Multiple Lysine Mutations in the C-Terminal Domain of p53 Interfere with MDM2-Dependent Protein Degradation and Ubiquitination (original) (raw)
Abstract
To investigate the effect of mutations in the p53 C-terminal domain on MDM2-mediated degradation, we introduced single and multiple point mutations into a human p53 cDNA at four putative acetylation sites (amino acid residues 372, 373, 381, and 382). Substitution of all four lysine residues by alanines (the A4 mutant) and single lysine-to-alanine substitutions were functional in sequence-specific DNA binding and transactivation; however, the A4 mutant protein was resistant to MDM2-mediated degradation, whereas the single lysine substitutions were not. Although the A4 mutant protein and the single lysine substitutions both bound MDM2 reasonably well, the single lysine substitutions underwent normal MDM2-dependent ubiquitination, whereas the A4 protein was inefficiently ubiquitinated. In addition, the A4 mutant protein was found in the cytoplasm as well as in the nucleus of a subpopulation of cells, unlike wild-type p53, which is mostly nuclear. The partially cytoplasmic distribution of A4 mutant protein was not due to a defect in nuclear import because inhibition of nuclear export by leptomycin B resulted in nuclear accumulation of the protein. Taken together, the data suggest that mutations in the putative acetylation sites of the p53 C-terminal domain interfere with ubiquitination, thereby regulating p53 degradation.
The p53 tumor suppressor gene has been implicated in the malignant progression of cancers, and the mutational inactivation of p53 is the most frequent genetic alteration in human cancers. Wild-type p53 plays a key role in tumor suppression by monitoring DNA damage and executing pathways that negatively control cell growth, either by blocking cells in the G1 phase of the cell cycle or by inducing apoptosis (26). Wild-type p53 regulates these processes, at least in part, by functioning as a transactivator of gene expression. Wild-type p53 is a transcription factor that recognizes a specific consensus DNA sequence and also interacts with a number of components of the transcription complex (17, 34, 37) to activate the expression of target genes associated with either growth arrest (Waf1 and GADD45) or cell death (Bax) (2, 12, 22).
The p53 protein normally exists in cells as a short-lived and latent protein. Upon exposure to stress, it is stabilized and activated mainly through posttranslational mechanisms. The p53 protein has also been reported to be modified by phosphorylation (21), acetylation (5, 27) and glycosylation (28). Phosphorylation within its N terminus is thought to disrupt the interaction between p53 and its negative regulator, MDM2, and therefore alleviate transcriptional inhibition mediated by MDM2 (29, 30). In contrast, acetylation of sites within the C terminus of p53 has been shown to stimulate the binding of DNA by p53 in vitro (5). The C-terminal domain of p53 maintains the whole protein in a non-DNA-binding latent form. One possible mechanism of this regulation may be allosteric, as shown in a model in which the C-terminal domain interacts with another region of p53 and retains the protein in a non-DNA-binding conformation (9–11). In addition to the modifications mentioned above, deletion of, point mutation in, and binding of specific antibody to this region disrupt the interaction, resulting in stimulation of p53's DNA binding (13). However, the biological significance of modification of the C terminus of p53 has yet to be clarified.
p53 has been reported to undergo degradation directed by MDM2 through a ubiquitin-dependent pathway (19). Indeed, MDM2 functions as a p53-specific E3 ubiquitin ligase in vitro (7), and ubiquitinated p53 is degraded on cytoplasmic 26S proteasomes. However, recent studies of p53 degradation revealed that its efficiency requires some portions of p53. For example, p53 interacts with MDM2 through its conserved box I in the N-terminal domain (16). Mutations in this area abolish MDM2 binding and render p53 resistant to MDM2-mediated degradation (15). In addition, the C-terminal domain of p53 has been shown to be important for MDM2-mediated degradation for two reasons. First, the binding of MDM2 to p53 and p53's concomitant ubiquitination require p53 oligomerization via a domain located in the C terminus of p53 (residues 324 to 355) (18), and yet MDM2 binds to the N terminus of p53. Thus, MDM2 probably requires a certain tertiary structure of p53 in order to function. Second, the extreme C-terminal region of p53 (residues 363 to 393) is apparently also required for efficient degradation directed by MDM2, since for some unknown reason deletion mutants of p53 lacking this region are resistant to MDM2-mediated degradation (15).
