Inhibition of Nuclear Import by Protein Kinase B (Akt) Regulates the Subcellular Distribution and Activity of the Forkhead Transcription Factor AFX (original) (raw)
Abstract
AFX belongs to a subfamily of Forkhead transcription factors that are phosphorylated by protein kinase B (PKB), also known as Akt. Phosphorylation inhibits the transcriptional activity of AFX and changes the steady-state localization of the protein from the nucleus to the cytoplasm. Our goal was threefold: to identify the cellular compartment in which PKB phosphorylates AFX, to determine whether the nuclear localization of AFX plays a role in regulating its transcriptional activity, and to elucidate the mechanism by which phosphorylation alters the localization of AFX. We show that phosphorylation of AFX by PKB occurs in the nucleus. In addition, nuclear export mediated by the export receptor, Crm1, is required for the inhibition of AFX transcriptional activity. Both phosphorylated and unphosphorylated AFX, however, bind Crm1 and can be exported from the nucleus. These results suggest that export is unregulated and that phosphorylation by PKB is not required for the nuclear export of AFX. We show that AFX enters the nucleus by an active, Ran-dependent mechanism. Amino acids 180 to 221 of AFX comprise a nonclassical nuclear localization signal (NLS). S193, contained within this atypical NLS, is a PKB-dependent phosphorylation site on AFX. Addition of a negative charge at S193 by mutating the residue to glutamate reduces nuclear accumulation. PKB-mediated phosphorylation of AFX, therefore, attenuates the import of the transcription factor, which shifts the localization of the protein from the nucleus to the cytoplasm and results in the inhibition of AFX transcriptional activity.
Protein kinase B (PKB), also known as Akt, promotes cell survival in many different cell types (24, 38, 40, 66). Following its initial cloning (17, 34), PKB was isolated as the transforming gene of v-Akt, hence the name c-Akt and its classification as a proto-oncogene (6, 67). Activation of PKB requires the lipid phosphatidylinositol 3,4,5-triphosphate (PIP3) (15) and phosphorylation by an upstream kinase, PDK1 (2, 69, 72). PIP3 is produced by phosphatidylinositol 3-kinase in response to signals from extracellular growth factors (for a review see reference 60). PKB has been shown to phosphorylate and regulate the activity of transcription factors in response to survival factors. Genetic studies of Caenorhabditis elegans have demonstrated that the PKB signal transduction pathway inhibits the activity of the Forkhead transcription factor, daf-16, a gene that regulates longevity (55). There are three human orthologues of daf-16, AFX (13), FKHR (27), and FKHRL1 (3), that were first identified as chromosomal breakpoints in human tumors.
AFX is phosphorylated by PKB in response to insulin and serum at three sites: T28, S193, and S258 (42). Phosphorylation of these residues by PKB leads to both inhibition of the transcriptional activity of AFX and cytoplasmic retention and/or nuclear exclusion of the protein. Withdrawal of serum or insulin results in AFX dephosphorylation, nuclear localization, and target gene activation. In the absence of survival factors, Forkhead family members have been shown to induce the transcription of proapoptotic genes, such as the FasL gene (14) and the Bim gene (22), thus triggering a cascade of events that lead to apoptosis. In addition, overexpression of AFX blocks cell cycle progression at G1 by a mechanism that is independent of functional retinoblastoma protein but dependent on the cell cycle inhibitor p27kip (49). Dysregulation of Forkhead proteins may, therefore, comprise an important step in oncogenic transformation by both inhibiting apoptosis and promoting progression through the cell cycle.
Efficient regulated nuclear localization of transcription factors in response to extracellular signals is essential for their function (41). For example, in unstressed cells, p53 continuously shuttles into and out of the nucleus, and its subcellular distribution varies throughout the cell cycle. In response to certain stresses, however, p53 relocalizes to the nucleus to promote gene transcription. In some p53-related tumors, however, there is a defect in p53 localization (52, 65). In these cells, p53 is constitutively cytoplasmic due to an increase in the export rate of the protein. As a result, p53 cannot accumulate in the nucleus and carry out its normal activities. This defect leads to unregulated cellular proliferation. Likewise, the transcriptional activity of Forkhead family members may be regulated not only by phosphorylation but also by changes in their subcellular localization.
Many proteins are transported constitutively into and out of the nucleus by members of the β-importin family of nuclear transport receptors (28, 48). These receptors recognize specific localization signals within their cargoes, and their association with these signal sequences is controlled by the small GTPase Ran. The regulators of Ran are distributed asymmetrically within the cell: the RanGTPase-activating protein (RanGAP) is cytoplasmic (10), whereas the Ran exchange factor (RanGEF) is nuclear (11). Therefore, most of the Ran present in the nucleus is predicted to be GTP bound, while cytoplasmic Ran is predicted to be GDP bound. The directionality of transport depends on this RanGTP gradient. Unloading of cargo from an import receptor in the nucleus is triggered by the binding of RanGTP to the receptor (29, 31, 61). Conversely, an export receptor can only load its cargo in the presence of RanGTP. Release of export cargo into the cytoplasm occurs by the hydrolysis of RanGTP to RanGDP, stimulated by RanGAP and other required cofactors (39).
In contrast to constitutive transport, regulated transport occurs only in response to specific cellular signals, such as phosphorylation (35). For example, mitogen-activated protein kinase-activated protein kinase 2 is a nuclear protein in unstimulated cells. In response to specific extracellular signals, it is phosphorylated, binds p38 stress-activated protein kinase, and is only then exported from the nucleus (7). Regulated transport mechanisms play critical roles in augmenting or attenuating signaling information by controlling the cellular localization of proteins. Many proteins that have regulated transport mechanisms are shuttling proteins that move continuously between the cytoplasm and nucleus. Thus, their steady-state localization is determined by the relative rate constants for import and export. Shuttling coordinates nuclear and cytoplasmic events by providing a rapid and reversible method to regulate a nuclear and/or cytoplasmic activity. Nuclear transport often works in tandem with other mechanisms to regulate the function of proteins. Some transcription factors require phosphorylation in addition to changes in nuclear localization to completely regulate their activity. This redundancy increases the strength of cellular switches and provides additional ways to integrate environmental cues with cellular signals. Finally, proteins that alter their cellular localization in response to extracellular signals need a mechanism for turning that signal off when it is no longer required. Protein degradation, dephosphorylation, and nuclear transport are all mechanisms used to terminate activity. Although the regulated transport mechanisms for several yeast proteins have been identified, including those for Pho4 (36, 37) and Mig1 (20, 21), few regulated transport mechanisms have been described for mammalian proteins.
Our goal, therefore, was to elucidate how PKB phosphorylation and the resulting cellular relocalization of AFX alter its transcriptional activity. In this study, we show that PKB phosphorylation of AFX in the nucleus followed by Crm1-dependent nuclear export is required to inhibit the transcriptional activity of the Forkhead family member. Although phosphorylation by PKB does not significantly alter AFX export from the nucleus, it appears that nuclear import of phosphorylated AFX is regulated. Amino acids 180 to 221 of AFX are necessary and sufficient for nuclear import but comprise a nonclassical nuclear localization signal (NLS). Addition of a negative charge at S193 by mutating the residue to a glutamate inhibits nuclear import. Therefore, phosphorylation of AFX by PKB likely reduces the nuclear import rate of AFX. This results in a shift in the steady-state localization of AFX from the nucleus to the cytoplasm, thereby inhibiting its transcriptional activity.
MATERIALS AND METHODS
Plasmids and recombinant proteins.
pMT2-HA-AFX, pMT2-HA-SASA, pMT2-HA-A3, pSG5-gagPKB, p1205-luc, and pCMV-lacZ have been described previously (42, 49). pMT2-HA-AFX (Δ198–216) was constructed using PCR-based mutagenesis. To construct the vectors encoding the fusions of green fluorescent protein (GFP) to the C termini of various AFX sequences, pKGFP3 was used. To create pKGFP3, GFP-GFP was PCR amplified and inserted as a _Bgl_II-_Bgl_II fragment into pKGFP. For pKGFP3-AFX(180–197), pKGFP3-AFX(198–221), and pKGFP3-AFX(180–221), the indicated residues of AFX were PCR amplified and ligated into pKGFP3 as _Xba_I-_Xba_I fragments. Site-directed mutagenesis of residues within pKGFP3-AFX(180–221) to produce pKGFP3-AFX(180–221)I-V, pKGFP3-AFX(180–221)S193A, and pKGFP3-AFX(180–221)S193E was carried out using the Quikchange mutagenesis kit (Stratagene) according to the manufacturer's recommendations. All mutations were verified by sequencing. pQE60-Crm1 was a gift from Iain Mattaj (EMBL, Heidelberg, Germany) (5). Crm-His6 was purified using Talon beads (Clontech) as described previously (39). The addition of 14 mM β-mercaptoethanol was required to retain the stability of the protein. pQE32-RanQ69L was a gift from Dirk Görlich (University of Heidelberg). His6-RanQ69L was prepared as described earlier (31).
