Polypyrimidine Tract Binding Protein Antagonizes Exon Definition (original) (raw)
The removal of introns from mRNA precursors (pre-mRNAs) involves two relatively straightforward chemical reactions. The recognition of intron-exon boundaries, the splice sites, however, requires the integration of information provided by many _cis_-acting elements and a complex splicing machinery (64). The _cis_-acting elements that define the borders between exons and introns are quite diverse and yet are recognized efficiently by the splicing machinery. This machinery is composed of general splicing factors (GSFs), which make up the spliceosome and its associated proteins, and of regulatory factors. The same machinery must also make cell-type-specific choices in cases in which pre-mRNAs are alternatively spliced. This is a monumental task given that it is estimated that transcripts from 30% of all genes in humans are alternatively spliced (http://devnull.lbl.gov:8888/alt).
The spliceosome, like many macromolecular machines, is not preassembled as an active enzyme but rather assembles on the substrate. The substrate, a functional pre-mRNA, is thought to first interact with U1 snRNP, hnRNP proteins, and SR proteins (5, 34). The interaction is determined by RNA-RNA base pairing between the 5′ end of U1 snRNA and the consensus sequence at the 5′ splice site and by interactions mediated by protein factors (34). The protein factors U2AF and SF1 also recognize the polypyrimidine tract, the branch point, and the 3′ splice site, thus bridging the two groups that subsequently will be involved in the first transesterification reaction. This leads to the formation of the commitment complex (CC). The interaction between the protein factors and the 3′ splice site of an internal exon is enhanced by the binding of U1 snRNP at the downstream 5′ splice site (35, 59). This interaction is the basis for exon definition, an idea discussed in greater detail below. The CC and the U2 snRNP interact to yield the prespliceosome, and the branch point sequence is recognized again, albeit differently in this complex. The prespliceosome interacts with a preformed U5-U4-U6 tri-snRNP to form the immature spliceosome, which then undergoes rearrangements that result in the formation of a fully competent enzyme. This interplay of multiple protein factors and RNA components sets the stage for numerous opportunities for and targets of regulation.
The complexity of constitutive and alternative splice site recognition suggests multiple layers of regulation, with each layer the result of combinatorial arrays of elements and factors (38, 48, 64). The first layer is direct sequence recognition that likely occurs early in the formation of the spliceosome. U1 snRNA can read the sequence at the 5′ splice site, and protein factors SF1, U2AF65, and U2AF35 recognize the branch point, the polypyrimidine tract, and the 3′ splice site, respectively (3, 5, 45, 61,75). These and other GSFs interact with each other and can act as molecular rulers sensing the relative locations of the_cis_-acting elements. Positional and distance information provides a second layer of discrimination that overlies the detection of individual binding sites. This type of information is transmitted via protein-protein interactions in the definition of exons (2). Another example of this type of distance detection is seen in the α-tropomyosin pre-mRNA (63), where the close proximity of the 5′ splice site of exon 2 to the branch point upstream of exon 3 precludes the inclusion of both exons into the mRNA. Modulation of splice site strength by proteins of the SR family provides yet another layer of regulation (33, 66,71). SR proteins play roles in constitutive splicing and can be considered GSFs; however, in some instances SR proteins have important roles in alternative splicing. These proteins can be recruited directly to the RNA by enhancer elements in exons or introns or indirectly by interactions with other GSFs (36, 43, 58, 67). hnRNP proteins, some of which bind all pre-mRNAs, can also influence splice site choices, possibly by counteracting SR proteins (6). hnRNP A1 and polypyrimidine tract binding protein (PTB), two proteins classified as hnRNP proteins, repress certain splicing events and thereby provide a layer of negative regulation. Very precise regulation is provided by the existence of cell-type-specific factors; several of these have been described in_Drosophila melanogaster_ (38). The integration of the information in these regulatory layers leads to splice site choice.