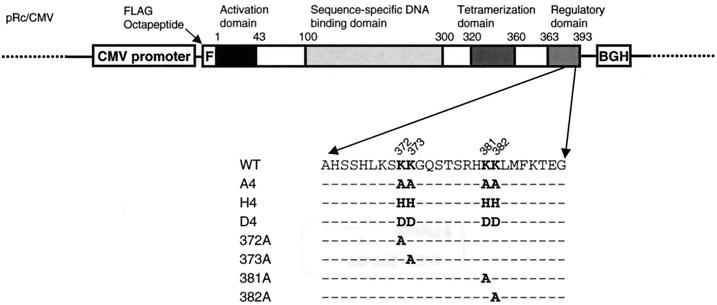
Together, these observations suggest that the C-terminal domain of p53 regulates the MDM2-mediated degradation of p53 in addition to its sequence-specific DNA binding activity. The purpose of this study therefore was to explore the effects of specific modification of the putative acetylation site of the C-terminal domain of p53 on its biological properties. To produce mutated p53, we chose to mutate four lysine residues (residues 372, 373, 381, and 382) in the C-terminal domain, because these lysine residues have been shown to be acetylated (5) and because basic amino acid residues in the extreme C-terminal region of p53 are supposed to be critical determinants of tetramerization (32) even though they are not located in the oligomerization domain. Here, we demonstrate that mutant p53 containing these altered lysine residues became more stable and resistant to MDM2-mediated degradation.
MATERIALS AND METHODS
Cell culture.
The p53 negative human non-small-cell lung cancer cell lines H1299 and H322 were obtained as a gift from Adi Gazdar and John Minna (The University of Texas Southwestern Medical Center, Dallas, Tex.). All cells were cultured in RPMI medium containing 5% (vol/vol) fetal bovine serum at 37°C in a humidified 5% CO2 atmosphere.
Plasmid construction and mutagenesis.
Full-length p53 was amplified by PCR using a wild-type p53 pBluescript plasmid as the template (24), forward primer 5′-GTAAGCTTACCATGGACTACAAGGACGACGATGACAA GATGGAGGAGCCGCAG-3′, and reverse primer 5′-TCAAAGCTTGTCGACAAGTG GAGAATG-3′. PCR was performed essentially as described earlier and consisted of 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 1 min. The PCR fragments were inserted into a pRc/CMV vector (Invitrogen) at the _Hin_dIII site. Point mutations in the C-terminal domain of wild-type p53 were introduced using an in vitro site-directed mutagenesis system (GeneEditor; Promega Corp.) according to the manufacturer's instructions. All constructs generated this way were sequenced to ensure that only the intended mutation occurred. The plasmids pVgRXR (which encodes the receptor subunits), pIND (which contains ecdysone response elements), and pIND/lacZ were all obtained from Invitrogen. For inducible expression, wild-type and mutated p53 cDNAs were inserted into pIND vectors. A plasmid expressing the mouse MDM2 gene (pCOC-mdm2 X2) (6) was kindly provided by Moshe Oren (Weizmann Institute of Science, Rehovot, Israel).
Transfection.
H1299 cells were seeded at a density of 4 × 105 cells per well in six-well plates (35 mm in diameter) or at 1 × 106 cells per 10-cm dish and transfected the following day with 1,2-dioleoyl-3-(trimethylammonium) propane (DOTAP; Boehringer Mannheim Corp.) according to the manufacturer's instructions. For Western blot or luciferase assay, a mixture containing 5 μg of the indicated plasmid(s) and 25 μl of DOTAP was added to the cells cultured in six-well plates. For nuclear extraction or immunoprecipitation, a mixture containing 10 μg of plasmid(s) and 50 μl of DOTAP was overlaid onto a 10-cm dish. In all cases, after a 4-h incubation, the transfection mixture was removed and replaced with a serum-containing medium. Unless otherwise indicated, transfected cells were harvested 24 h after transfection.