Cell culture.
Insulin receptor-overexpressing NIH 3T3 cells (A14) were grown as described previously (15). All other cells were passaged in Dulbecco's modified Eagle medium (supplemented with 5% [vol/vol] fetal calf serum and penicillin-streptomycin). Baby hamster kidney cells (BHK21), human embryonic kidney cells (HEK293), and HeLa cells were cultured in a humidified, 37°C–5% CO2 incubator. tsBN2 cells were grown at 33.5°C. Where indicated, the tsBN2 cells were shifted to 39.5°C for 3 h after the addition of 50 μM cycloheximide. The leptomycin B (LMB) used in some cell-based assays was a gift from Barbara Wolff (Novartis).
Transfections.
Hemagglutinin (HA)-tagged AFX constructs were transfected into A14, BHK21, HEK293, HeLa, and tsBN2 cells by the calcium phosphate method. At 24 h posttransfection, the transfection medium was replaced with fresh medium and the cells were incubated at 37°C for an additional 24 h. Transfected HEK293 cells were harvested for immunoprecipitations at 48 h posttransfection. Transfected A14 cells were serum starved overnight where indicated. For immunofluorescence, A14 cells were either untreated, treated with 10 μM LY294002 (Calbiochem) for 10 min, or treated with 2 ng of LMB per ml for 30 min prior to the addition of 1 μg of insulin per ml. The cells were then incubated with insulin for an additional 30 min before fixation. Transfected tsBN2, HeLa, and BHK21 cells were plated on poly-l-lysine-coated coverslips. Transfected tsBN2 and HeLa cells, where indicated, were serum starved for 1.5 h prior to fixation. BHK21 cells were transfected with all pKGFP3 constructs using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. These cells were processed 20 h after transfection. After the indicated treatments, the transfected cells were fixed and permeabilized with 4% paraformaldehyde–2% sucrose in phosphate-buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) and ice-cold methanol as described previously (54, 58, 73). Fixed cells were then prepared for analysis by fluorescence microscopy.
Fluorescence microscopy.
After fixation and permeabilization, cells expressing GFP fusions were incubated with 4′,6-diamidino-2-phenylindole (DAPI; 10 ng/ml) to stain the nuclei, and then the coverslips were mounted on glass slides by using GelMount (Biomeda). Images of the cells were captured using a 60× water-immersion objective lens on a Nikon inverted microscope equipped with a Hamamatsu charge-coupled device camera. All immunofluorescence data were obtained and processed using Openlab (Improvision) and Adobe Photoshop software. Images for each set of experiments were obtained using the same camera settings. The relative nuclear and cytoplasmic fluorescence levels of the GFP3 constructs were measured using Openlab (Improvision). Nuclear fluorescence was calculated as a percentage of the total cellular fluorescence [N/(N+C)]. All fluorescence measurements were corrected for background fluorescence levels. Each data point represents the mean fluorescence obtained from 12 randomly chosen cells. Error is expressed as the standard deviations of the means. All cells expressing HA-tagged AFX constructs were blocked in 10% bovine serum albumin (BSA)–PBS at room temperature incubated with monoclonal antibody 12CA5 (2 μg/ml) and Texas red-conjugated donkey anti-mouse immunoglobulin G (1:1,500 dilution; Jackson ImmunoResearch Laboratories), both in 3% BSA; stained with DAPI; and mounted and viewed as described above. Cells expressing gag-PKB were detected using anti-gag and anti-PKB antisera (15).
Immunodetection.
BHK21 cells expressing GFP3 constructs were lysed with an equal volume of boiling Laemmli sample buffer. Equal volumes of lysate (10 μl) were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting for GFP3 with polyclonal anti-GFP (1:1,000 dilution; Molecular Probes) and horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (1:20,000 dilution; Jackson Laboratories). Proteins were revealed by chemiluminescence (Kirkegaard & Perry Laboratories). Subcellular fractionation of insulin-treated (1 μg/ml) A14 cells was carried out according to published protocols (47, 76). Anti-phosphoT32 FKHR-L1, anti-c-cbl, and anti-RNA pol II were gifts from Anne Brunet (Harvard University), Kris Reedquist (University Medical Center, Utrecht, The Netherlands), and Marc Timmers (University Medical Center), respectively. Anti-phosphoS193 AFX and anti-phosphoS473 PKB were obtained from New England Biolabs.
Luciferase assays.
A14 cells were cotransfected with the 1205-luc reporter, pMT2-HA-AFX, and pCMV-lacZ. Cells were also transfected where indicated with pSG5-gagPKB. Cells were preincubated with LMB where noted for 30 min prior to the addition of insulin. Cells were then treated with insulin for 16 h in the absence or presence of LMB. Transcriptional activity was measured 48 h after transfection. Luciferase and β-galactosidase measurements were performed as described (42).
Crm1 binding assays.
HEK293 cells were either mock transfected or transfected with pMT2-HA-AFX or pMT2-HA-A3. Where indicated, cells were treated with LY294002 (10 μM) for 2 h prior to lysis. All cells were then washed twice with ice-cold PBS, placed on ice, and lysed by the addition of 400 μl of lysis buffer (25 mM HEPES [pH 7.4], 300 mM NaCl, 1.5 mM MgCl2, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 0.1% Triton X-100, 1 mM okadaic acid, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of leupeptin per ml, and 20 μg of aprotinin per ml). Lysates were cleared by centrifugation (5 min at 14,000 × g, 4°C). The lysate was used for immunoprecipitation with 12CA5 at 4°C for 1 h. Protein A-Sepharose beads were added to the samples for 1 h at 4°C. Immunoprecipitates were washed three times with buffer A (PBS, 1% NP-40, 20 mM β-glycerophosphate, 2 mM sodium orthovanadate, 1 mM okadaic acid) and once with buffer B (50 mM morpholine propanesulfonic acid [pH 7.5], 500 mM lithium chloride, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM okadaic acid). To release endogenous 14-3-3 protein bound to HA-AFX, immunoprecipitates were washed three times with PBS containing 1 M MgCl2 where indicated. Controls were washed three times with PBS without added MgCl2.
The immunoprecipitates were resuspended in binding buffer (20 mM HEPES [pH 7.3], 150 mM potassium acetate, 2 mM magnesium acetate, 0.1% Tween 20, 28 mM β-mercaptoethanol, 0.05% ovalbumin, 1 mM sodium orthovanadate, 20 mM β-glycerophosphate, 1 mM okadaic acid, and 1 mM PMSF). Crm1-His6 was added to each assay to yield a final concentration of 500 nM. His6-RanQ69L was added as indicated to a final concentration of 3 μM. Samples were incubated for 2 h at 4°C and then were washed three times with binding buffer. Beads were resuspended in Laemmli sample buffer and the proteins were separated by SDS-PAGE and immunoblotted with HRP-conjugated 12CA5 (1:5,000 dilution), monoclonal anti-His6 (1:2,000 dilution; BabCo), or polyclonal anti-14-3-3β (1:100 dilution; Santa Cruz Biotechnology), and with HRP-conjugated goat anti-mouse or anti-rabbit secondary antibody (1:20,000 dilution; Jackson Laboratories). Proteins were revealed by chemiluminescence (Kirkegaard & Perry Laboratories).
Heterokaryon fusion assays.
BHK21 cells were transfected with pMT2-HA-A3. An acceptor cell line, GSN2, was a gift from Bryce Paschal (University of Virginia) (12). This stably transfected HeLa cell line expresses the nondiffusible nuclear protein GFP-streptavidin-SV40NLS. BHK21 and GSN2 cells were coplated onto poly-l-lysine-coated coverslips overnight. Cells were treated with 50 μM cycloheximide for 30 min. The plasma membranes were then fused for 2 min with 50% polyethylene glycol (molecular weight, 8,000) prewarmed to 37°C. Cells were washed four times with medium and were incubated at 37°C for an additional 1 h in the presence of cycloheximide. Cells were then fixed and processed for immunocytochemistry as described above.
RESULTS
PKB-dependent phosphorylation of AFX triggers relocalization of AFX from the nucleus to the cytoplasm.
