Protein Binding Protects Sites on Stable Episomes and in the Chromosome from De Novo Methylation (original) (raw)
Abstract
We have utilized the Escherichia coli lac repressor-operator system to test whether protein binding can interfere with de novo DNA methylation in mammalian cells. We find that a DNA binding protein can protect sites on the episome as well as in the genome from the de novo methylation activity of Dnmt3a. Transcriptional machinery moving through the binding sites does not affect the de novo methylation of these sites, and it does not affect the binding protein protection of these sites from de novo methylation. This study and previous studies provide a possible mechanism for the observation that an Sp1 site can serve as a _cis_-acting signal for demethylation and for preventing de novo methylation of the CpG island upstream of the mouse adenine phosphoribosyltransferase (Aprt) gene. These findings also support the hypothesis that protein binding may play a crucial role in changes of CpG methylation pattern in mammalian cells.
It has been proposed that protein binding may affect the DNA methylation pattern during differentiation of mammalian cells (26) and may protect CpG islands from methylation (1). It has also been hypothesized that transcription may interfere with maintenance methylation (20). Studies using prokaryotic methylases to map chromatin structure imply that sequences bound by the histones are protected from methylation (12, 13). It has also been postulated that DNA remains unmethylated at or near sequences bound by nonhistone proteins (25). Protein-DNA contacts in the promoter region of the phosphoglycerate kinase 1 (PGK1) gene have been detected on the active X chromosome but not on the inactive X chromosome (19, 21). It is likely that the proteins observed at the PGK1 promoter are involved in the methylation-free status of the region on the active X chromosome (21). Sp1 binding has been proposed to prevent methylation and cause or preserve the hypomethylated state of the Aprt gene (2, 17). When Sp1 sites are present in an in vitro-methylated construct, the CpG island upstream of the hamster APRT gene in the integrated construct becomes demethylated in mouse ES cells (2). Also, the presence of Sp1 sites in a transgene can prevent methylation of the Aprt CpG island in the unmethylated transgene (2, 17). Although the absence of Sp1 sites and the absence of transcription cannot be clearly distinguished in this system, it is clear that the Sp1 site is a _cis_-acting element for keeping the CpG island upstream of the Aprt gene free of methylation. It is possible that Sp1 binding can protect Sp1 sites and that Sp1 binding may recruit other factors to the region, thereby protecting the local region from de novo methylation under circumstances where the island is initially unmethylated. When the island is methylated in vitro before integration into ES cells, Sp1 binding can lead to demethylation of the Sp1 sites (2). There have been studies demonstrating that protein binding may protect sites from prokaryotic methylases in Escherichia coli and Saccharomyces cerevisiae, and some of these methylases have been used to map chromatin structure or potential protein binding sites in these organisms (12, 13, 27, 29, 31, 36). However, there is no direct evidence as to whether protein binding can protect DNA sites from a mammalian methyltransferase in mammalian cells. We have demonstrated previously that protein binding can specify sites for demethylation (9, 14). If protein binding can also protect sites from de novo methylation, it would provide an explanation for events observed at the CpG island upstream from the Aprt gene (2, 17). We are interested in establishing a system to test whether protein binding can protect sites from de novo methylation in human cells.
Two murine de novo methyltransferases, Dnmt3a and Dnmt3b, have been reported recently (18). We have shown that Dnmt3a and Dnmt3b function in human cells (10), and others have shown that Dnmt3a can methylate DNA in Drosophila cells (16). Although the levels of endogenous DNMT1, DNMT3A, and DNMT3B were not measured, we have previously shown that de novo methylation was not detected using stable episomes as methylation targets in 293/EBNA1 cells (10). Interestingly, exogenously expressed murine Dnmt3a methylates some regions more than others on the episome (10). Based on this, we reasoned that this episomal system could be used to test the protein protection hypothesis.
In a previous study, we showed that the E. coli lac repressor (LacI) binds to the lac operator (lacO) sequence and that isopropyl-β-d-thiogalactopyranoside (IPTG) can inhibit LacI binding to lacO in 293/EBNA1 cells (14). We have also shown that LacI binding to methylated lacO sites can lead to demethylation at these sites and that the demethylation process is primarily determined by protein occupancy of both episomal and chromosomal DNA binding sites (14, 15). Here, we tested episomes containing the lacO sequences to determine whether the lacO sequence is a target of murine Dnmt3a. We found that the lacO sequences on the episomes can be methylated by Dnmt3a. Moreover, active transcription does not affect de novo methylation of the lacO sites by Dnmt3a in 293/EBNA1 cells. We further tested whether LacI can protect lacO sites from de novo methylation by Dnmt3a. We found that LacI can protect lacO sites on the episomes from de novo methylation in cells overexpressing Dnmt3a and that IPTG can abolish this protection. Furthermore, LacI binding to chromosomal lacO sites can also protect these sites from de novo methylation by Dnmt3a. This is the first direct indication that protein binding can protect sites from de novo methylation in human cells.
MATERIALS AND METHODS
Plasmids.
Plasmids pOLucOriP, pOLucRLTR, and pOLucΔLTR (14) bearing the lacO sites were used as targets for de novo methyltransferase in the episomal experiments. Plasmids pMT3aMyc and pMT3aMut (10) were used to supply the normal and the mutant murine Dnmt3a, respectively. A 4.1-kb _Sac_II/Nhe_I fragment from pOLucOriP (14) was used for lacO integration. This fragment contains the Rous sarcoma virus (RSV) long terminal repeat (LTR) promoter, the simian virus 40 (SV40) intron with three copies of the lacO sites, and the luciferase gene (Fig. 1). pCMV_lacI was used to supply LacI in human cells (3). Plasmid LXSP, bearing the puromycin N_-acetyltransferase gene driven by the SV40 early promoter, was used to provide puromycin resistance for lacO integration. The hygromycin resistance gene used for pCMV_lacI integration was obtained from plasmid pHyg (28) by a _Sal_I and _Nru_I double digestion.