Protein extraction and immunoblotting.
To prepare the whole-cell lysates, the medium was removed, and the cells were then washed twice with phosphate-buffered saline (PBS) and lysed with 300 μl of 1× sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris, pH 6.8; 2% SDS, 10% glycerol; 0.01% bromophenol blue). The lysed cells were transferred into 1.5-ml microcentrifuge tubes, which were then sonicated for 3 s at 20% output in an ultrasonic cell disruptor (Heat Systems, Inc.) and centrifuged at 15,000 rpm for 15 min at 4°C. The protein concentration of the extracts was measured using the bicinchoninic acid (BCA) protein assay kit (Pierce). Then, 50 μg of protein was separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 10 or 7.5% polyacrylamide gels and transferred to a nitrocellulose membrane (Hybond ECL; Amersham Pharmacia Biotech). p53 was detected using either p53 monoclonal antibody PAb1801 or monoclonal antibody PAb421 (Oncogene Research Products). p21/WAF1 was detected using the WAF1 monoclonal antibody EA10 (Oncogene Research Products). Monoclonal anti-actin antibody (Sigma) was used as an internal loading control. Finally, the specific proteins were detected using the ECL protocol (Amersham Pharmacia Biotech).
Preparation of nuclear extracts.
To prepare nuclear extracts, the transfected cells were washed twice with ice-cold PBS, scraped away from their culture dishes, and transferred to microcentrifuge tubes. The cells were then centrifuged at 1,000 rpm for 5 min at 4°C. After centrifugation, the supernatant was discarded, and the pellet was resuspended in 400 μl of cold buffer A (10 mM HEPES-KOH [pH 7.9]–10 mM KCl–1 mM dithiothreitol [DTT] containing 1 × protease inhibitor cocktail; Boehringer Mannheim Corp.). After incubation on ice for 15 min, 12.5 μl of 10% Nonidet P-40 was added, and the mixture was vortexed briefly. The nuclei were pelleted by centrifugation at 4,000 rpm for 4 min at 4°C and then resuspended in 50 μl of ice-cold buffer B (20 mM HEPES-KOH, pH 7.9; 0.4 M NaCl; 1 mM DTT; 1× protease inhibitor cocktail) followed by incubation on ice for 30 min. The mixture was then centrifuged at 15,000 rpm for 5 min, and the supernatant was collected as a nuclear extract. The protein concentration was determined using the BCA protein assay kit.
Electrophoretic mobility shift assay (EMSA).
The oligonucleotide encoding the p53 consensus binding site (p53CON) (4), GGACATGCCCGGGCATGTCC, was end labeled with 32P and used as a p53-specific probe. The DNA-binding reaction mixture (20 μl) contained 25 mM HEPES-KOH (pH 7.9), 1 mM DTT, 1% Nonidet P-40, 5% glycerol, 100 ng of poly(dI-dC), 80 mM NaCl, 10 mg of bovine serum albumin per ml, 1 ng of 32P-labeled probe DNA, and nuclear extract as indicated. Reaction mixtures were preincubated on ice for 20 min before the probe DNA was added and further incubated at room temperature for 30 min. For the supershift assay, 0.2 μg of PAb421 or PAb1801 was added to the reaction mixture, and the mixture was incubated at room temperature for 20 min. Each reaction mixture was then loaded onto a native 4% polyacrylamide gel (acrylamide:bisacrylamide, 29:1) containing 0.5× Tris-borate (TBE) and electrophoresed in 0.5× TBE at 100 to 150 V for 2.5 h at 4°C. After being dried, the gels were exposed to a phosphor screen and analyzed using a Storm 860 imager (Molecular Dynamics).