Negative regulation of exon inclusion is emerging as a critical layer in splice site choice. Fairbrother and Chasin considered why certain exons are selected, while others, which seem perfectly competent, are ignored (21). These authors suggest that many, and possibly all, exons are under a global repressive influence mediated by many intronic sequences (21). Thus, splice site utilization can be described as a function of both splice site strength and the intensity of the repressive field within a specific region of a pre-mRNA. This global repressive influence can also contribute to the outcome of regulated alternative splicing events, setting the stage for cell-type-specific derepression of exons (1, 8, 12,46–49, 72, 73). In mammalian cells PTB has been identified as a key splicing repressor. In this review we critically evaluate the role of PTB in exon silencing and speculate on possible mechanisms of its action. We also provide a brief discussion of potential ways in which selective exon inclusion could be achieved by cell-type-specific derepression.
PTB: A REPRESSOR OF EXON DEFINITION
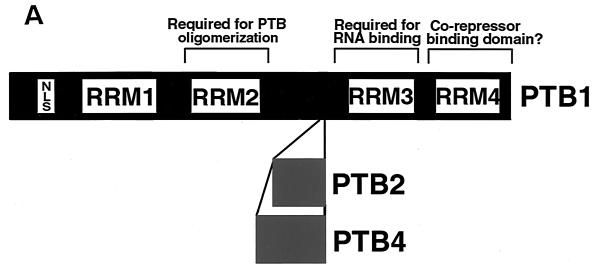
PTB was discovered as a protein that bound the U-rich polypyrimidine tract of several introns (22, 70). Multiple forms of PTB were observed and at least three different cDNAs, which are the result of alternative splicing, were cloned and sequenced (24, 54, 60) (Fig. 1A). Numerous homologs have been subsequently identified, suggesting highly conserved function (Fig. 1B). Sequence alignment reveals that the highest homology lies within the second RNA recognition motif (RRM2) and a flanking region that has been suggested to be required for PTB dimerization (53, 56). RRM4, which is required for function (P. J. Grabowski, personal communication) is also highly conserved. The binding to active polypyrimidine tracts (22) and biochemical complementation assays (54) suggested a role in constitutive splicing; however, this view did not withstand the test of time. Mulligan et al. (51) noted that PTB bound sequences that repressed inclusion of exon 7 in the β-tropomyosin pre-mRNA and proposed that PTB was a repressor of splicing. This insightful proposal was supported by subsequent data from several laboratories.
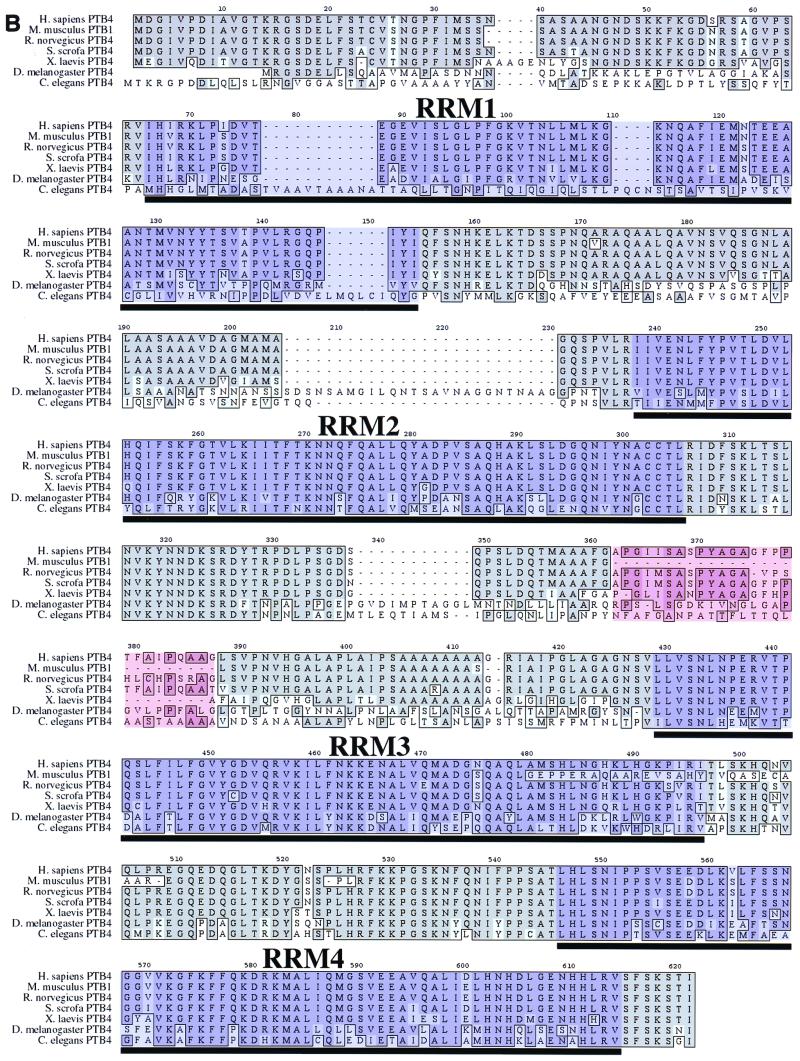
FIG. 1.