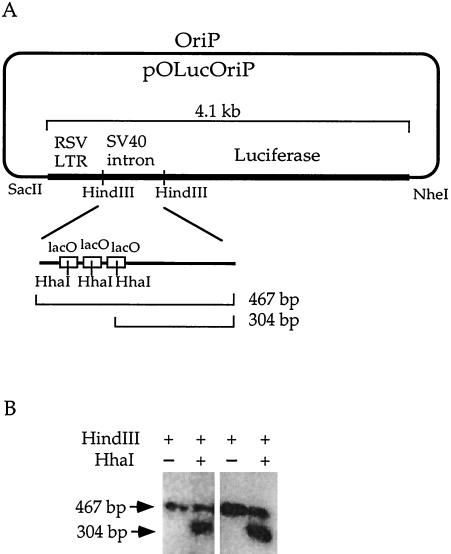
FIG. 1.
De novo methyltransferase Dnmt3a methylates lacO sequences. (A) Diagram of pOLucOriP and lacO sites. The _Sac_II/_Nhe_I fragment of 4.1 kb includes the RSV LTR, the SV40 intron harboring three copies of the lacO sequence, and the luciferase gene. (B) Methylation of the lacO sequences by Dnmt3a. A Southern blot of _Hin_dIII- or _Hin_dIII- and _Hha_I-digested pOLucOriP DNA harvested 7 days after transfection showed a 467-bp band when it was probed with the 467-bp _Hin_dIII fragment, indicating methylation at the _Hha_I sites in the lacO sequence. The two panels show plasmid DNAs harvested from two independent transfections.
Cell lines and transfection.
The 293/EBNA1 cell line (8) was used in all experiments utilizing integrated lacO sites. 293/ElacI cells, derived by integrating LacI into 293/EBNA1 cells (14), and 3a-5 cells, derived by integrating Dnmt3a into 293/EBNA1 cells (10), were used in experiments with lacO sites on the stable episome. Throughout the study, the calcium phosphate transfection method was used to introduce DNA into the human cells (8, 34). The lacO sites (Fig. 1) were integrated by cotransfecting 1 μg of the _Sac_II/Nhe_I fragment and 150 ng of the puromycin expression plasmid pLXSP into approximately 106 293/EBNA1 cells. Puromycin-resistant cell clones were selected by adding puromycin to a final concentration of 2.5 μg/ml in the tissue culture medium 48 h after transfection. Twenty-four puromycin-resistant clones were isolated, and the successful integration and the methylation status of the lacO sites were confirmed by Southern blotting. These clones are designated LacO1 through LacO24. Two of these cell clones, LacO13 and LacO21, had clearly unmethylated inserts and easily detectable lacO fragments by Southern blotting, and these were used for further transfection experiments. LacI-expressing pCMV_lacI was integrated by cotransfecting a 2-kb _Sal_I/_Nru_I DNA fragment containing the hygromycin resistance gene at a 10:1 ratio into LacO13 and LacO21 cells. Hygromycin-resistant cell clones from each cell line were selected using 200 μg of hygromycin per ml beginning at 2 days after transfection. Twenty-four cell clones from each of the LacO13 and LacO21 cell lines were isolated. These clones were designated with an “I” and a number following “LacO13” and “LacO21,” for example, LacO13I1 and LacO21I1. The expression of LacI was tested by immunofluorescent staining as described previously (6). The cell clones LacO13I12, LacO13I17, and LacO13I18 from the LacO13 cell line and LacO21I21, LacO21I29, and LacO21I36 from the LacO21 cell line had high levels of LacI expression by immunofluorescent staining and, therefore, were used in further experiments.
DNA recovery and analysis.
For experiments involving episomes, plasmid DNA was harvested by the Hirt method (7) when the cells reached confluence after transfection. A small fraction of the cells was replated for later harvests. The plasmid DNA harvested from the transfected cells was singly digested with _Hin_dIII or doubly digested with _Hin_dIII and _Hha_I, fractionated on a 1% agarose gel, and analyzed by Southern blot analysis using the 467-bp _Hin_dIII fragment as a probe, unless otherwise specified. When cells with integrated lacO sites reached confluence in the experiments, genomic DNA was harvested by the sodium dodecyl sulfate lysis-proteinase K-phenol-chloroform method from the remaining cells. Approximately 10 μg of the genomic DNA harvested from each cell line in the experiment was digested with _Hin_dIII or doubly digested with _Hin_dIII and _Hha_I to assess the methylation status, as described above. The amount of radioactivity in various fragments on the Southern blots was quantitated using a phosphorimager (Bio-Rad FX). The radioactivity in the 304-bp band (previously described in the work of Lin et al. [14] as 338 bp due to a clerical error) was corrected for the size of the fragment to reflect that only a portion of the probe can hybridize to this fragment. The percentage of plasmids that become methylated in each lane is determined by dividing the amount of radioactivity in the 467-bp band by the total radioactivity in the 467- and the 304-bp bands, after normalization for size.
RESULTS
Murine Dnmt3a can methylate lacO sites on episomes in human cells.