Luciferase assay.
The luciferase reporter plasmid pGUP.PA.8-53CON, a gift from Jerry Shay (The University of Texas Southwestern Medical Center), was used to assay p53 transactivation activity. The plasmid contains a p53CON sequence and a basal promoter element of the human heat shock protein gene upstream of the firefly luciferase coding sequence. For each transfection, 1 μg of pGUP.PA.8-p53CON plasmid and 10 ng of a p53-expressing plasmid were transfected into cells along with DOTAP. After 24 h, the transfected cells were lysed by scraping them from their culture dishes into 250 μl of reporter buffer (Promega Corp.) and then spinning the mixture for 30 s at 15,000 rpm and 4°C. The total protein concentration was determined using the BCA protein assay kit (Pierce). Then, 5 to 20 μl of each sample was first mixed with 100 μl of luciferase assay substrate that had been reconstituted with luciferase assay buffer (Promega Corp.) and then assayed for light emission using a luminometer (Monolight 2010; Analytical Luminescence Laboratory). The resulting activity was normalized to protein concentration.
Immunoprecipitation.
Immunoprecipitation was performed as described by Maki (18) to detect p53-ubiquitin conjugates and p53-bound MDM2. For detection of p53-ubiquitin conjugates, transfected cells were washed with PBS and scraped from their culture dishes into 1 ml of radioimmunoprecipitation assay (RIPA) buffer (2 mM Tris, pH 7.4; 5 mM EDTA; 150 mM NaCl; 1.0% Nonidet P-40; 1.0% deoxycholate; 0.025% SDS; 1 mM phenylmethylsulfonyl fluoride). The cells were then sonicated for 10 s at 20% output in an ultrasonic cell disruptor (Heat Systems, Inc.) and centrifuged at 15,000 rpm for 15 min. For detection of MDM2 binding to p53, cells were rinsed with PBS, scraped from their culture dishes into 1 ml of TBE buffer (50 mM Tris, pH 7.5; 5 mM EDTA; 150 mM NaCl; 0.5% Nonidet P-40; 1 mM phenylmethylsulfonyl fluoride), and incubated on ice for 30 min. The cells were then centrifuged at 15,000 rpm for 15 min. p53 was immunoprecipitated from extracts using the agarose-conjugated p53 antibody DO-1 (Santa Cruz). The antibody-conjugated agarose was collected by centrifugation and then washed four times with ice-cold RIPA or TBE buffer. Next, the immunoprecipitates were dissolved in 2× SDS sample buffer (100 mM Tris, pH 6.8; 4% SDS; 5% 2-mercaptoethanol; 20% glycerol; 0.02% bromophenol blue) and boiled for 5 min. The immunoprecipitates were then subjected to PAGE and transferred to a nitrocellulose membrane. The ubiquitin-p53 conjugates and MDM2 coprecipitated with p53 in p53 immunoprecipitates were detected using the ubiquitin monoclonal antibody P1A6 (Santa Cruz) or the MDM2 polyclonal antibody C-18 (Santa Cruz) whose epitope was located within the C-terminal domain of MDM2 of human origin (identical to corresponding mouse sequence), respectively. Each blot was stripped and reprobed with the p53 monoclonal antibody Bp53-12 (Santa Cruz).
Establishment of ecdysone-inducible cell lines.
To establish ecdysone-inducible cell lines, H1299 cells were first transfected with pVgRXR and then selected with zeocin (200 μg/ml). Twenty-four clones isolated in this way were then transiently transfected with pIND/lacZ, cultured in the presence of the ecdysone analogue ponesterone A (PA; Invitrogen), and stained with X-Gal (5-bromo-4-chloro-3-indoly-β-d-galactopyranoside). The most inducible clone was R-11. Then, to generate cell lines that would express wild-type p53 or its A4 mutant under the control of an ecdysone-inducible system, R-11 cells were transfected with pIND plasmids carrying either the wild-type p53 gene or A4 mutant gene and then selected with G418 (300 μg/ml) and zeocin. Immunohistostaining with PAb1801 was used to screen the lines for inducible p53 expression. Clones WT-18 and A4-38 were found to express comparable levels of p53 and so were used in further experiments.
