu X, Hill A, Packer J, Lin D, Ma Y-A, Trapnell C, Single-cell mRNA quantification and differential analysis with Census, Nat Meth 14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larbi A, Fulop T, From “truly naïve” to “exhausted senescent” T cells: when markers predict functionality, Cytometry A 85, 25–35 (2014). [DOI] [PubMed] [Google Scholar]
- 51.De Jong R, Brouwer M, Hooibrink B, Van der Pouw-Kraan T, Miedema F, Van Lier RA, The CD27- subset of peripheral blood memory CD4+ lymphocytes contains functionally differentiated T lymphocytes that develop by persistent antigenic stimulation in vivo, Eur. J. Immunol 22, 993–999 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Schiott A, Lindstedt M, Johansson-Lindbom B, Roggen E, Borrebaeck CAK, CD27- CD4+ memory T cells define a differentiated memory population at both the functional and transcriptional levels, Immunology 113, 363–370 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD, Characterization of CD4(+) CTLs ex vivo, The Journal of Immunology 168, 5954–5958 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Griffiths GM, Alpert S, Lambert E, McGuire J, Weissman IL, Perforin and granzyme A expression identifying cytolytic lymphocytes in rheumatoid arthritis, Proc. Natl. Acad. Sci. U.S.A 89, 549–553 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Namekawa T, Wagner UG, Goronzy JJ, Weyand CM, Functional subsets of CD4 T cells in rheumatoid synovitis, Arthritis Rheum 41, 2108–2116 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Mattoo H, Mahajan VS, Maehara T, Deshpande V, Della-Torre E, Wallace ZS, Kulikova M, Drijvers JM, Daccache J, Carruthers MN, Castelino FV, Stone JR, Stone JH, Pillai S, Clonal expansion of CD4(+) cytotoxic T lymphocytes in patients with IgG4- related disease, J. Allergy Clin. Immunol 138, 825–838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohem CL, Brezinschek RI, Wisbey H, Tortorella C, Lipsky PE, Oppenheimer-Marks N, Enrichment of differentiated CD45RBdim,CD27- memory T cells in the peripheral blood, synovial fluid, and synovial tissue of patients with rheumatoid arthritis, Arthritis Rheum 39, 844–854 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Lee YC, Bingham CO, Edwards RR, Marder W, Kristine P, Bolster MB, Clauw DJ, Moreland LW, Lu B, Wohlfahrt A, Zhang Z, Neogi T, Pain Sensitization is Associated with Disease Activity in Rheumatoid Arthritis Patients: A Cross-Sectional Study, Arthritis Care Res (Hoboken) (2017), doi: 10.1002/acr.23266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, Bendall SC, Normalization of mass cytometry data with bead standards, Cytometry Part A 83A, 483–494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Der Maaten L, Accelerating t-SNE using tree-based algorithms, The Journal of Machine Learning Research 15, 3221–3245 (2014). [Google Scholar]
- 61.Aletaha D, Nell VP, Stamm T, Uffmann M, Pflugbeil S, Machold K, Smolen JS, Acute phase reactants add little to composite disease activity indices for rheumatoid arthritis: validation of a clinical activity score, Arthritis Res Ther 7, 796–806 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, de Silva AD, Harris E, Peters B, Seumois G, Weiskopf D, Sette A, Vijayanand P, Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis, Sci Immunol 3(19) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Simbert P, Simultaneous epitope and transcriptome measurement in single cells, Nat Methods 14(9), 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA, Multiplexed quantification of proteins and transcripts in single cells, Nat Biotechnol 35(10), 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Ellis B, Haaland P, Hahne F, Le Meur N, Gopalakrishnan N, Spidlen J, Jiang M, flowCore: Basic structures for flow cytometry data. R package version 1.46.1 (2018). [Google Scholar]
- 66.Bates D, Maechler M, Bolker B, Walker S, Fitting Linear Mixed-Effects Models Using lme4, Journal of Statistical Software 67(1), 1–48 (2015), doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 67.Wickham H, ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag; New York: (2016) [Google Scholar]
- 68.Kolde R, pheatmap: Pretty Heatmaps . R package version 1.0.10 (2018). [Google Scholar]
- 69.Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J, Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline, PLOS Computational Biology 12(9) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification, Nature Biotechnology 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Luo W, Freidman M, Shedden K, Hankenson K, Woolf P, GAGE: generally applicable gene set enrichment for pathway analysis, BMC Bioinformatics 10, 161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan J, Kharchenko P, liger: Lightweight Iterative Geneset Enrichment, R package version 0.1 (2018). [Google Scholar]
- 73.LeDell E, Gill N, Aiello S, Fu A, Candel A, Click C, Kraljevic T, Nykodym T, Aboyoun P, Kurka M, Malohlava M, h2o: R Interface for ‘H2O’, R package version 3.20.0.2 (2018). [Google Scholar]
- 74.Scrucca L, Fop M, Murphy TB, Raftery AE, mclust 5: clustering, classification and density estimation using Gaussian finite mixture models, The R Journal 8/1, 205–233 (2016). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement
Figure S1. MASC Type 1 Error.
Figure S2. t-SNE Projection Density.
Figure S3. Cluster Informativeness Metric Analysis of Clustering Approaches.
Figure S4. DensVM Clustering Elbow Plots.
Figure S5. Association Permutation Testing and Cluster Alignment.
Figure S6. Phenograph and FlowSOM Clustering.
Figure S7. Association Testing with Citrus.
Figure S8. Flow Cytometry and RNA-seq Gating Strategies.
Figure S9. CD27 and HLA-DR Expression in Flow Cytometry Cohort.
Figure S10. CD4+ T Cell Populations in a Clinical Response Cohort.
Figure S11. RNA-seq Analysis of CD4+ T Cell Subsets.
Figure S12. Flow Cytometry Expression Quantification.
Figure S13. Auto-encoder Clustering Method.
Figure S14. CD27- HLA-DR+ Frequency and Clinical Characteristics.
Table S1. Panel design for mass cytometry experiments.
Table S2. MASC analysis of the 19 clusters identified in the resting dataset.
Table S3. MASC analysis of the 21 clusters identified in the stimulated dataset.
Table S4. Gene set enrichment analysis of genes differentially expressed in CD27- HLA-DR+ cells.
Supplemental Figure s15
Figure S15. Marker Expression Distribution Plots for DensVM Clusters.