Functional domains and motifs of PTB. (A) Three variants of human PTB exist: PTB1 (protein accession no. CAA43973), PTB2 (protein accession no. CAA46443), and PTB4 (protein accession no.CAA46444). The PTB4 isoform was also identified as a member of the hnRNP family and named hnRNP I (23). Each of the isoforms has four RRMs (24, 54), as well as an N-terminal nuclear localization signal (56). RRM2, as well as the flanking sequence, has been demonstrated to be necessary for PTB oligomerization (53, 56). RRM3 has been shown to be required for RNA binding (53, 56). RRM4 appears to be required for function but not for RNA binding and thus may bind to an unknown corepressor (Grabowski, personal communication). (B) Alignment of PTB protein sequences from seven species using the MacVector CLUSTALW Alignment function. Dark-gray regions within the alignment represent identity, while the light-gray regions denote conserved amino acid changes. Areas of purple shading are the RRMs, and the segment shaded in light red highlights the region of presumed alternative splicing. The NCBI protein accession numbers for these proteins are as follows: Homo sapiens PTB4, CAA46444; S. scrofa PTB4, CAA63597;Mus musculis PTB1, P17225; _Rattus norvegicus_PTB4, Q00438; Xenopus laevis PTB4, AAF00041; D. melanogaster PTB4, AAF22979; and _Caenorhabditis elegans_PTB4, T20381.
PTB has also been implicated in other processes that may be unrelated to its role in pre-mRNA splicing, such as a regulation of cap-independent translation, localization of cytoplasmic RNA, and poly(A) site cleavage (see, for example, references 13, 27, and50 and references therein). These activities are not discussed here. We address here evidence that PTB is a key regulator of splicing, and we argue that its major, if not only, effect on pre-mRNA splicing is exon silencing.
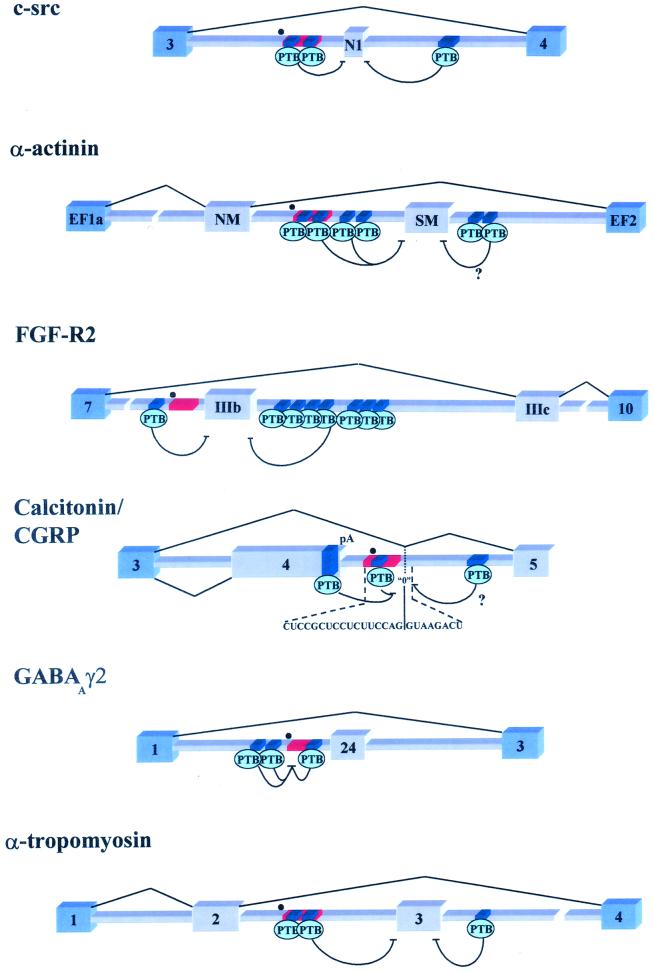
There are compelling, if not definitive, data that PTB mediates exon silencing. Here we define exon silencing as the opposite of exon definition (2). PTB binds to intronic sequences that mediate splicing repression (intronic splicing silencers [ISS]) in a long list of alternatively spliced pre-mRNAs (1, 8, 10,26, 51, 52, 65) (Fig. 2). Mutation of the PTB binding sites within ISS sequences reverses both exon silencing in vivo and PTB binding in vitro (1, 8, 10, 12, 25, 26,55, 65). PTB has also been found to bind an exonic splicing silencer that represses use of a suboptimal 3′ splice site in bovine papillomavirus type 1 (74). Competition for PTB with exogenous RNAs (1, 10) and, more importantly, depletion of PTB (12, 65) leads to derepression of regulated exons in vitro. Readdition of purified or recombinant PTB can reconstitute repression (1, 12, 65). Finally, in vivo overexpression of PTB enhances the silencing of exon IIIb in FGF-R2 pre-mRNAs (8), and this effect depends on the presence of a PTB binding site upstream of exon IIIb (Fig. 2). Overexpression of PTB did not result in an overall decrease in splicing. Similar results have been observed with an α-actinin splicing precursor (C. Smith, personal communication). These data suggest that PTB mediates exon silencing.