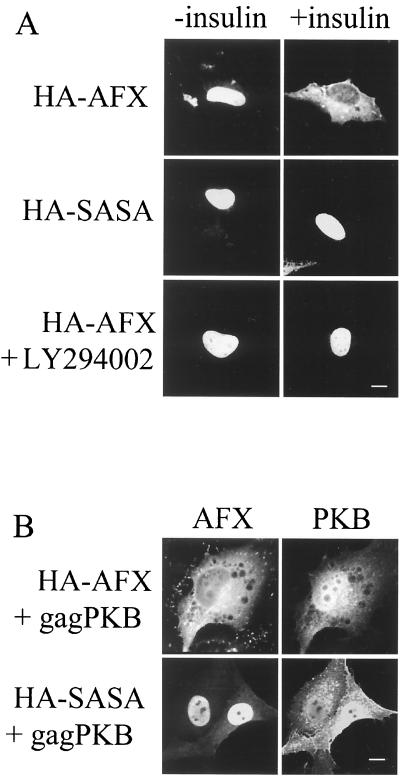
The subcellular distribution of Forkhead family members is altered on addition of insulin or serum to cells (9, 14, 71). This relocalization is dependent on the phosphorylation of the Forkhead protein by PKB. As a basis for subsequent experiments, we confirmed that the addition of insulin results in a change in the steady-state distribution of HA-AFX in serum starved cells from the nucleus to the cytoplasm within 30 min (Fig. 1A). Mutation of PKB phosphorylation sites S193 and S258 to alanine (HA-SASA) inhibited this relocalization, as did treatment with the phosphatidylinositol 3-kinase inhibitor LY294002 prior to the addition of insulin (Fig. 1A). Additionally, the coexpression of constitutively active PKB (gagPKB) resulted in the redistribution of HA-AFX but not HA-SASA to the cytoplasm (Fig. 1B). A significant fraction of gagPKB, however, is localized to the nucleus (Fig. 1B). This result is consistent with previous studies that have shown gagPKB localization at the plasma membrane (40%), in the cytoplasm (30%), and within the nucleus (30%) (1, 16). These results confirm that phosphorylation of AFX by PKB results in a rapid redistribution of AFX from the nucleus to the cytoplasm.
FIG. 1.
PKB-dependent phosphorylation of AFX triggers relocalization of AFX from the nucleus to the cytoplasm. (A) A14 cells were transfected with pMT2-HA-AFX or pMT2-HA-SASA. At 24 h posttransfection, serum was withdrawn for 18 to 24 h. Insulin (1 μg/ml) was then added as indicated, and these cells were incubated for 30 min. Cells treated with LY294002 (10 μM) to inhibit PIP3K were preincubated for 10 min prior to the addition of insulin. Cells were fixed, and then HA-AFX and HA-SASA were stained with 12CA5 and Texas red-conjugated secondary antibody. (B) A14 cells were transfected with pMT2-HA-AFX or pMT2-HA-SASA and with constitutively active PKB (pSG5-gagPKB). At 48 h posttransfection, the cells were fixed and HA-AFX and HA-SASA were stained as described for panel A. gagPKB was stained with anti-gag antiserum and fluorescein isothiocyanate-conjugated secondary antibody. Bars, 10 μm.
Nuclear export subsequent to phosphorylation by PKB in the nucleus is required for the inhibition of AFX transcriptional activity.
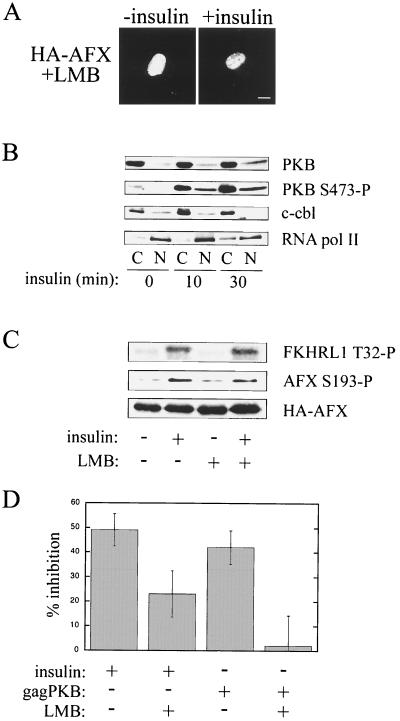
We wanted to test the hypothesis that PKB-dependent redistribution of AFX to the cytoplasm is required to regulate the transcriptional activity of the protein. The best-characterized nuclear export pathway uses a leucine-rich nuclear export signal (NES), which is bound by the export receptor Crm1 in the presence of RanGTP in the nucleus (25, 26, 43, 56, 68). The trimeric complex is then exported to the cytoplasm and is disassembled. Crm1-dependent export can be inhibited by LMB (74), which specifically inactivates Crm1 by covalent modification of a key cysteine residue in the NES-binding region of the protein (44). We tested, therefore, whether LMB could inhibit the cytoplasmic relocalization of HA-AFX in response to insulin (Fig. 2A) and serum (data not shown). As shown in Fig. 2A, LMB completely blocked the nuclear export of AFX. Thus, nuclear export of AFX in response to insulin or serum proceeds via a Crm1-dependent pathway.
FIG. 2.
Nuclear export subsequent to phosphorylation by PKB in the nucleus is required for the inhibition of AFX transcriptional activity. (A) A14 cells were transfected with pMT2-HA-AFX. At 24 h posttransfection, serum was withdrawn for 18 to 24 h. The cells were treated with LMB (2 ng/ml) for 30 min to inhibit Crm1-dependent export. Then insulin (1 μg/ml) was added as indicated, and the cells were incubated for an additional 30 min. Cells were fixed, and then HA-AFX was stained as described previously. Bar, 10 μm. (B) Serum-starved A14 cells were treated with insulin (1 μg/ml) and then fractionated at the indicated times. Equal amounts of nuclear (N) and cytoplasmic (C) lysates (50 μg) were analyzed by SDS-PAGE and Western blotting. An anti-PKB antibody was used to detect endogenous PKB protein levels. An anti-PKB S473-P antibody was used to detect activated PKB. c-cb1 and RNA pol II represent cytoplasmic and nuclear protein markers, respectively. (C) A14 cells were transfected and treated as described for panel A prior to cell lysis. Equal amounts of cellular lysate (50 μg) were analyzed by SDS-PAGE and Western blotting. An anti-pT32-specific antibody was used to detect endogenous FKHRL1 phosphorylated by PKB (14). Since A14 cells do not express endogenous AFX, an anti-pS193-specific antibody was used to detect HA-AFX phosphorylated by PKB. 12CA5 was used to visualize HA-AFX expression. (D) A14 cells were cotransfected with the 1205-luc reporter, pCMV-LacZ, pMT2-HA-AFX, and where indicated, pSG5-gagPKB. Cells were preincubated with LMB where noted for 30 min prior to the addition of insulin. Cells were then treated with insulin for 16 h in the absence or presence of LMB. Luciferase activity was measured 48 h after transfection, and luciferase levels were corrected for β-galactosidase expression. pMT2-HA-AFX-transfected control cells were serum starved, and the inhibition of AFX activity was normalized to 0% (i.e., 100% relative promoter activity). Data were obtained from five separate experiments.
Activated PKB may enter the nucleus to phosphorylate its target proteins (4, 50); however, this view is controversial. In support of the idea that PKB translocates into the nucleus, we have observed by subcellular fractionation that A14 cells show an increase in endogenous activated PKB within the nucleus 10 min after the addition of insulin (Fig. 2B; PKB S473-P). To examine this issue within the context of Forkhead transcription factors, we used the inhibition of AFX export by LMB to establish the cellular compartment in which this protein is phosphorylated by PKB. The addition of insulin to serum-starved cells expressing HA-AFX resulted in phosphorylation of S193 (Fig. 2C). Importantly, an equivalent level of S193 phosphorylation was observed when cells were treated with LMB prior to the addition of insulin. This result suggests that PKB can translocate into the nucleus in order to phosphorylate target proteins and that PKB-dependent phosphorylation of AFX occurs in the nucleus. This conclusion is supported by our results obtained for the PKB-mediated phosphorylation of endogenous FKHRL1 in the presence of LMB (Fig. 2C). FKHRL1 is another Forkhead family member that also has a steady-state nuclear localization in serum starved cells (14). T28 is a reported PKB-dependent phosphorylation site on FKHRL1 (14). This site was phosphorylated by PKB to similar levels upon the addition of insulin in either the absence or the presence of LMB pretreatment (Fig. 2C). We conclude that Forkhead family members can be phosphorylated by PKB in the nucleus, although we are unable to rule out the possibility that some phosphorylation of AFX by PKB may occur in the cytoplasm.