Immunohistochemical staining.
Cells were grown on a glass chamber slide and treated with 5 μM PA for 24 h. They were then washed with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. Next, after permeabilization with 0.1% Nonidet P-40 in PBS for 10 min, cells were incubated for 1 h with PAb1801 and then incubated for 1 h with goat anti-mouse secondary antibody (Santa Cruz) and processed for immunohistochemical staining using an ABC kit.
Preparation of nuclear and cytosolic extracts.
Nuclear and cytosolic extracts were separated as described previously (33). In brief, cells were seeded at 106 per 10-cm dish and treated with 5 μM PA for 24 h. They were then washed twice with ice-cold PBS, scraped into 500 μl of lysis buffer (10 mM Tris-HCl, pH 7.4; 10 mM NaCl; 6 mM MgCl2; 1 mM DTT) containing 1× protease inhibitor cocktail (Boehringer Mannheim Corp.), and homogenized. After confirmation by trypan blue staining that >95% of cells had ruptured, the cells were spun at 10,000 rpm for 1 min. The resulting supernatant was saved as the cytosolic extract. Next, the pellet was washed with lysis buffer, dissolved in 500 μl of 1× SDS sample buffer, and centrifuged. This supernatant was saved as the nuclear extract.
RESULTS
Expression and DNA-binding activity of p53 mutants.
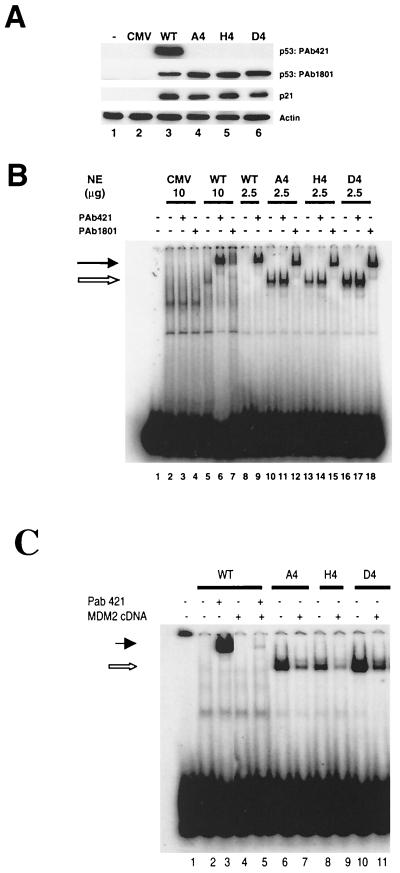
To investigate the function of putative acetylation sites in the C-terminal domain of p53, a series of constructs was established (Fig. 1). At first, the lysine residues at amino acid residues at 372, 373, 381, and 382 were altered to alanine residues to neutralize positively charged lysine residues. The resulting mutant p53 was named A4. Also, the same lysine residues were substituted with positively charged histidine (H4) or negatively charged aspartic acid (D4). Plasmids carrying either a wild-type p53 gene or one of the mutants were transfected into a p53-null H1299 cell line, and p53 expression was analyzed using Western blotting. The levels of all three mutant proteins were higher than that of wild-type p53 (Fig. 2A, lanes 3 to 6). Since it is possible that different transfection efficiencies give rise to variations in expressed protein levels, these differences in protein levels were confirmed in three independent experiments in which the densities of the bands on the exposed films were measured and normalized to the density of actin bands. The respective densities of the A4, H4, and D4 mutants were 1.9 ± 0.42, 1.9 ± 0.66, and 1.6 ± 0.35 (mean ± the standard deviation) times higher than the density of wild-type p53. These data indicated that the transfected cells accumulated more mutant proteins than wild-type p53 protein. A4 and H4 mutant proteins were as electrophoretically mobile as the wild-type p53 protein (Fig. 2A), while the D4 mutant protein was slightly less mobile, probably because of its altered amino acid residues. Although all three mutants were detected by PAb1801, which recognizes amino acids 46 to 55 of the p53 protein, they were not detected by PAb421, which recognizes the C-terminal domain of p53. In addition, compared with p53, all three mutants induced expression of endogenous p21. Since the level of p53 was different but the level of transactivation was the same, the implication was that there must have been a difference in transactivation activity.