FIG. 2.
Schematic representation of six pre-mRNAs that are regulated by PTB through exon definition antagonism. Blue exons represent constitutively spliced exons, while the gray exons are regulated. Dark-blue segments represent PTB binding sites; red rectangles are the branch point-associated polypyrimidine tract, while the black dots represent identified or putative branch points. (c-src) The N1 exon is repressed in nonneural cell types by intronic PTB binding sites flanking the exon (9, 10, 12). (α-actinin) The SM exon may be repressed in tissues other than smooth muscle through a network of PTB binding sites located on both sides of the SM exon (65). It should be noted that in vitro the intron downstream of SM is not required for PTB repression. (FGF-R2) Exon IIIb is also repressed in mesenchymal tissues through multiple PTB binding sites found on either side of IIIb (8; E. J. Wagner and M. A. Garcia-Blanco, unpublished results). (Calcitonin/CGRP) The calcitonin-CGRP fourth exon is included and subsequently polyadenylated in the majority of tissues, possibly due to PTB-mediated repression of a zero-length exon located in the downstream intron. Repression of this exon blocks a potential recursive-splicing pathway to exon 5 (14, 39–42). Splicing to exon 5 occurs in neural cell types where PTB levels may be reduced. (GABAAγ2) The small 24-nucleotide exon is repressed in nonneural cell types through PTB binding near the branch point sequence (1, 72, 73). (α-tropomyosin). Exon 3 is specifically repressed in smooth muscle tissue by PTB or a novel PTB variant (25, 26, 55) (Smith, unpublished).
PTB overexpression was also found to activate polyadenylation and concomitant inclusion of exon 4 as the 3′-terminal exon in the CT/CGRP pre-mRNA (Fig. 2) (40). Lou et al. (40) posit that PTB binding to exon 4 promotes polyadenylation by stabilizing an interaction with U1 snRNP at the intronic enhancer of polyadenylation. PTB binding to sites both within exon 4 and the intronic enhancer are believed to be required to activate polyadenylation. A potential problem with this model is that mutations of the PTB binding site within exon 4 have no effect on polyadenylation. An alternative model predicts dual and competing roles for U1 snRNP bound to the enhancer: U1 snRNP could activate polyadenylation through enhancement of exon 4 recognition or it could be involved in a recursive splicing pathway leading to exon 5 inclusion (30) (Fig. 2). PTB would repress the use of the zero-length exon predicted in recursive splicing, thus freeing U1 snRNP to activate polyadenylation. Overexpression of PTB would therefore increase the likelihood of this event. In this case, as in those mentioned above, PTB can be best thought of as an antagonist of exon definition (Fig. 2).