To test whether nuclear export is essential for the inhibitory effect of PKB on Forkhead transcriptional activity, we performed a luciferase reporter gene assay. Forkhead transcription factors can bind to and regulate the activity of the IGFBP-1 promoter in vitro. In cotransfection assays using a chloramphenicol acetyltransferase reporter under the control of the IGFBP-1 promoter, it was shown previously that Forkhead binding results in increased transcriptional activity and that insulin represses this activity (42). HA-AFX activity in serum-starved cells was normalized to 0% inhibition of transcriptional activity (i.e., the relative promoter activity was 100%). PKB phosphorylation of HA-AFX resulting from the addition of insulin or coexpression of gagPKB inhibited the reporter gene activity by ∼50% (Fig. 2D). Pretreatment with LMB to inhibit HA-AFX export, however, attenuated the effect of insulin to approximately 20% and abolished the effect of gagPKB on AFX activity. Since LMB does not affect the ability of PKB to phosphorylate AFX (Fig. 2C), the effect of LMB cannot be explained by a lack of PKB phosphorylation. Nor can it be explained by a global effect on the cellular transcription machinery, since LMB has no effect on the transcriptional activity of AFX mutants, such as HA-SASA and HA-A3 (see below), that do not relocalize to the cytoplasm on insulin treatment or the cotransfection of gagPKB (data not shown). The reduced effect of LMB observed in insulin-treated cells most likely is due to the activation of other identified signaling pathways that regulate AFX activity independent of cellular localization (42). The complete inhibition observed in gagPKB cotransfected cells suggests that PKB-dependent regulation of AFX activity requires the relocalization of the protein to the cytoplasm. We conclude, therefore, that both PKB phosphorylation in the nucleus and nuclear export mediated by Crm1 are essential for the full inhibition of AFX transcriptional activity. These data strongly suggest that a major component of the transcriptional regulation of AFX occurs at the level of nuclear transport.
Regulation of localization of AFX by PKB does not occur at the level of export.
There are several distinct mechanisms by which PKB-dependent phosphorylation of AFX could trigger cytoplasmic accumulation of the protein. First, PKB phosphorylation could facilitate nuclear export by promoting the binding of Crm1 to AFX. In a second model, AFX may be exported from the nucleus constitutively but be reimported rapidly when it is unphosphorylated, so that its steady-state distribution is predominantly nuclear. PKB phosphorylation would then inhibit or decrease the rate of import, leading to a redistribution to the cytoplasm. Third, both nuclear import and export of AFX could be constitutive, and phosphorylation would permit cytoplasmic retention by increasing the affinity of AFX for a cytoplasmic anchor protein. Finally, phosphorylation by PKB could release AFX from a nuclear anchor.
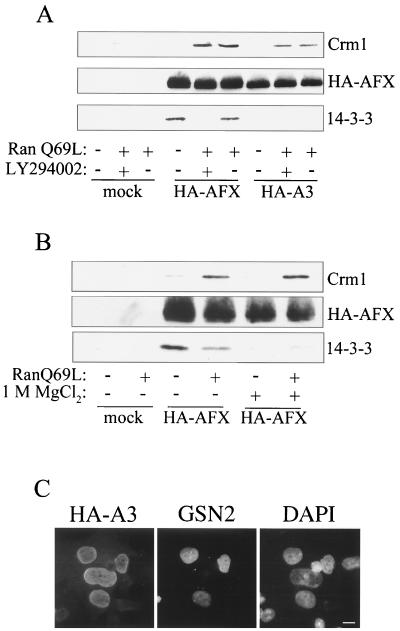
To distinguish between these possibilities, we performed Crm1 binding assays. AFX cannot be expressed in appreciable amounts as a recombinant protein in Escherichia coli. Therefore, HA-AFX and HA-A3 (HA-AFX with the following mutations: T28A, S193A, and S258A) were immunoprecipitated from mammalian cell lysates and incubated with recombinant Crm1 in the absence or presence of RanQ69L, a mutant of Ran locked in its GTP bound conformation. RanQ69L increases the affinity of Crm1 for its export cargoes. Interestingly, both PKB-phosphorylated (HA-AFX) and -unphosphorylated (HA-AFX plus LY294002 and HA-A3) AFX bound to Crm1 in the presence of RanQ69L (Fig. 3A). These data demonstrate that binding of Crm1 to AFX is independent of phosphorylation by PKB.
FIG. 3.
Both phosphorylated and unphosphorylated AFXs bind Crm1 and are exported from the nucleus. (A) HEK293 cells were transfected with either pMT2-HA-AFX or pMT2-HA-A3 (T28A, S193A, S258A), incubated in the presence of serum for 48 h, and where indicated, incubated with LY294002 (10 μM) for 2 h prior to cell lysis. The HA-tagged proteins were immunoprecipitated with 12CA5 and immobilized on protein A-Sepharose. The beads were then incubated with recombinant Crm1 (500 nM) in the absence or presence of RanQ69L (3 μM), a Ran mutant locked in the GTP bound conformation. Proteins that bound to AFX were analyzed by SDS-PAGE and Western blotting. An anti-His6 antibody was used to visualize Crm1, which possesses a carboxy-terminal His6 tag. Directly conjugated HRP-12CA5 was used to assess HA-AFX and HA-A3 immunoprecipitation. An anti-14-3-3 β antibody was used to detect 14-3-3 binding. (B) HEK293 cells were transfected with pMT2-HA-AFX and were treated as described for panel A. Prior to incubation with Crm1 and RanQ69L, immobilized HA-AFX was washed with 1 M MgCl2 where indicated to remove bound 14-3-3 proteins. Proteins that bound AFX were analyzed as described for panel A. (C) Heterotypic cell fusions were performed between BHK21 cells transiently transfected with pMT2-HA-A3 and a stably transfected HeLa cell line expressing GFP-streptavidin-NLS (GSN2). Cell cultures were trypsinized 24 h after transfection, mixed, allowed to adhere to coverslips overnight, and then fused by using polyethylene glycol. After incubation for 1 h with cycloheximide (50 μM), cells were fixed and stained for HA-A3 with 12CA5 and a Texas red-conjugated secondary antibody. Nuclei were visualized by staining the DNA with DAPI. Bar, 10 μm.
To confirm that HA-AFX was phosphorylated in this experiment, we examined its interaction with 14-3-3. The 14-3-3 proteins are a family of proteins reported to bind Forkhead transcription factors in a phosphorylation-dependent manner (14). As shown in Fig. 3A, endogenous 14-3-3 coimmunoprecipitated with HA-AFX from cells grown in the presence of serum, thereby confirming that phosphorylation of HA-AFX by PKB had occurred (Fig. 3A). These data raised the possibility that Crm1 binding to phosphorylated AFX is indirect and is mediated by 14-3-3. It has been reported previously that 14-3-3 contains a leucine-rich NES-like motif (63). It is not clear, however, that this sequence can function as an NES, especially when binding proteins are associated with 14-3-3. To test this hypothesis, we stripped 14-3-3 proteins from AFX using 1 M MgCl2. This treatment substantially decreased the amount of 14-3-3 associated with AFX but did not reduce the amount of Crm1-RanQ69L that was bound (Fig. 3B). It should be noted that although 14-3-3 proteins have been reported to bind Crm1 in the presence of mammalian cell lysates (63), we have been unable to detect a direct interaction between recombinant 14-3-3ζ, Crm1, and RanQ69L (data not shown).
The observation that Crm1 binding to AFX is independent of PKB phosphorylation is supported by the results of heterokaryon fusion assays. HA-A3 has a steady-state nuclear localization but nonetheless may actively shuttle into and out of the nucleus. Nucleocytoplasmic shuttling can be observed by the use of cell fusion assays in which the donor and acceptor nuclei of the fused cells can be distinguished. Redistribution of a tagged protein from one nucleus to another can only occur if the protein exits the donor nucleus and is reimported into the acceptor nucleus. To perform heterokaryon fusion assays, we used BHK21 cells expressing HA-A3 as the donor and a reporter cell line, GSN2, that expresses GFP-streptavidin-NLS as the acceptor. The GFP-streptavidin-NLS fusion protein, which is constitutively nuclear and does not shuttle, acts as a marker for the acceptor nuclei. HA-A3 shuttling would lead to its equilibration into the nucleus of a fused GSN2 cell. On the other hand, if HA-A3 does not shuttle, no HA-A3 would be detected in the nuclei of the GSN2 cells. Figure 3C shows an AFX donor nucleus surrounded by three GSN2 acceptor nuclei that also stained positive for HA-A3. Therefore, even though unphosphorylated AFX is predominantly nuclear in its steady-state distribution, it is constitutively shuttling into and out of the cytoplasm. AFX phosphorylation by PKB, however, results in a shift in the steady-state distribution of AFX to the cytoplasm. Taken together these results suggest that the regulation of AFX localization in response to PKB phosphorylation occurs not at the level of nuclear export or nuclear retention but instead at the level of nuclear import or through cytoplasmic retention of phosphorylated AFX.