We learned from previous experiments that, in the absence of expression of LacI and Dnmt3a, the unmethylated lacO sequences introduced into 293/EBNA1 cells remained free of methylation over long intervals, indicating the absence of de novo methylation (14). We initially wanted to test whether the lacO sites are targets of the de novo methyltransferase Dnmt3a. Specifically, we wished to test whether the _Hha_I sites in the lacO sequences can be methylated in 293/EBNA1 cells overexpressing Dnmt3a. Plasmid pOLucOriP, which bears three lacO sites (Fig. 1A), was transfected into Dnmt3a-overexpressing cells, called 3a-5 cells (10), to test whether Dnmt3a can methylate the lacO sequences on this plasmid. A Southern blot of pOLucOriP DNA harvested 7 days (Fig. 1B) and 11 days after transfection and singly digested with _Hin_dIII or doubly digested with _Hin_dIII and _Hha_I was probed with the 467-bp _Hin_dIII fragment. If all three _Hha_I sites in the lacO sequences remained free of methylation, a 304-bp band would be detected on the Southern blot. If these three _Hha_I sites in the lacO sequences were methylated by Dnmt3a, a 467-bp fragment would be observed on the Southern blot. Methylation at one or more of the three _Hha_I sites within the lacO sequences would lead to the detection of fragments larger than 304 bp, up to a maximum of 467 bp on the Southern blot. Two bands of 467 and 304 bp were detected in the pOLucOriP DNA harvested 7 days after transfection (Fig. 1B). The plasmid DNA harvested 11 days after transfection showed similar results (data not shown). This finding shows that the _Hha_I sites within the lacO sequences were methylated by Dnmt3a on a substantial fraction of the plasmids. Complete methylation of a given site has not been observed on any of the episomes used here or previously (9, 14). This may be because the expression of Dnmt3a is not the same in all cells even though these cells were derived from a single cellular clone. This may also be a reflection of how Dnmt3a functions in the cell. Dnmt3a may not be as efficient as Dnmt1, and it may not methylate all of its targets. Nevertheless, the finding that _Hha_I sites within the lacO sequences are efficient targets of Dnmt3a demonstrates the possibility of using this system to test whether protein binding can protect sites from de novo methylation.
Transcription does not affect lacO methylation by Dnmt3a.
The lacO sequences on pOLucOriP are located in an SV40 intron placed immediately downstream of the RSV LTR promoter. Active transcription through the lacO sites may play a role in lacO methylation. To test this, we used constructs in which the RSV LTR was either deleted (pOLucΔLTR) or in the reverse orientation relative to that of the luciferase gene (pOLucRLTR). Both of these constructs result in an approximately 100-fold reduction of luciferase gene transcription to near background levels in cells without LacI expression (14). Plasmids pOLucΔLTR and pOLucRLTR were transfected individually into 3a-5 cells to test whether lack of transcription through the lacO sites can alter the methylation of these sites. Plasmid DNA was harvested 7 and 11 days after transfection, doubly digested with _Hin_dIII and _Hha_I, fractionated on a 1% agarose gel, transferred to a Southern blot, and probed with the 467-bp _Hin_dIII fragment. The majority of the plasmid population was free of methylation at the three _Hha_I sites in the lacO sequences, as reflected by the strong hybridization at the 304-bp band in the DNA harvested 7 days after transfection (Fig. 2A). The presence of the 467-bp band (Fig. 2A) indicates that a fraction of the plasmid population acquired methylation at the three _Hha_I sites in the lacO sequences. A Southern blot of plasmid DNA harvested at 11 days after transfection showed similar results, with nearly equal levels of hybridization to the 467- and 304-bp bands (Fig. 2B). No obvious difference was observed between the three plasmids at 7 or 11 days after transfection.
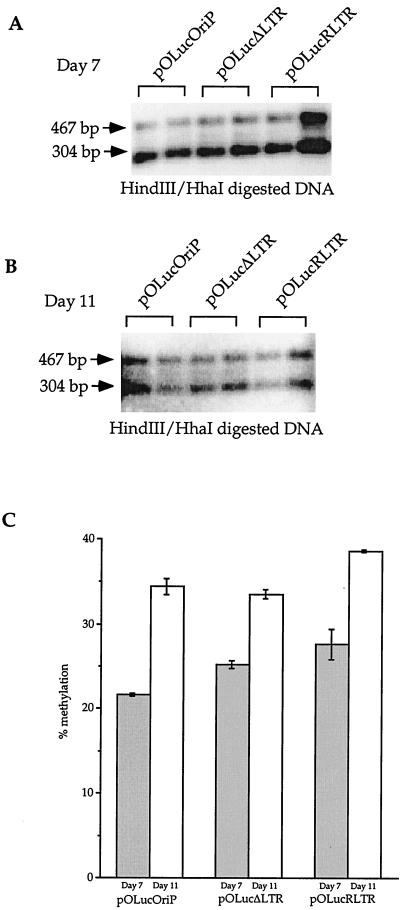
FIG. 2.
Transcriptional activity does not affect de novo methylation of the lacO sites. Plasmid DNAs harvested from transfections were doubly digested with _Hin_dIII and _Hha_I, fractionated on a 1% agarose gel, transferred to a Southern blot, probed with the 467-bp _Hin_dIII fragment. (A) Plasmid DNAs harvested 7 days after transfection. The two lanes in each bracket are independent transfections. The 467-bp band was observed in DNAs from all three plasmids, indicating lacO methylation on all three plasmids. (B) Plasmid DNAs harvested 11 days after transfection. The two lanes in each bracket reflect results of independent transfections. The 467-bp band is stronger than the same band in panel A relative to the 304-bp band. (C) Quantitation of the fraction of the plasmid that became methylated. The radioactivity in the 467-bp band was divided by the total radioactivity in the lane after correction of the fragment sizes.