Although PTB has the ability to interfere with exon definition, it seems likely that the presence of PTB binding sites is not sufficient to silence otherwise robust exons (25, 26, 37, 55). This may be the case for exon 3 of the rat α-tropomyosin pre-mRNA, which is silenced in smooth muscle cells but not in many other tissues. Although silencing of exon 3 requires PTB binding sites, it cannot be mediated by PTB alone, given that this protein is found in cells where exon 3 is included. Regulated weak exons require other silencer sequences or weak splice sites in order to achieve repression of exon inclusion. This is clearly the case with the IIIb exon of FGF-R2 in which, in addition to elements that bind PTB, silencing requires a weak polypyrimidine tract and an exonic silencer that interacts with hnRNP A1 (8, 16, 19). PTB also appears to be a component of a multiprotein complex that assembles on regulatory elements in c-src and β-tropomyosin pre-mRNAs (10, 12, 29, 64). Together, these data suggest that PTB acts in concert with corepressors to mediate exon silencing. Given its ubiquitous distribution in cell lines and tissues, it is likely that PTB provides global repression of weak exons. PTB may play a dual role by not only discriminating between splice sites and pseudo-splice sites during constitutive splicing but also setting the stage for cell-type-specific selection of regulated exons during alternative splicing.
MECHANISMS OF PTB REPRESSION
PTB binding sites sometimes overlap binding sites for U2AF65, and simple competition could account for the inhibitory action of PTB (37, 62). A well-characterized example of a potential competition model is the repression of the 24-nucleotide exon of the GABAAγ2 pre-mRNA. In this case, there are four silencer sequences that bind PTB and act in concert to inhibit the recognition of this exon. Three of these PTB binding sites surround the branch point, with one sitting within the associated polypyrimidine tract (Fig. 2). In this particular example, PTB may be acting in a manner similar to the _D. melanogaster_female-specific splicing factor SXL on TRA pre-mRNA (28,68). SXL has but one site on the TRA pre-mRNA and excludes access to U2AF65 when bound, thus repressing the use of the non-sex-type-specific exon. In most other instances, PTB binding sites do not directly overlap the binding sites of GSFs (9, 10, 12, 25, 26, 55, 65). In fact, it has recently been demonstrated for the SM exon in α-actinin that the critical sites do not overlap with the branch point and polypyrimidine tract of intron 3 (J. Southby and C. Smith, personal communication). Therefore, in this case and many others, PTB-mediated exon silencing is clearly not caused by direct competition with GSFs (see discussion about SXL below). Although these observations suggest that PTB may act by two distinct mechanisms, we argue below for a more parsimonious explanation.
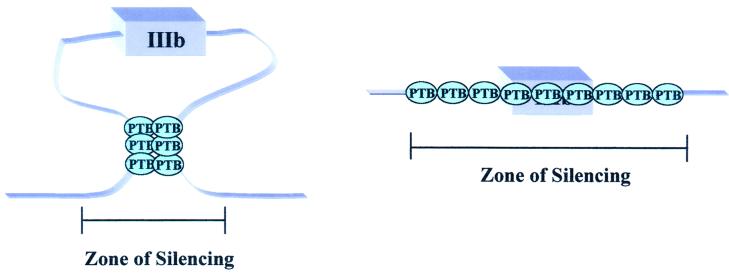
The majority of exons silenced by PTB are flanked by PTB binding sites on both adjacent introns (Fig. 2). Given that PTB can multimerize, it has been postulated that PTB proteins can interact across the exon (12, 53, 56) (Fig. 3). Chou et al. (12) have shown evidence for such an interaction in vitro, demonstrating that mutations in the upstream binding site affected binding of PTB to the downstream site and vice versa. A very similar interaction was proposed for hnRNP A1 proteins binding on either side of a regulated exon of the hnRNP A1 pre-mRNA (4). In that case the effect was postulated to be activation of the downstream exon by approximation. The idea that PTB can interact across exons fits well with the evidence that PTB is an antagonist of exon definition. The PTB sites flanking silenced exons could define a repressive zone within a pre-mRNA. Binding on both flanks is most reminiscent of the postulated mechanism for SXL autoregulation (31, 32). The SXL protein binds in numerous positions flanking the male-specific exon 3 in its own pre-mRNA and prevents its inclusion. SXL interacts with the_Drosophila_ U1A/U2B" homolog, a component of both U1 and U2 snRNPs, and is likely to interfere with both splice sites (20,38). It is not known, however, whether SXL multimerizes to promote an exon sequestration event or if it instead inhibits the recognition of the 5′ and 3′ splice sites independently. The data of Chou et al. (12) make it unlikely that PTB is independently inhibiting recognition of the 3′ and 5′ splice sites. We favor exon sequestration mediated in one of two ways: PTB-PTB interactions at a distance or PTB multimerization (Fig. 3). The zone of repression formed by PTB may or may not involve direct competition with U2AF.