AFX import into the nucleus proceeds via an active, Ran-dependent mechanism.
On account of the results described above, it was important to identify the mechanism by which AFX is imported into the nucleus. In principle, AFX could accumulate in the nucleus either by an active mechanism or by diffusion followed by nuclear retention. Most active nuclear transport pathways studied to date require energy and an intact Ran gradient across the nuclear envelope. To determine whether the nuclear accumulation of AFX observed in serum-starved cells proceeds via an active, Ran-dependent mechanism, we used tsBN2 cells. These cells have a temperature-sensitive RanGEF (RCC1) allele (70). When grown at the permissive temperature (33.5°C), tsBN2 cells have an intact Ran gradient. However, incubation at the nonpermissive temperature (39.5°C) causes collapse of the Ran gradient and prevents Ran-dependent nuclear transport. As we have shown previously, incubation for 3 h at 39.5°C in the presence of cycloheximide (to prevent new RanGEF synthesis) completely inhibits the import of other substrates known to utilize a classical import pathway, such as the glucocorticoid receptor (62).
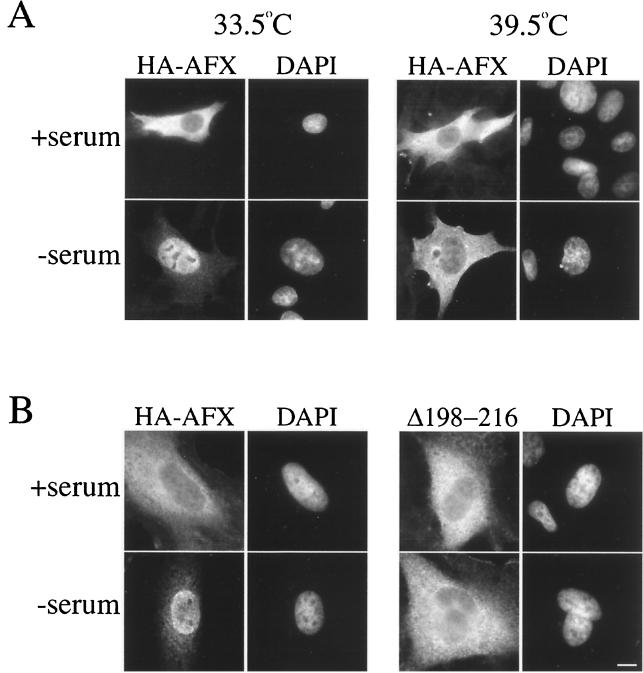
To test the Ran dependence of AFX import, we transfected tsBN2 cells with a plasmid encoding HA-AFX. In the presence of cycloheximide, cells were grown at either 33.5°C or were shifted to 39.5°C for 3 h. The cells were then serum starved. Immunofluorescence revealed that at the permissive temperature HA-AFX entered the nucleus on serum starvation (Fig. 4A, 33.5°C). In cells incubated at 39.5°C, however, HA-AFX did not accumulate in the nucleus (Fig. 4A, 39.5°C). We conclude, therefore, that import of AFX into the nucleus proceeds via an active, Ran-dependent mechanism.
FIG. 4.
AFX import into the nucleus requires a Ran gradient and basic residues downstream from S193. (A) tsBN2 cells were transfected with pMT2-HA-AFX and were incubated at 33.5°C in the presence of serum for 48 h. Cycloheximide (50 μM) was then added to the medium to prevent new protein synthesis, and the cells were incubated at either 33.5°C (right) or 39.5°C (left) for 3 h. Where indicated, the serum was then withdrawn and the cells were returned to 33.5°C or 39.5°C for 1.5 h. Cells were fixed and HA-A3 was stained as described previously. (B) HeLa cells were transfected with pMT2-HA-AFX or pMT2-HA-AFX(Δ198–216) and were incubated in the presence of serum for 48 h. Serum was then withdrawn for 1.5 h. Cells were fixed, and HA-AFX and HA-AFX(Δ198–216) were stained as before. Nuclei were visualized by staining the DNA with DAPI. Bar, 10 μm.
Basic residues on both sides of S193 are necessary and sufficient for nuclear import of AFX.
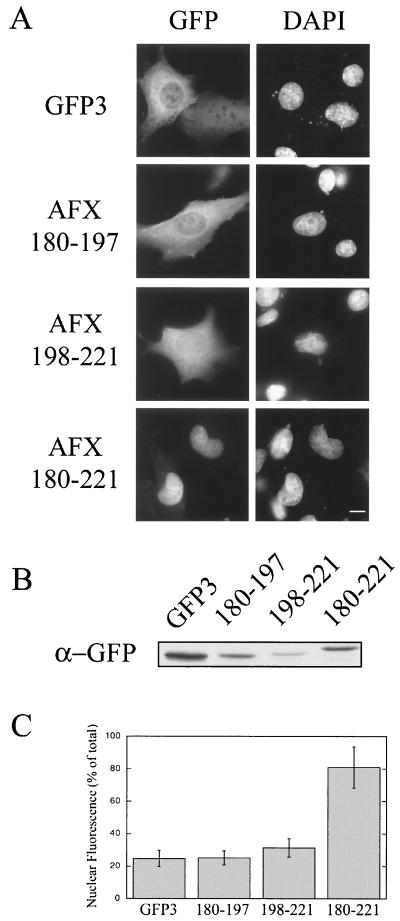
To determine whether PKB phosphorylation regulates the nuclear import of AFX, we needed to identify the NLS within the AFX sequence. Clusters of basic residues are often predictive that a region of a protein may act as an NLS. AFX contains two clusters of basic residues flanking the PKB phosphorylation site S193. Therefore, deletion of these clusters would test whether they are required for AFX nuclear import. Deletion of residues 198 to 216 from HA-AFX that are C terminal to S193 results in a protein that is unable to enter the nucleus under the same conditions of serum starvation that result in HA-AFX accumulation in the nucleus (Fig. 4B). Thus, this region is necessary for nuclear import of AFX. To test whether this region is sufficient to mediate import, we expressed residues 198 to 221 as an amino-terminal fusion to GFP-GFP-GFP [AFX(198-221)-GFP3] in mammalian cells. The triple GFP construct (∼80 kDa) was used to prevent passive diffusion through the nuclear pore complex (NPC) which has a diffusion limit of ∼60 kDa. (These fusion proteins could not be expressed in E. coli, because the NLS is cleaved stoichiometrically during the induction of expression under all conditions tested). As shown in Fig. 5A, AFX(198-221)-GFP3 did not accumulate in the nucleus. The basic region upstream of S193 was also not able to act as an NLS in isolation [Fig. 5; AFX(180–197)-GFP3]. However, a construct containing both basic regions accumulated efficiently in the nucleus [AFX(180–221)-GFP3]. Expression of these constructs as carboxy-terminal fusions to GFP-GFP-GFP resulted in the same localization (data not shown). We conclude, therefore, that residues 180 to 221 of AFX constitute an NLS and that basic residues on both sides of S193 are required for the nuclear import of AFX.
FIG. 5.
Basic residues on both sides of S193 are sufficient for nuclear import. Basic residues on either side of PKB phosphorylation site S193 were expressed as N-terminal fusions to GFP-GFP-GFP (GFP3). pKGFP3, pKGFP3-AFX(180–197), pKGFP3-AFX(198–221), and pKGFP3-AFX(180–221) were transfected into BHK21 cells. At 18 h posttransfection, the cells were fixed and the DNA was stained with DAPI (A) or the cells were lysed and analyzed by SDS-PAGE (10 μl of lysate/lane) and immunoblotting using an anti-GFP antibody (B). (C) The relative nuclear and cytoplasmic fluorescence levels of the constructs were obtained using Openlab (Improvision). Nuclear fluorescence was calculated as a percentage of the total cellular fluorescence corrected for the background fluorescence. Each data point represents the mean fluorescence obtained from 12 randomly chosen cells. Error is expressed as standard deviations of the means. Bar, 10 μm.