Quantitation of the radioactivity in the bands detected in two independent transfection experiments showed that an average of 21.7% ± 0.2% of pOLucOriP was methylated at the three _Hha_I sites within the lacO sequences at 7 days after transfection and that an average (± deviation) of 34.5% ± 1.0% of the plasmids were methylated at these sites at 11 days after transfection (Fig. 2C). Similarly, 25.3% ± 0.5% of pOLucΔLTR and 27.7% ± 2% of pOLucRLTR became methylated at the _Hha_I sites in the lacO sequences at 7 days after transfection (Fig. 2C). The same analysis showed that 33.6% ± 0.6% of pOLucΔLTR and 38.6% ± 0.1% of pOLucRLTR became methylated at these sites at 11 days after transfection (Fig. 2C). Repeated experiments showed similar results (data not shown). These findings indicate that the lacO sequences became more methylated over time and that a 100-fold difference in transcriptional activity (14) did not enhance or inhibit de novo methylation of the lacO sequences by Dnmt3a in human cells. These findings also suggest that de novo methylation can occur within the transcriptionally inactive region as well as within the transcriptionally active regions. This indicates that active transcription is not a requirement for de novo methylation as some have recently postulated (11) and that the lacO sites are mostly occupied by the LacI protein instead of the transcription machinery.
LacI binding specifically protects lacO sites on stable episomes from de novo methylation by Dnmt3a in human cells.
It has been shown that deletion of the Sp1 site upstream of the murine Aprt gene leads to methylation of the CpG sites in the surrounding region. It is likely that protein binding can protect sites from methylation in mammalian cells as reported for E. coli and yeast (12, 13, 29, 31, 36), but this has not been previously shown experimentally. It is clear that lacO is a target of Dnmt3a in vivo. Therefore, it can be used to test the hypothesis that protein binding can protect sites from de novo methylation in human cells. To test this hypothesis, a Dnmt3a expression vector and _lacO_-bearing plasmid were cotransfected into LacI-expressing cells. The experiment has to be done in this way rather than by cotransfecting a LacI expression vector and the _lacO_-bearing plasmid into 3a-5 cells. The latter design risks the possibility that lacO sites can be methylated by Dnmt3a before LacI starts to express, and the LacI-expressing plasmid may become methylated in 3a-5 cells before it starts to transcribe and express less LacI. In contrast, in the former design the LacI protein is expressed constitutively in 293/ElacI cells, as described previously (14). If LacI binding to lacO can protect the _Hha_I sites in the lacO sequences from methylation by Dnmt3a, the lacO sites on pOLucOriP should remain free of methylation upon cotransfection with the Dnmt3a expression vector pMT3aMyc into 293/ElacI cells. When IPTG is present in the tissue culture medium to inhibit LacI binding to the lacO sites, overexpression of Dnmt3a should lead to methylation of the lacO sites.
To test this, plasmid pOLucOriP was transfected into 293/ElacI cells with either pMT3aMyc (wild-type Dnmt3a expression vector) or pMT3aMut (mutant Dnmt3a expression vector). Although transcription through the lacO sites is not expected to play a role in lacO site methylation, plasmid pOLucRLTR was trasfected into 293/ElacI cells with pMT3aMyc to test that. A parallel experiment was carried out at the same time with IPTG added to a final concentration of 5 mM in the tissue culture media prior to transfection and through the time of first harvest at 9 days after transfection. Plasmid DNA was harvested and analyzed as described in Materials and Methods. On the Southern blot, a very faint band at 467 bp and a very strong band at 304 bp were detected in pOLucOriP and pOLucRLTR DNAs harvested from 293/ElacI cells overexpresing Dnmt3a and without IPTG treatment (Fig. 3A). In contrast, a very strong 467-bp band and a much weaker 304-bp band were detected in the pOLucOriP DNA and the pOLucRLTR DNA harvested from IPTG-treated 293/ElacI cells with Dnmt3a overexpression (Fig. 3A). A single 304-bp band was detected in the pOLucOriP DNA harvested from 293/ElacI cells after cotransfection with the mutant Dnmt3a, regardless of the presence or absence of IPTG (Fig. 3A). The 467-bp band observed in this experiment is much stronger than what was observed in the experiment described above using 3a-5 cells, indicating a higher level of methylation in the lacO sequences with the transiently transfected Dnmt3a expression vector. This result is most likely due to the higher expression of Dnmt3a in the cells transiently transfected with the Dnmt3a expression vector than in cells with stable expression of Dnmt3a.
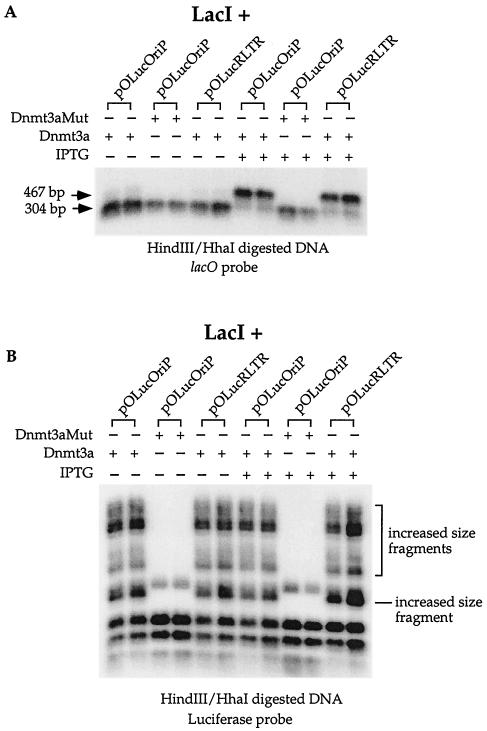
FIG. 3.
Protection of the lacO sites from de novo methylation by LacI. Shown is a Southern blot of plasmid DNA doubly digested with _Hin_dIII and _Hha_I harvested from LacI-expressing cells after cotransfection with the Dnmt3a or the mutant Dnmt3a expression vector. (A) The 467-bp fragment was absent when LacI was present in the cells, and this band was detected in the cells treated with IPTG. In contrast, the 467-bp fragment was absent in the DNA cotransfected into the LacI-expressing cells with the mutant Dnmt3a expression vector, regardless of the presence or absence of IPTG, indicating no methylation at the lacO sites by the mutant Dnmt3a. The two lanes in each bracket reflect the results of two independent transfections. (B) LacI protection is specific for lacO sequences. The same Southern blot shown in panel A was stripped and rehybridized with a probe containing the luciferase coding region downstream from the lacO sequence. No difference in the increased-size fragments was observed with or without the presence of IPTG. The two lanes in each bracket reflect the results of two independent transfections.