FIG. 3.
Two potential mechanisms to define a zone of silencing. The left model predicts that PTB-PTB interactions between binding sites flanking an exon sequesters an exon, thus precluding the definition of this exon. The model on the right suggests that PTB can oligomerize across an exon, resulting in the coating of the exon, which will also antagonize its definition. The model on the right could also explain the silencing of exons that have a PTB site only on one flank; PTB could multimerize, covering a region of the RNA determined by interactions with other factors.
CELL TYPE-DEPENDENT DEREPRESSION OF EXONS
PTB is one of the mediators of global silencing of weak or otherwise highly regulated exons. It is interesting to examine how this layer of negative regulation is specifically derepressed in some cells and tissues. The question of how PTB action is counteracted probably separates PTB from SXL in terms of mechanistic similarities. In_Drosophila_, counteracting SXL is simple; its absence in males results in the inclusion of exons repressed by SXL in females. PTB, however, is expressed in most cell types, albeit at different levels (54).
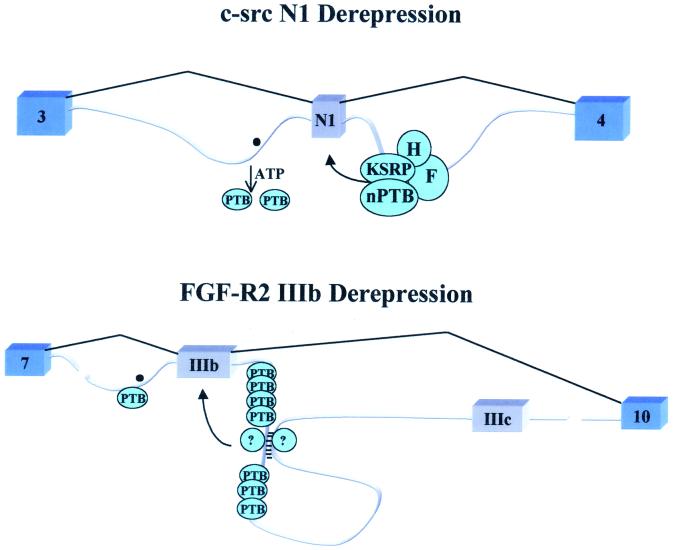
There are several examples of exons included only in neural cells; silencing in nonneural cells is attributed to PTB action (1, 10,73). How do neural tissues overcome the repressive effect of PTB? Lower levels of PTB in neural tissues and the presence of a neural type of PTB (nPTB) or brPTB (for brain PTB) have been proposed as explanations (10, 73). Indeed nPTB can compete with PTB for binding of the RNA but has a weaker repressive effect (44). Overexpression of nPTB, however, can inhibit neuron-specific exon activation by the Nova protein (57), suggesting that nPTB may act both as an activator and as a repressor depending on the context. Levels of PTB per se may not be the whole answer, but a combination of lower levels of PTB and antagonizing factors could result in neuron-specific exon inclusion (11, 46,47). Recently, in splicing extracts from cells that include the c-src N1 exon, but not in extracts that exclude it, PTB binding was shown to be inhibited in the presence of ATP (Fig.4) (12). It is clear that PTB is not displaced by the ATP-dependent binding of U2 snRNP, and the identity of the process that mediates PTB dissociation is unknown.