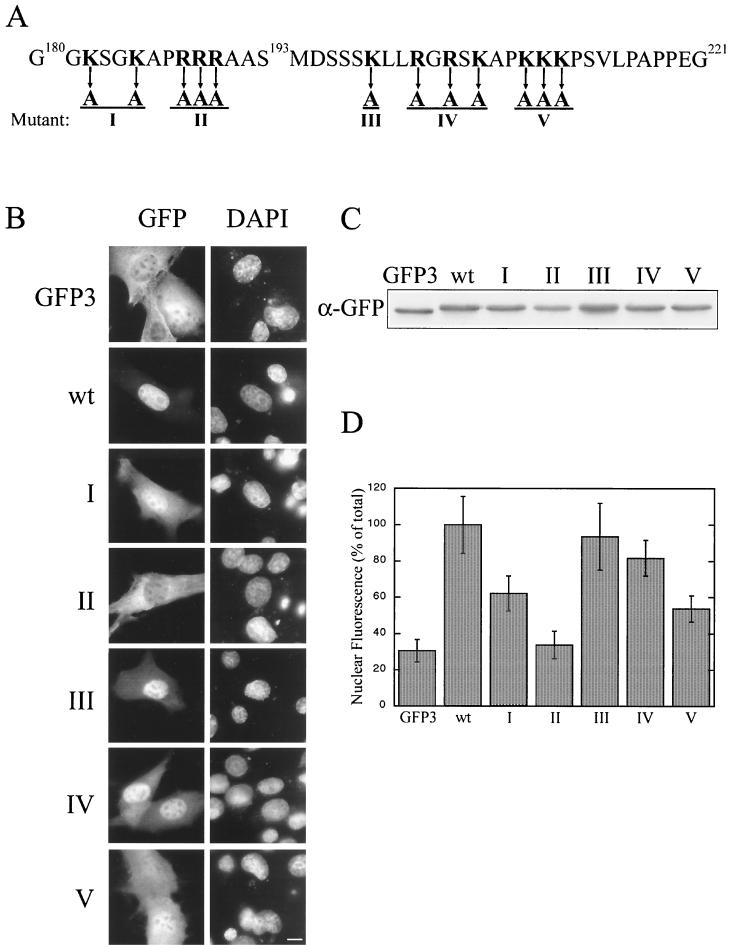
Although the 180 to 221 region contains 12 lysine and arginine residues, it does not comprise a classical monopartite or bipartite NLS. In addition, binding of the classical import receptor importin α1, importin α3, or importin β to this region of the protein could not be detected (data not shown). Therefore, we produced a series of mutations in AFX(180–221)-GFP3 to identify residues required for import (Fig. 6). Quantification of relative nuclear and cytoplasmic fluorescence levels demonstrated that mutation of arginines 188 to 190 to alanine had the most deleterious effect on import (Fig. 6, mutant II), followed by the mutation of lysines 209 and 210 (Fig. 6, mutant V). This result reinforces the requirement for basic residues both upstream and downstream of S193 for nuclear import and confirms that the sequence is an atypical NLS.
FIG. 6.
Basic residues on both sides of S193 are required for nuclear import, and the sequence is not a classical NLS motif. (A) There are 12 lysine and arginine residues that surround S193. Five constructs with mutations of basic residues within AFX(180–221)-GFP were made (I through V). pKGFP3, pKGFP-AFX(180–221), pKGFP3-AFX(180–221)-I, pKGFP3(180–221)-II, pKGFP3-AFX(180–221)-III, pKGFP3-AFX(180–221)-IV, and pKGFP-AFX(180–221)-V were transfected into BHK21 cells. At 18 h posttransfection, the cells were either fixed (B) or lysed (C) and analyzed as described for Fig. 5 (D). Bar, 10 μm. wt, wild type.
Phosphorylation of S193 reduces the rate of nuclear import.
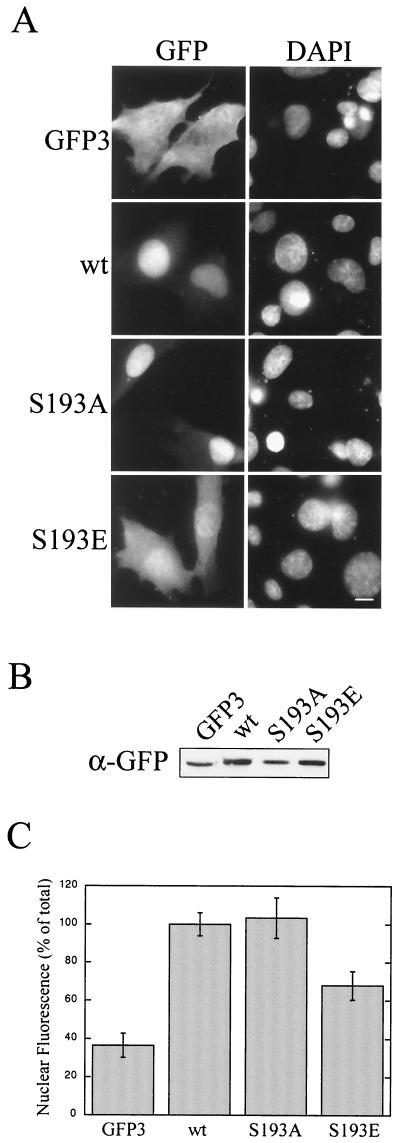
Phosphorylation of S193 adds a negative charge to a positively charged region of AFX. This may affect the binding of AFX to its nuclear import receptor and reduce its rate of import. To test this hypothesis, we created two AFX(180–221)-GFP3 mutants: S193A, which cannot be phosphorylated, and S193E, which may mimic phosphorylation at this site. Nuclear import of S193A was comparable to that of wild-type AFX(180–221)-GFP3 (Fig. 7A and C). This demonstrates that mutation of S193 results in a stable fusion protein that is able to enter the nucleus. The S193E mutation, however, substantially reduced nuclear accumulation (Fig. 7A and C). This result suggests that PKB phosphorylation of S193 reduces the rate of nuclear import of AFX. It is important to note that these GFP3 constructs do not contain an NES. Therefore, over a 20-h transfection period, even with a greatly reduced rate of import some nuclear accumulation would be expected, since the construct cannot be reexported. Therefore, a phosphorylation-dependent reduction in the import rate of AFX, combined with constitutive nuclear export, would shift the steady-state distribution of the protein from the nucleus to the cytoplasm. This mechanism is consistent with the known redistribution of AFX to the cytoplasm on addition of insulin or serum.
FIG. 7.
Phosphorylation of S193 reduces the rate of nuclear import. Two AFX(180–221)-GFP3 mutants were created, S193A and S193E. pKGFP3, pKGFP3-AFX(180–221), pKGFP3-AFX(180–221)S193A, and pKGFP3-AFX(180–221)S193E were transfected into BHK21 cells. At 18 h posttransfection, the cells were either fixed (A) or lysed (B) and analyzed as described for Fig. 5 (C). Bar, 10 μm. wt, wild type.
DISCUSSION
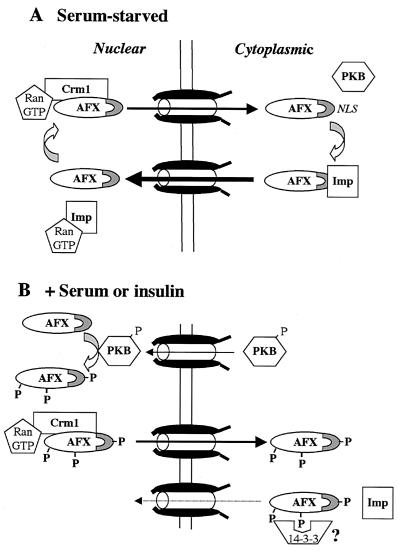
Nuclear transport of the Forkhead transcription factor, AFX, plays a critical role in the regulation of its transcriptional activity. Phosphorylation of AFX by PKB results in a rapid change in the steady-state distribution of the protein from the nucleus to the cytoplasm. However, the mechanism by which this relocalization occurs has not been determined, although it has been proposed by others that export may be regulated by PKB and that 14-3-3 may function as a chaperone for export, as has also been suggested for the regulation of cdc25 export (19). Based on the data presented in this paper, we propose a different model, in which nuclear import, not export, of AFX is the principal target of regulation by PKB (Fig. 8).
FIG. 8.
Nuclear import, not export, of AFX is regulated by PKB. (A) Unphosphorylated AFX appears nuclear at steady state but is actually shuttling rapidly between the nucleus and the cytoplasm. Therefore, the rate of import of unphosphorylated AFX exceeds its rate of export. Nuclear export of both phosphorylated and unphosphorylated AFX is likely mediated by the exportin, Crm1, since both HA-AFX and HA-A3 bind Crm1 in the presence of RanGTP. Imp, importin. (B) The addition of insulin to cells activates PKB and causes it to translocate into the nucleus, where it phosphorylates Forkhead family members. Phosphorylated AFX exits the nucleus by binding Crm1. Phosphorylation of AFX at S193 attenuates nuclear import, perhaps by reducing the affinity of AFX for its nuclear import receptor. Therefore, phosphorylation by PKB decreases the import rate constant without altering the export rate constant. Since the steady-state localization of a protein is determined by its relative rates of import and export, the localization of AFX would shift from the nucleus to the cytoplasm as observed. This alteration in transport rates in conjunction with proposed cytoplasmic retention by binding 14-3-3 proteins would result in exclusion of AFX from the nucleus. Since AFX requires nuclear localization to carry out transcription, this mechanism of regulated transport would inhibit the activity of AFX in response to phosphorylation by PKB.