The fraction of the plasmid that became methylated at the three _Hha_I sites in the lacO sequences can be measured by dividing the radioactivity in the 467-bp band by the total radioactivity in both the 304- and the 467-bp bands after correction for the fraction of the probe that can hybridize to the 304-bp band. The average amounts of the DNAs that became methylated at these _Hha_I sites from two independent transfections when no IPTG was added to the tissue culture medium were 9.4% ± 0.7% and 6.6% ± 3% of pOLucOriP and pOLucRLTR, respectively. In contrast, 61.9% ± 1.5% and 63.4% + 1% of pOLucOriP and pOLucRLTR, respectively, became methylated at these _Hha_I sites when IPTG was added to a concentration of 5 mM in the tissue culture media. Differences found in the absence and the presence of IPTG were very reproducible (data not shown).
It is conceivable that the presence of LacI may interfere with the overall methyltransferase activity of Dnmt3a. If this is the case, the episome should exhibit an overall lack of methylation when LacI is present and it should acquire methylation when both IPTG and LacI are present upon Dnmt3a cotransfection into the cells with the episome. To examine this possibility, the same Southern blot used for Fig. 3A was stripped and rehybridized with a probe containing the luciferase coding region. The luciferase coding region is immediately downstream from the 467-bp _Hin_dIII fragment containing the lacO sequences (Fig. 1). There is no _Hin_dIII site within the entire luciferase coding region. DNAs doubly digested with _Hin_dIII and _Hha_I from cells cotransfected with the episome and the mutant Dnmt3a showed the complete digestion pattern (Fig. 3B). Episomes cotransfected with wild-type Dnmt3a yielded increased-size _Hha_I fragments with or without IPTG treatment (Fig. 3B). This finding indicates that Dnmt3a methylates the luciferase coding region equally well in the presence and the absence of LacI protein. This determination supports the conclusion that LacI protection of sites from de novo methylation by Dnmt3a is specific to the lacO sites despite the fact that LacI binds to DNA nonspecifically at a lower affinity. It is noteworthy that the majority of the plasmids became methylated in the lacO sequences when IPTG and Dnmt3a were both present (Fig. 3A) but that a majority of the plasmids remained free of methylation in the luciferase coding region (Fig. 3B). These findings are consistent with the previous finding that Dnmt3a has preferred targets on the episome (10).
As shown by our previous study, the methylated _Hha_I sites within the lacO sequences on the episome can become demethylated when LacI is present in the cells (14). We wanted to test whether the protection of unmethylated lacO sites from de novo methylation and the targeting of methylated lacO sites for demethylation can occur in the same cells. The Dnmt3a expression vector pMT3aMyc was nearly completely lost from the human cells after several rounds of cell division because it does not have a replication origin that allows it to replicate in human cells. We observed a decrease in methylation in the lacO sites after IPTG was withdrawn from the treated cells after the first harvest at 9 days after transfection (data not shown). Conversely, the transfected cells not treated with IPTG before the first harvest were treated with a final concentration of 5 mM IPTG in the tissue culture media after the first harvest. We observed no changes in the small amount of lacO site methylation in these cells (data not shown). This is consistent with the observation that, in many previous experiments, the methylation pattern of the episomes was maintained in 293/EBNA1 cells. This observation is also consistent with our previous finding that LacI binding can lead to demethylation of methylated lacO sites since no change in methylation of lacO was observed when LacI was not allowed to bind to the lacO sequences.
These findings confirm that methylation of the three _Hha_I sites in the lacO sequences is a result of the de novo methyltransferase activity of Dnmt3a because mutant Dnmt3a fails to methylate this sequence. These results also demonstrate that the presence of LacI can protect lacO sites from the de novo methylation activity of Dnmt3a and that IPTG can inhibit this protection. LacI binding to lacO sites can protect these sites from de novo methylation by Dnmt3a and target these sites for loss of methylation in the same cells in the presence of LacI after Dnmt3a is no longer expressed. Furthermore, transcriptional activity through the lacO sites did not interfere with de novo methylation of these sites by Dnmt3a, as shown above, and it did not change the role of LacI in protecting the lacO sites from Dnmt3a. The small fraction of plasmids that became methylated at the lacO sites when LacI was present indicates that the amount of LacI in 293/ElacI cells may not be sufficient to protect all of the lacO sites from Dnmt3a. However, it is clear that LacI binding is essential for protecting the lacO sites from being methylated, regardless of the transcriptional activity through the region. Most importantly, these results clearly suggest that LacI protection of sites from de novo metylation by Dnmt3a is very specific to lacO sites.
Dnmt3a can methylate the lacO sites integrated into the chromosome, and LacI can protect these lacO sites from de novo methylation by Dnmt3a.
To test whether Dnmt3a can methylate lacO sites in the chromosome, cell lines with unmethylated lacO sites integrated into the chromosome were generated. For this, a 4.1-kb _Sac_II/_Nhe_I fragment containg the RSV LTR, an SV40 intron containing three lacO sites, and the luciferase gene from plasmid pOLucOriP was integrated into 293/EBNA1 cells using puromycin as a selection marker. Twenty-four puromycin-resistant cell clones were analyzed for methylation status by probing the Southern blot of _Hin_dIII- and _Hha_I-digested genomic DNAs from these cell clones with the 467-bp _Hin_dIII fragment. A single 304-bp band should have been observed if the inserts remained unmethylated at all three _Hha_I sites within the lacO sequence. As described earlier, methylation of any copy of the insert at one or more of the three _Hha_I sites within the lacO sequences would lead to the detection of fragments larger than 304 bp and up to 467 bp on the Southern blot. A single 304-bp fragment was detected in DNAs doubly digested with _Hin_dIII and _Hha_I from 23 of the 24 cell clones (data not shown). Deletions in this 467-bp _Hin_dIII fragment larger than 40 bp can be detected on the Southern blot of _Hin_dIII-digested DNA. No smaller bands were detected on the Southern blot of _Hin_dIII-digested DNA from all 24 clones (data not shown), indicating that the SV40 intron containing lacO sites was not grossly rearranged in these cell clones.