FIG. 4.
Two possible mechanisms of PTB derepression. (c-src N1 Derepression) In neural cell types it has been suggested that an activity that requires ATP is capable of displacing PTB from the intronic binding sites. This could allow the formation of a neuron-specific complex on the downstream control sequence enhancer resulting in the further activation of the exon (11, 44,46–49). (FGF-R2 IIIb Derepression) In epithelial cells, exon IIIb is included. The repressive effect of PTB is overcome at least in part through the counteracting activity of yet-to-be-identified proteins, which could potentially stabilize a secondary structure between the two cis elements ISAR and IAS2 (7, 8, 15,17; Wagner and Garcia-Blanco, unpublished).
In the case of the FGF-R2 pre-mRNA, exon IIIb is silenced in cells of mesenchymal origin, and this silencing is mediated in part by PTB (7, 8). In epithelial cells, exon IIIb is included efficiently even though PTB is plentiful (69). The activation of exon IIIb is mediated by at least two cell-type-specific_cis_ elements and others that appear to be activated in all cells (18). Two of the cell-type-specific elements can form a predicted RNA secondary structure in which one stem would be located between seven consensus PTB binding sites in a downstream intronic silencer sequence (17) (Fig. 4). Formation of this structure might be expected to interfere with PTB binding and exon silencing.
SUMMARY
PTB appears to be a global repressor of weak or regulated exons. We propose here that PTB multimerization sequesters these exons to prevent exon definition. This is likely critical not only to prevent inclusion of pseudo-exons but also to set up cell-type-specific exon definition. What remains unclear about PTB can probably be broken down into two basic questions. First, what is the precise mechanism of repression? Second, how is this mechanism circumvented? Most of the research to resolve the first question has focused primarily on identifying instances of PTB repression but has done little to understand how that repression is achieved. Recently, both in vivo and in vitro assays for PTB repression have been developed (8,65); thus, a detailed structure-function analysis can be done. Information from this approach may address mechanistic questions such as if PTB multimerization is required for repression or if there are PTB cofactors. Understanding how this repression is lifted will probably be a more complicated issue. Overwhelming PTB may occur by numerous mechanisms, such as strengthening weak splice sites via activators such as TIA-1 (18), causing the enhancement of inclusion via a tissue-specific expression of antagonizing RNA-binding proteins, or simply by modulating the expression of a PTB cofactor.
ACKNOWLEDGMENTS
We thank B. Cullen, A. Goldstrohm, P. Grabowski, J. Keene, and C. Smith for critically reading the manuscript and for insightful suggestions. We thank members of the Garcia-Blanco laboratory, most especially R. Carstens, for helpful discussions.
This work was supported by a grant from the American Cancer Society. E.J.W. acknowledges the support of a predoctoral fellowship from the D.O.D. M.A.G.-B. acknowledges the support of the Raymond and Beverly Sackler Foundation.
REFERENCES
- 1.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 2.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 3.Berglund J A, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burge C T, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R, editor. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 6.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 7.Carstens R P, McKeehan W L, Garcia-Blanco M A. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstens R P, Wagner E J, Garcia-Blanco M A. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan R C, Black D L. Conserved intron elements repress splicing of a neuron-specific c-srcexon in vitro. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. . (Erratum, 17:2970, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-srcexon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou M Y, Rooke N, Turck C W, Black D L. hnRNP H is a component of a splicing enhancer complex that activates a c-srcalternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou M Y, Underwood J G, Nikolic J, Luu M H, Black D L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 13.Cote C A, Gautreau D, Denegre J M, Kress T L, Terry N A, Mowry K L. A Xenopusprotein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 14.Cote G J, Nguyen I N, Berget S M, Gagel R F. Calcitonin exon sequences influence alternative RNA processing. Mol Endocrinol. 1990;4:1744–1749. doi: 10.1210/mend-4-11-1744. [DOI] [PubMed] [Google Scholar]