Unphosphorylated AFX is predominantly nuclear at steady state. However, heterokaryon fusion assays (Fig. 3C) demonstrated that the protein is, in fact, continually shuttling between the nucleus and the cytoplasm. The rate constant for unphosphorylated AFX import, therefore, must exceed the rate constant for its export (Fig. 8A). Importantly, the nuclear export of AFX appears to be unaltered by PKB phosphorylation. Nuclear export of both phosphorylated and unphosphorylated AFXs is likely mediated by the export receptor, Crm1. Both wild-type HA-AFX and the triple mutant, HA-A3, which cannot be phosphorylated by PKB, bind Crm1 in the presence of RanGTP (Fig. 3A). In support of the identification of Crm1 as the export receptor, we show that AFX export from the nucleus in response to insulin is blocked by the addition of LMB (Fig. 2A), a potent and specific inhibitor of Crm1 function.
The addition of insulin to cells results in the activation of PKB and its translocation into the nucleus, where it phosphorylates Forkhead family members (Fig. 8B). Treatment of cells with LMB traps AFX and FKHRL1 in the nucleus. However, both are phosphorylated in response to insulin to the same extent and at the same residues as in cells not treated with LMB (Fig. 2B). This result, therefore, strongly indicates that PKB enters the nucleus when activated by insulin to phosphorylate Forkhead transcription factors and other target proteins. Phosphorylation alone, however, is not sufficient to inhibit the transcriptional activity of AFX. We have shown, using transcriptional activation assays, that blockade of AFX nuclear export by the addition of LMB results in a loss of insulin-induced transcriptional control (Fig. 2B). Therefore, both phosphorylation by PKB and nuclear export mediated by Crm1 are essential for complete inhibition of AFX transcriptional activity.
We have found that AFX enters the nucleus by an active, Ran-dependent mechanism (Fig. 4A) and that import requires a basic region in AFX encompassing PKB phosphorylation site S193 (Fig. 5). Importantly, the addition of a negative charge at S193 substantially attenuates nuclear import (Fig. 7), most likely by reducing the affinity of AFX for its nuclear import receptor. We propose, therefore, that phosphorylation by PKB at S193 reduces the rate of AFX import. Since the steady-state localization of a protein is determined by its relative rate constants for import and export, the localization of AFX would shift from the nucleus to the cytoplasm (Fig. 8B). This alteration in the transport rate constants in response to phosphorylation by PKB, in conjunction with proposed cytoplasmic retention by binding 14-3-3 proteins (14), would result in both the efficient nuclear exclusion of AFX and the inhibition of its transcriptional activity. In conclusion, we propose that this nuclear exclusion mechanism is required to regulate the activity of AFX.
These data are consistent with the results of transcriptional activation assays reported previously for FKHR (30). The PKB phosphorylation site S256 in FKHR is analogous to S193 in AFX. Mutation of S256 to an alanine in the context of full-length FKHR abolished the ability of insulin and PKB to inhibit FKHR activity. In contrast, mutation of S256 to an aspartate resulted in a substantial inhibition of its transcriptional activity. These effects were not observed when the other two PKB sites were mutated independently. Although the localization of the S256D mutant was not assayed, our data suggest that the inhibition of FKHR activity may be due, in part, to a deficit in nuclear import.
“Classical” nuclear localization sequences are characterized by short amino acid stretches that are enriched in basic amino acids. The NLS of the large T antigen of simian virus 40 was identified by deletion analysis that resulted in mislocalization of the protein to the cytoplasm (45). It was later defined as a seven-amino-acid sequence (PKKKRKV) sufficient to confer nuclear localization when conjugated to a carrier protein (46). Some proteins contain similar sequences that are referred to as monopartite NLSs. Other proteins, such as nucleoplasmin, contain bipartite NLSs that consist of two patches of basic residues separated by a 10-amino-acid spacer (23). Generally, proteins carrying classical or bipartite NLSs bind a cytoplasmic receptor, importin α (28, 48). Importin α associates with importin β, a protein that docks import complexes at the NPC and translocates import cargo into the nucleus. There are, however, many exceptions to this type of import, and there is a large family of importins and exportins that can recognize distinct NLSs and mediate transport of different subsets of cargo. For example, ribosomal proteins (33) and histones (32) have been shown to bind directly to several different importin β family members and dock at the NPC independently of importin α. In addition, other proteins, such as hnRNP K (51) and β-catenin (75), can translocate through the NPC in the absence of any soluble factors.
We have delineated residues 180 to 221 of AFX as a novel type of nuclear import signal. This region contains a small portion of the DNA binding domain. Therefore, like other Forkhead family members, the DNA binding domain of AFX contributes to DNA binding and nuclear localization (59). This region of AFX contains 12 lysine and arginine residues. Although several groups of these basic residues could act as monopartite or bipartite NLSs, the information provided by mutational analysis suggests that AFX contains a nonclassical NLS. Mutation of arginines 188 to 190 in the NLS of AFX has the greatest inhibitory effect on nuclear import, followed by the mutation of lysines 209 to 211. These groups of basic residues are separated by 18 amino acids, and both are contained within similar sequence repeats (KAPRRR and KAPKKK). These repeats may be important for import receptor binding.
The identity of the nuclear import receptor of AFX remains to be established. Since we do not observe binding of AFX to several classical import receptors, including importins α1, α3, and β, it may bind to a novel member of the importin family (which comprises >20 members in mammalian cells). Alternatively, AFX may enter the nucleus in other ways, for instance, by piggybacking on another protein that contains a classical NLS. The role of PKB-mediated phosphorylation and 14-3-3 binding in altering AFX import can be more thoroughly studied once the receptor is identified.
Binding of Forkhead family members to 14-3-3 proteins has been proposed to play a role in the retention of phosphorylated Forkheads in the cytoplasm (14). Phosphorylated FKHRL1 (14) and AFX (Fig. 3A) both bind 14-3-3 proteins. However, we have never observed any specificity of AFX binding to a particular 14-3-3 isoform. In fact, we have observed binding to 14-3-3β, 14-3-3ɛ, and 14-3-3ζ (data not shown). Recently, it has been proposed that 14-3-3 protein binding may impart no specific information about subcellular targeting (53). Instead, a 14-3-3 protein may obscure the NLS or NES of the protein to which it binds. In this way, 14-3-3 proteins would affect the subcellular localization of their target proteins by interfering with the binding of transport receptors. 14-3-3 binding to Forkhead family members may sterically inhibit import receptor binding and, thereby, act to prevent import.
The proposal has also been made that 14-3-3 possesses an NES that is recognized by Crm1 and participates directly in the export of binding partners through a Crm1 interaction (63). In our hands, however, 14-3-3 does not bind Crm1 directly, nor is its association with AFX necessary for the interaction of AFX with Crm1. Therefore, in this case at least, 14-3-3 isoforms appear to play no part in regulating AFX nuclear export.
What is the AFX NES? Leucine-rich regions found in several Forkhead family members conform to the consensus sequence for a Crm1-dependent NES (9). This region in AFX corresponds to residues 300 to 308 (LELLDGLNL). Deletion of residues 298 to 308 results in a protein that is unable to relocalize from the nucleus to the cytoplasm on stimulation with insulin (unpublished data). This region, therefore, likely represents the Crm1-dependent NES.
Distinct chromosomal translocations in pediatric alveolar rhabdomyosarcoma and acute lymphoblastic leukemia (ALL) involve two human Forkhead genes. Alveolar rhabdomyosarcomas are associated with unique chromosomal translocations that arise from fusion of PAX3 or PAX7 to the FKHR gene (27). The PAX3-FKHR fusion protein is a more potent transcriptional activator than PAX3 alone (8) and transforms primary cells (64). Several chromosomal translocations in ALL occur at the 11q23 locus, and all convey a poor prognosis that is usually associated with a high rate of treatment failure and relapse. These breakpoints affect the MLL gene (also called HRX, ALL, or Htrx1) that is disrupted midway through the coding region (18, 77). A well-documented translocation arises from the fusion of MLL to the AFX gene on chromosome X (13).