To assess whether LacI can protect the lacO sites in the chromosome from de novo methylation by Dnmt3a, the cell clones LacO13 and LacO21, which contain unmethylated lacO integrants, were used to integrate linearized pCMV_lacI_ with a hygromycin selection marker. Twenty-four hygromycin-resistant cell clones were isolated from each of the LacO13 and LacO21 cell lines. LacI expression in these cell clones was verified by immunofluoresence staining using polyclonal rabbit anti-LacI antibody (Stratagene). The level of LacI expression may vary due to the integration site and copy number differences in different cell clones. Therefore, three cell clones from each of the LacO13 and LacO21 cell lines (LacO13I12, LacO13I17, LacO13I18, LacO21I27, LacO21I29, and LacO21I36) expressing LacI at high levels were used for further experiments. Southern blot analysis of genomic DNAs from these six cell clones confirmed that the _Hha_I sites in the lacO sequence remained unmethylated and that no rearrangement occurred in the 467-bp _Hin_dIII fragment (data not shown).
The Dnmt3a expression vector pMT3aMyc was transfected into the six cell clones with or without IPTG treatment. Genomic DNAs from these transfected cell clones were harvested 9 to 19 days after transfection. The DNA was singly digested with _Hin_dIII or doubly digested with _Hin_dIII and _Hha_I and analyzed by Southern blotting. A single 467-bp band was detected in the _Hin_dIII-digested DNA from untransfected LacO21I27 cells, and a single 304-bp band was observed in the DNA doubly digested with _Hin_dIII and _Hha_I from untransfected LacO21I27 cells (Fig. 4A). A single 304-bp band and a very faint 467-bp band were detected in genomic DNA harvested from the LacO21I27 cells transfected with Dnmt3a and not treated with IPTG (Fig. 4A). A 304-bp band and a 467-bp band were clearly detected in the DNA harvested from LacO21I27 cells transfected with Dnmt3a and treated with IPTG (Fig. 4A). The other two cell clones, LacO21I29 and LacO21I36, showed similar results only with no detectable 467-bp band in the DNA harvested from transfected cells not treated with IPTG (Fig. 4A and data not shown). The three clones from the LacO13 cell line also showed similar results; however, the 467-bp band is much weaker, indicating that lacO is less methylated in these cells than in the LacO21 cell clones (Fig. 4A and data not shown). A faint band at 467 bp was also observed in one of the LacO13 clones, LacO13I12 not treated with IPTG after transfection (Fig. 4A). The fraction of plasmid that became methylated at the three _Hha_I sites within lacO can be estimated based on the quantitation of the radioactivity in the 467- and the 304-bp bands from each transfection as described above. The average levels of methylation were 35.9% ± 1.5%, 53.6% ± 2.2%, and 43.8% ± 0.6% from independent transfections of each of the LacO21I27, LacO21I29, and LacO21I36 cell clones, respectively. The average levels of methylation were 28.5% ± 1.3%, 12.5% ± 1.3%, and 14.6% ± 1.1% in LacO13I12, LacO13I17, and LacO13I18 cell clones, respectively, from two independent transfections of each cell line. Repeated experiments showed similar results (data not shown). The fractions of lacO sites that became methylated in the three cell clones derived from the same cell line were very consistent. This suggests that the integration site may play an important role in the accessibility of the lacO sites to Dnmt3a.
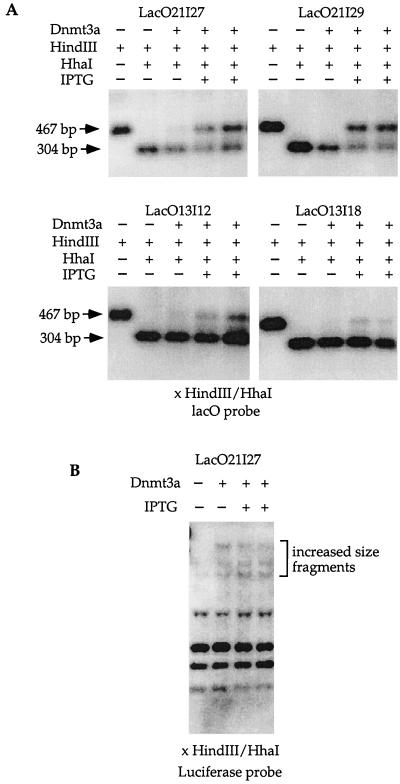
FIG. 4.
LacI protection of lacO sites from de novo methylation in the chromosome. (A) The Dnmt3a expression vector was transfected into independent LacI-expressing cell clones containing the lacO sequences treated or not treated with IPTG. A single 304-bp band was detected in the DNA doubly digested with _Hin_dIII and _Hha_I harvested from untransfected cells, indicating no methylation at the lacO sites without the presence of Dnmt3a. A single 304-bp band was detected in the _Hin_dIII- and _Hha_I-digested DNA from transfected cell clones not treated with IPTG. This indicates that no methylation occurred at the _Hha_I sites in the lacO sequences when IPTG was absent and that LacI was allowed to bind to lacO in the cells. The 467-bp band was present in the DNA doubly digested with _Hin_dIII and _Hha_I from these cell clones when IPTG was present. This result demonstrates that Dnmt3a can methylate lacO when LacI binding to lacO is inhibited by IPTG. (B) LacI protection is specific for lacO sequences. No difference in the increased-size fragments was observed when the Southern blot shown in panel A was stripped and rehybridized with a probe containing the luciferase coding region, regardless of the presence or absence of IPTG.