- 15.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Gatto F, Gesnel M C, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Gatto F, Plet A, Gesnel M C, Fort C, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Gatto-Konczak F, Bourgeois C F, Le Guiner C, Kister L, Gesnel M C, Stevenin J, Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Gatto-Konczak F, Olive M, Gesnel M C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande G, Samuels M E, Schedl P D. Sex-lethal interacts with splicing factors in vitro and in vivo. Mol Cell Biol. 1996;16:5036–5047. doi: 10.1128/mcb.16.9.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fairbrother W G, Chasin L A. Human genomic sequences that inhibit splicing. Mol Cell Biol. 2000;20:6816–6825. doi: 10.1128/mcb.20.18.6816-6825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Blanco M A, Jamison S F, Sharp P A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 23.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 25.Gooding C, Roberts G C, Moreau G, Nadal-Ginard B, Smith C W. Smooth muscle-specific switching of alpha-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gooding C, Roberts G C, Smith C W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 27.Gosert R, Chang K H, Rijnbrand R, Yi M, Sangar D V, Lemon S M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol Cell Biol. 2000;20:1583–1595. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granadino B, Penalva L O F, Green M R, Valcarcel J, Sanchez L. Distinct mechanisms of splicing regulation in vivo by the Drosophila protein Sex-lethal. Proc Natl Acad Sci USA. 1997;94:7343–7348. doi: 10.1073/pnas.94.14.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman J S, Meyer M I, Wang Y C, Mulligan G J, Kobayashi R, Helfman D M. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatton A R, Subramaniam V, Lopez A J. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon-exon junctions. Mol Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 31.Horabin J I, Schedl P. Regulated splicing of the Drosophilasex-lethal male exon involves a blockage mechanism. Mol Cell Biol. 1993;13:1408–1414. doi: 10.1128/mcb.13.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horabin J I, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol Cell Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 34.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 35.Kuo H C, Nasim F H, Grabowski P J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- 36.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 37.Lin C H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 39.Lou H, Gagel R F, Berget S M. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 40.Lou H, Helfman D M, Gagel R F, Berget S M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lou H, Neugebauer K M, Gagel R F, Berget S M. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lou H, Yang Y, Cote G J, Berget S M, Gagel R F. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol Cell Biol. 1995;15:7135–7142. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophiladoublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 44.Markovtsov V, Nikolic J M, Goldman J A, Turck C W, Chou M Y, Black D L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 46.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 47.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 48.Modafferi E F, Black D L. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-srcpre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulligan G J, Guo W, Wormsley S, Helfman D M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 52.Norton P A. Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res. 1994;22:3854–3860. doi: 10.1093/nar/22.19.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh Y L, Hahm B, Kim Y K, Lee H K, Lee J W, Song O, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang S K. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem J. 1998;331:169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 55.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 56.Perez I, McAfee J G, Patton J G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 57.Polydorides A D, Okano H J, Yang Y Y, Stefani G, Darnell R B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romanelli M G, Lorenzi P, Morandi C. Organization of the human gene encoding heterogeneous nuclear ribonucleoprotein type I (hnRNP I) and characterization of hnRNP I related pseudogene. Gene. 2000;225:267–272. doi: 10.1016/s0378-1119(00)00331-0. [DOI] [PubMed] [Google Scholar]
- 61.Shaoping W, Romfo C M, Nilson T W, Green M R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 62.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 63.Smith C W, Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 64.Smith C W, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 65.Southby J, Gooding C, Smith C W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutually exclusive exons. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 68.Valcarcel J, Singh R, Zamore P D, Green M R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 69.Wagner E J, Carstens R P, Garcia-Blanco M A. A novel isoform ratio switch of the polypyrimidine tract binding protein. Electrophoresis. 1999;20:1082–1086. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1082::AID-ELPS1082>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, Pederson T. A 62,000 molecular weight spliceosome protein crosslinks to the intron polypyrimidine tract. Nucleic Acids Res. 1990;18:5995–6001. doi: 10.1093/nar/18.20.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 72.Zhang L, Ashiya M, Sherman T G, Grabowski P J. Essential nucleotides direct neuron-specific splicing of gamma 2 pre-mRNA. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, Liu W, Grabowski P J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng Z M, Huynen M, Baker C C. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc Natl Acad Sci USA. 1998;95:14088–14093. doi: 10.1073/pnas.95.24.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zorio D A, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]