FIG. 1.
Structure of an expression plasmid carrying wild-type p53 cDNA. Functional domains of human p53 and positions of mutations (highlighted) are displayed. BGH, BGH poly(A) signal; WT, wild-type p53.
FIG. 2.
Expression and DNA-binding activity of wild-type and mutant p53 proteins. (A) Western blot analysis of p53 and p21 protein expression in H1299 cells transiently transfected with wild-type or mutated p53 cDNAs as indicated. At 24 h after transfection, cell extracts were prepared, resolved by SDS–10% PAGE, and transferred to a nitrocellulose membrane. The membrane was probed with the indicated antibodies. (B) Nuclear extracts were prepared from H1299 cells transfected with the indicated plasmids and used for EMSA. The 32P-labeled p53CON oligonucleotide was incubated in the absence (lane 1) or presence of the indicated nuclear extracts (lanes 2 to 18). Incubations were performed either in the absence of PAb421 and PAb1801 (lanes 1, 2, 5, 8, 10, 13, and 16) or in the presence of either PAb421 (lanes 3, 6, 9, 11, 14, and 17) or PAb1801 (lanes 4, 7, 12, 15, and 18). (C) Overexpression of exogenous mouse MDM2 inhibited p53 DNA binding activity. Mutants also showed inhibition of their DNA binding activity (lanes 7, 9, and 11). The mixtures were resolved by PAGE and visualized using autoradiography. The open and closed arrows indicate the positions of the shifted and supershifted complexes, respectively.
Next, the DNA-binding activity of the mutants was examined by EMSA using the p53CON oligonucleotide. The p53-expressing plasmids were transfected into H1299 cells, and nuclear extracts were prepared as described above. The nuclear extracts were then mixed with radiolabeled p53CON in the presence or absence of anti-p53 monoclonal antibody. Wild-type p53 showed weak binding to p53CON, and the addition of the anti-p53 monoclonal antibody to the mixture increased its DNA binding activity and showed a supershift of the p53CON-p53 protein complex (Fig. 2B, lanes 5 to 7). In particular, PAb421 greatly increased the binding activity of wild-type 53, a result consistent with previous studies (10), while PAb1801 produced only a supershift. In contrast, the mutants A4, H4, and D4 showed strong DNA-binding activity (Fig. 2B, lanes 10 to 18), while all of the nuclear extracts contained comparable levels of p53 (data not shown). The addition of PAb421 did not produce supershifted bands of all the mutants because the epitope of PAb421 was altered. On the other hand, PAb1801 supershifted the mutant-p53CON complexes, indicating that these complexes actually contained p53 mutant protein.
H1299 cells were cotransfected with MDM2 cDNA plasmids and wild-type or the mutant p53 constructs. Nuclear extracts were analyzed for DNA binding activity using 10 μg of nuclear extracts from wild-type-transfected cells and 2.5 μg of nuclear extracts from mutant-transfected cells. MDM2 overexpression significantly inhibited the DNA binding ability of wild-type p53. In agreement with earlier reports, wild-type p53 DNA binding activity was strongly enhanced in the presence of PAb421 antibody, and the wild-type p53 band was strongly supershifted. Addition of MDM2 antibody caused no further shift. However, when wild-type p53 was cotransfected with MDM2-expressing plasmids, its DNA binding ability was significantly impaired and the supershifted band was reduced (Fig. 2C). In contrast, C-terminal mutants showed strong DNA binding activity. As was the case for the wild-type p53, the DNA binding activity of the A4, H4, and D4 mutants was reduced in the presence of MDM2 overexpression.