To examine whether LacI expression can protect sites other than lacO from de novo methylation by Dnmt3a, the same Southern blot described above was stripped and rehybridized with a probe containing the luciferase coding region as described above in the episomal experiment. DNA doubly digested with _Hin_dIII and _Hha_I from untransfected LacO21I27 showed the complete digestion pattern (Fig. 4B). LacO21I27 cells transfected with pMT3aMyc showed increased-size _Hha_I fragments regardless of the presence or absence of IPTG (Fig. 4B). This finding is consistent with the results using the episome. Similar to what occurred in the episomal experiment, the lacO sequences were more methylated (Fig. 4A) than the luciferase coding region (Fig. 4B) when IPTG and Dnmt3a were both present. This result is consistent with the previous finding that Dnmt3a has preferred targets on the episome (10).
These findings indicate that Dnmt3a can methylate the _Hha_I sites in the lacO sequences in a human chromosome and that LacI binding to lacO can prevent Dnmt3a methylation of the _Hha_I sites within lacO. However, the protection of the lacO site by LacI may vary depending upon LacI expression or the amount of Dnmt3a in the cells. These results also indicate that Dnmt3a can methylate the luciferase coding region integrated in the chromosome and that the presence of LacI does not protect this region from de novo methylation. As observed on the episome, LacI protection of sites from de novo methylation by Dnmt3a is specific to lacO sites. These findings further support the conclusion that methylation events occurring on the episome are a close reflection of those occurring in the chromosome.
DISCUSSION
Utilizing the well-characterized E. coli lac repressor-operator system, this study showed for the first time that protein binding protects DNA sites on the stable episome as well as in the genome from de novo methylation by methyltransferase, Dnmt3a, in human cells. The lac operator DNA, lacO, can be methylated by Dnmt3a in vivo, and this methylation activity is not enhanced or inhibited by transcription through the region containing the binding sites. When the LacI protein was present in the same cells, the lacO sequences were protected from methylation by Dnmt3a and remained free of methylation. The same protection by LacI protein was also observed when lacO sites were integrated into the chromosome. The presence of LacI protein specifically protected the lacO sequences from methylation because the presence of LacI did not affect the methylation of the luciferase gene that is adjacent to the lacO sites. This is the first direct evidence that protein binding to DNA can protect specific sites from de novo methylation in human cells.
De novo methylation occurs in the lacO sequence on the episome regardless of the transcriptional activity through the region. It has been described that transcriptionally silent regions harbor hypoacetylated histones, and hyperacetylated histones accumulate in transcriptionally active regions (for a review, see reference 35). The transcriptionally active chromatin has been proposed as being more accessible, and the transcription complex movement may target CpG islands for de novo methylation (11). This study showed that active transcription through the lacO sites did not inhibit the de novo methylation of this sequence and that active transcription did not increase the level of methylation in the lacO sites. It is possible that the near background level of transcription on the plasmids with no promoter or a reversed promoter is sufficient to specify a chromatin state that is different than a repressed region. If the accessibility dictated by basal transcription were sufficient to target de novo methylation, all DNA with basal transcription through the region should become methylated when Dnmt3a is expressed highly in the cells. This is obviously not the case because the luciferase gene is being actively transcribed, and de novo methylation of the region is very minimal. This suggests that de novo methylation does not occur during the time that the LacI protein is displaced by the transcriptional machinery and that the lacO sites are occupied mostly by LacI and not by the transcriptional machinery.
In two independent cell clones in the present study, the protection against methylation by Dnmt3a at the lacO DNA sites was incomplete even though LacI protein was present in the cells. This suggests that the protection of lacO sites against CpG methylation may be the result of LacI protein and Dnmt3a competing for binding at these sites. The binding affinity and protein concentration may be crucial as they relate to the methylation state of these protein binding sites. The binding of LacI can protect unmethylated lacO sites from de novo methylation, and it can also lead to methylation loss at the methylated lacO sites in the same cells. This conclusion is consistent with the observations that the presence of an Sp1 site can lead to loss of methylation of a methylated CpG island upstream of the Aprt gene in ES cells and that its presence can also prevent DNA methylation at this CpG island (2, 17). It is likely that Sp1 binding plays an essential role in the methylation state of the CpG island at the Aprt gene. No protein factors other than Sp1 have been reported to bind to the promoter of the Aprt gene (17). However, it is possible that Sp1 binding allows the recruitment of other factors to the promoter, forming a complex without the direct binding of the other factors to the DNA; the presence of such a complex anchored by Sp1 may be sufficient to protect the region. It is impossible to distinguish whether the absence of active transcription of the Aprt gene or the absence of the Sp1 site is important to the methylation of the Aprt gene, because Sp1 is part of the promoter and the absence of the Sp1 site eliminates transcription. The present study clearly demonstrates that protein binding, and not active transcription, protects sites from de novo methylation.