Importantly, the fusions of AFX and FKHR to their breakpoint partners occur at identical amino acid positions within the Forkhead proteins. The resulting fusion proteins contain the N-terminal DNA binding region of the fusion partner and the C-terminal transactivation domain of the Forkhead protein. The MLL-AFX fusion protein expresses residues 148 to 501 of AFX (57). This preserves the NLS of AFX identified in this study (residues 180 to 221) but deletes the T28 phosphorylation site. We have shown that deletion of residues N terminal to S193 causes a loss of transcriptional regulation by insulin (42). In addition, potential loss of S193 phosphorylation in the context of the fusion protein would allow constitutive import into the nucleus. Therefore, both the deregulation of Forkhead activity and a constitutive nuclear localization likely contribute to the oncogenic properties of these fusion proteins.
ACKNOWLEDGMENTS
We thank all members of our laboratories for their continued advice throughout the experimental and preparatory phases of the manuscript. In particular, we thank Mark Lindsay, Michael Nemergut, Kendra Plafker, Scott Plafker, Alicia Smith, Katie Welch, Nancy de Ruiter, and Hans Bos. We also thank Iain Mattaj and Dirk Görlich for their generosity in providing expression plasmids used in this study (Crm1, RanQ69L), Barbara Wolff for her gift of LMB, Anne Brunet for the anti-phospho T32 FKHRL1 antibody, Kris Reedquist for the c-cbl antibody, Marc Timmers for the RNA pol II antibody, and Bryce Paschal for the GSN2 cell line.
This work was supported by a grant awarded to I.G.M. from the National Institutes of Health, DHHS (GM-50526). A.M.B. is supported by a Postdoctoral National Service Award from the National Institutes of Health, DHHS (GM-20017). G.K. and B.B. are supported by grants from NWO and the Dutch Cancer Foundation (KWF).
REFERENCES
- 1.Ahmed N N, Franke T F, Bellacosa A, Datta K, Gonzalez-Portal M E, Taguchi T, Testa J R, Tsichlis P N. The proteins encoded by c-akt and v-akt differ in post-translational modification, subcellular localization and oncogenic potential. Oncogene. 1993;8:1957–1963. [PubMed] [Google Scholar]
- 2.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B α. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 3.Anderson M J, Viars C S, Czekay S, Cavenee W K, Arden K C. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 4.Andjelkovic M, Alessi D R, Meier R, Fernandez A, Lamb N J, Frech M, Cron P, Cohen P, Lucocq J M, Hemmings B A. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 5.Arts G J, Fornerod M, Mattaj I W. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 6.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Levy R, Hooper S, Wilson R, Paterson H F, Marshall C J. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr Biol. 1998;8:1049–1057. doi: 10.1016/s0960-9822(98)70442-7. [DOI] [PubMed] [Google Scholar]
- 8.Bennicelli J L, Edwards R H, Barr F G. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1996;93:5455–5459. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biggs W H, Meisenhelder J, Hunter T, Cavenee W K, Arden K C. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischoff F R, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischoff F R, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 12.Black B E, Levesque L, Holaska J M, Wood T C, Paschal B M. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol Cell Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borkhardt A, Repp R, Haas O A, Leis T, Harbott J, Kreuder J, Hammermann J, Henn T, Lampert F. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- 14.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 15.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J Q, Altomare D A, Klein M A, Lee W C, Kruh G D, Lissy N A, Testa J R. Transforming activity and mitosis-related expression of the AKT2 oncogene: evidence suggesting a link between cell cycle regulation and oncogenesis. Oncogene. 1997;14:2793–2801. doi: 10.1038/sj.onc.1201121. [DOI] [PubMed] [Google Scholar]
- 17.Coffer P J, Woodgett J R. Molecular cloning and characterisation of a novel putative protein-serine kinase related to the cAMP-dependent and protein kinase C families. Eur J Biochem. 1991;201:475–481. doi: 10.1111/j.1432-1033.1991.tb16305.x. [DOI] [PubMed] [Google Scholar]
- 18.Corral J, Forster A, Thompson S, Lampert F, Kaneko Y, Slater R, Kroes W G, van der Schoot C E, Ludwig W D, Karpas A, et al. Acute leukemias of different lineages have similar MLL gene fusions encoding related chimeric proteins resulting from chromosomal translocation. Proc Natl Acad Sci USA. 1993;90:8538–8542. doi: 10.1073/pnas.90.18.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta S R, Brunet A, Greenberg M E. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 20.DeVit M J, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 21.Devit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dijkers P F, Medemadagger R H, Lammers J J, Koenderman L, Coffer P J. Expression of the pro-apoptotic bcl-2 family member bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 23.Dingwall C, Sharnick S V, Laskey R A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 24.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 25.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Crm1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Crm1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 27.Galili N, Davis R J, Fredericks W J, Mukhopadhyay S, Rauscher F J, Emanuel B S, Rovera G, Barr F G. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 28.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 29.Görlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 30.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 31.Izaurralde E, Kutay U, Vonkobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakel S, Albig W, Kutay U, Bischoff F R, Schwamborn K, Doenecke D, Görlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakel S, Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P F, Jakubowicz T, Pitossi F J, Maurer F, Hemmings B A. Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc Natl Acad Sci USA. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaffman A, O'Shea E K. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 36.Kaffman A, Rank N M, O'Neill E M, Huang L S, O'Shea E K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 37.Kaffman A, Rank N M, O'Shea E K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 39.Kehlenbach R H, Dickmans A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of Crm1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 41.Komeili A, O'Shea E K. Nuclear transport and transcription. Curr Opin Cell Biol. 2000;12:355–360. doi: 10.1016/s0955-0674(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 42.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 43.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian Crm1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 44.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates Crm1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanford R E, Butel J S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- 46.Lanford R E, Kanda P, Kennedy R C. Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal. Cell. 1986;46:575–582. doi: 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- 47.Lee K A, Bindereif A, Green M R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 48.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 49.Medema R H, Kops G J, Bos J L, Burgering B M. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 50.Meier R, Alessi D R, Cron P, Andjelkovic M, Hemmings B A. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase B β. J Biol Chem. 1997;272:30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 51.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moll U M, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci USA. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muslin A J, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 54.Nemergut M E, Macara I G. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol. 2000;149:835–850. doi: 10.1083/jcb.149.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. The Forkhead transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 56.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of Crm1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 57.Parry P, Wei Y, Evans G. Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer. 1994;11:79–84. doi: 10.1002/gcc.2870110203. [DOI] [PubMed] [Google Scholar]
- 58.Plafker K, Macara I G. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol Cell Biol. 2000;20:3510–3521. doi: 10.1128/mcb.20.10.3510-3521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian X, Costa R H. Analysis of hepatocyte nuclear factor-3 β protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 1995;23:1184–1191. doi: 10.1093/nar/23.7.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rameh L E, Cantley L C. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 61.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 62.Richards S A, Carey K L, Macara I G. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 63.Rittinger K, Budman J, Xu J, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 64.Scheidler S, Fredericks W J, Rauscher F J, Barr F G, Vogt P K. The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci USA. 1996;93:9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlamp C L, Poulsen G L, Nork T M, Nickells R W. Nuclear exclusion of wild-type p53 in immortalized human retinoblastoma cells. J Natl Cancer Inst. 1997;89:1530–1536. doi: 10.1093/jnci/89.20.1530. [DOI] [PubMed] [Google Scholar]
- 66.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Interleukin 3-dependent survival by the Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staal S P. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc Natl Acad Sci USA. 1987;84:5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 69.Stokoe D, Stephens L R, Copeland T, Gaffney P R, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 70.Tachibana T, Imamoto N, Seino H, Nishimoto T, Yoneda Y. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J Biol Chem. 1994;269:24542–24545. [PubMed] [Google Scholar]
- 71.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, Kitamura T, Ogawa W, Kasuga M, Kikkawa U, Nishizuka Y. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA. 1999;96:11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walker K S, Deak M, Paterson A, Hudson K, Cohen P, Alessi D R. Activation of protein kinase B β and γ isoforms by insulin in vivo and by 3-phosphoinositide-dependent protein kinase-1 in vitro: comparison with protein kinase B α. Biochem J. 1998;331:299–308. doi: 10.1042/bj3310299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welch K, Franke J, Kohler M, Macara I G. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-α3. Mol Cell Biol. 1999;19:8400–8411. doi: 10.1128/mcb.19.12.8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 75.Yokoya F, Imamoto N, Tachibana T, Yoneda Y. β-Catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zerivitz K, Akusjarvi G. An improved nuclear extract preparation method. Gene Anal Tech. 1989;6:101–109. doi: 10.1016/0735-0651(89)90016-2. [DOI] [PubMed] [Google Scholar]
- 77.Ziemin-van der Poel S, McCabe N R, Gill H J, Espinosa III R, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]