It is known that DNA methylation patterns are different in different cell types and that changes occur in cancer cells. Recently, nonrandom CpG island methylation has been documented (4). It has been described that the methylation pattern is determined by the rate of de novo methylation and the rate of methylation maintenance (20, 24). It has also been postulated that a “determinator protein” may specify sites for loss of methylation or protect sites from methylation (22, 23). We have provided evidence in this study and previous studies that the interaction between a DNA binding protein and its binding site plays a critical role in targeting sites for loss of methylation and protecting sites from de novo methylation. It is clear that protein binding can affect both loss of methylation and de novo methylation, and it is reasonable to propose that the concentration and affinity of the DNA binding proteins, their interaction with other proteins, and the available binding sites in the cell can dictate the methylation pattern in the cell. A previously methylated site can become demethylated if the concentration of the binding protein increases. Conversely, a previously protected and unmethylated site can become methylated if the availability of the binding protein decreases. Therefore, when the balance between the number of DNA binding sites and DNA binding proteins or the balance between different proteins is altered during differentiation or carcinogenesis, it might lead to methylation changes. The dynamics of protein-DNA and protein-protein interaction may also be different in subtle ways that can lead to differences in DNA methylation patterns in the same cell type. This may explain the minor variations of methylation pattern observed by the bisulfite genomic sequencing method, in which single molecules can be assessed from cells of the same tissue type (5, 30, 32, 33).
REFERENCES
- 1.Bird A P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 3.Brown M, Figge J, Hansen U, Wright C, Jeang K-T, Khoury G, Livingston D M, Roberts T M. lac repressor can regulate expression from a hybrid SV40 early promoter containing a lac operator in animal cells. Cell. 1987;49:603–612. doi: 10.1016/0092-8674(87)90536-8. [DOI] [PubMed] [Google Scholar]
- 4.Costello J F, Fruhwald M C, Smiraglia D J, Rush L J, Robertson G P, Gao X, Wright F A, Feramisco J D, Peltomaki P, Lang J C, Schuller D E, Yu L, Bloomfield C D, Caligiuri M A, Yates A, Nishikawa R, Su Huang H, Petrelli N J, Zhang X, O'Dorisio M S, Held W A, Cavenee W K, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 5.Devereux T R, Horikawa I, Anna C H, Annab L A, Afshari C A, Barrett J C. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- 6.Grawunder U, Finnie N, Jackson S P, Riwar B, Jessberger R. Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. Eur J Biochem. 1996;241:931–940. doi: 10.1111/j.1432-1033.1996.00931.x. [DOI] [PubMed] [Google Scholar]
- 7.Hirt B. Selective extraction of polyoma DNA from infected mouse cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh C-L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh C-L. Evidence that protein binding specifies sites of DNA demethylation. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh C-L. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmat3b. Mol Cell Biol. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones P A. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 12.Kladde M P, Simpson R T. Positioned nucleosomes inhibit Dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kladde M P, Xu M, Simpson R T. Direct study of DNA-protein interactions in repressed and active chromatin in living cells. EMBO J. 1996;15:6290–6300. [PMC free article] [PubMed] [Google Scholar]
- 14.Lin I G, Tomzynski T J, Ou Q, Hsieh C-L. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol Cell Biol. 2000;20:2343–2349. doi: 10.1128/mcb.20.7.2343-2349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin I G, Hsieh C-L. Chromosomal DNA demethylation specified by protein binding. EMBO Rep. 2001;2:108–112. doi: 10.1093/embo-reports/kve023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyko F, Ramasahoye B H, Kashevsky H, Tudor M, Mastrangelo M-A, Orr-Weaver T L, Jaenish R. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat Genet. 1999;23:363–366. doi: 10.1038/15551. [DOI] [PubMed] [Google Scholar]
- 17.Macleod D, Charlton J, Mullins J, Bird A. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 18.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer G P, Riggs A D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991;5:1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- 20.Pfeifer G P, Steigerwald S D, Hansen R S, Gartler S M, Riggs A D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island; methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeifer G P, Tanguay R L, Steigerwald S D, Riggs A D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 22.Riggs A D. DNA methylation and cell memory. Cell Biophys. 1989;15:1–13. doi: 10.1007/BF02991574. [DOI] [PubMed] [Google Scholar]
- 23.Riggs A D, Jones P A. 5-Methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 24.Riggs A D, Xiong Z, Wang L, LeBon J M. Methylation dynamics, epigenetic fidelity and X chromosome structure. Novartis Found Symp. 1998;214:214–232. doi: 10.1002/9780470515501.ch13. [DOI] [PubMed] [Google Scholar]
- 25.Selker E C. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990;15:103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- 26.Singer J, Roberts-Ems J, Luthardt E W, Riggs A D. Methylation of DNA in mouse early embryos, teratocarcinoma cells and adult tissues of mouse and rabbit. Nucleic Acids Res. 1979;20:2369–2385. doi: 10.1093/nar/7.8.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh J, Klar A J S. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 28.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavazoie S, Church G M. Quantitative whole-genome analysis of DNA-protein interaction by in vivo methylase protection in E. coli. Nat Biotechnol. 1998;16:566–571. doi: 10.1038/nbt0698-566. [DOI] [PubMed] [Google Scholar]
- 30.Vu T H, Li T, Nguyen D, Nguyen B T, Yao X M, Hu J F, Hoffman A R. Symmetric and asymmetric DNA methylation in the human IGF2–H19 imprinted region. Genomics. 2000;64:132–143. doi: 10.1006/geno.1999.6094. [DOI] [PubMed] [Google Scholar]
- 31.Wang M X, Church G M. A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature. 1992;360:606–610. doi: 10.1038/360606a0. [DOI] [PubMed] [Google Scholar]
- 32.Warnecke P M, Mann J R, Frommer M, Clark S J. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics. 1998;51:182–190. doi: 10.1006/geno.1998.5371. [DOI] [PubMed] [Google Scholar]
- 33.Warnecke P M, Clark S J. DNA methylation profile of the mouse skeletal alpha-actin promoter during development and differentiation. Mol Cell Biol. 1999;19:164–172. doi: 10.1128/mcb.19.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wigler M, Sweet R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Transformation of mammalian cells with genes from prokaryotes and eukaryotes. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 35.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 36.Xu M, Simpson R T, Kladde M P. Gal4p-mediated chromatin remodelling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]