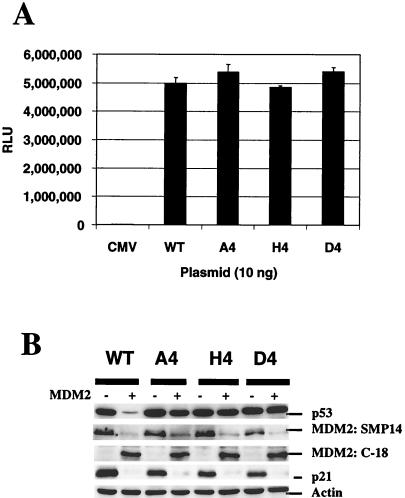
In addition, the transactivation property of wild-type p53 and the mutants were determined using the p53-responsive promoter, pGUP.PA.8-p53CON. Compared with wild-type p53, these mutants retained the same ability to activate transcription from this promoter (Fig. 3A). Thus, although the p53 proteins accumulated to a much higher level, the transactivation property did not correlate in this experiment.
FIG. 3.
Transactivation and stability of mutant p53 in the presence of MDM2 overexpression. (A) H1299 cells were transiently transfected with 10 ng of each p53 construct and 1 μg of pGUP.PA.8-p53CON plasmid DNA. After 24 h, cells were lysed and assayed for luciferase activity. The values presented here are representative data from triplicate experiments. Assays were performed twice independently and showed similar results. (B) H1299 cells were transfected with 0.5 μg of p53 plasmid and 4.5 μg of pRc/CMV empty vector or MDM2 cDNA plasmid. At 24 h after transfection, cell extracts were prepared, and expression of p53, expression of MDM2, and induction of p21 were analyzed by Western blot analysis using PAb1801, MDM2 mouse monoclonal SMP14, and rabbit polyclonal antibody C-18 (Santa Cruz), and p21 monoclonal antibody, respectively.
Effect of mutations at putative p53 acetylation sites on MDM2-mediated degradation.
Recently, it has been shown that the intact C-terminal domain of p53 is required for MDM2-mediated degradation (15). To determine the stability of these mutant p53s in the presence of MDM2, plasmid pCOC-mdm2 X2 was transiently cotransfected along with different p53 constructs into H1299 cells. As shown previously (14), wild-type p53 protein was sensitive to MDM2-mediated degradation (Fig. 3B). In contrast, these mutants were resistant to MDM2-mediated degradation (Fig. 3B), suggesting that alterations in these sites prevented p53 protein from undergoing MDM2-mediated degradation. Interestingly, p21 induction by transiently expressed wild-type and mutant p53s was effectively abolished by cotransfection of an MDM2-expressing plasmid.
Since MDM2 is another target gene for p53, the transactivation of MDM2 by these mutants was also examined. H1299 cells were cotransfected with p53 and MDM2 constructs, and the total protein was then analyzed for MDM2 and p21 expression. SMP14 antibody was used to recognize the epitope of human MDM2. Results indicated that wild-type p53 and most of the C-terminal p53 mutants could efficiently transactivate endogenous MDM2 to comparable levels; the exception was the D4 mutant, which, by comparison, showed somewhat reduced expression (Fig. 3B). However, overexpression of exogenous mouse MDM2 apparently inhibited the transactivation of both p21 and endogenous MDM2.
Reduced ubiquitination of A4 mutant.
To explore how these mutations contributed to p53's enhanced stability, the effects of changes in the ubiquitination-proteosome pathway were examined. We cotransfected the H1299 cells with equal amounts of MDM2 and p53 plasmids and, under these conditions, easily detected ubiquitinated p53 protein, even though the level of wild-type p53 protein decreased due to the high level of MDM2 protein expression.
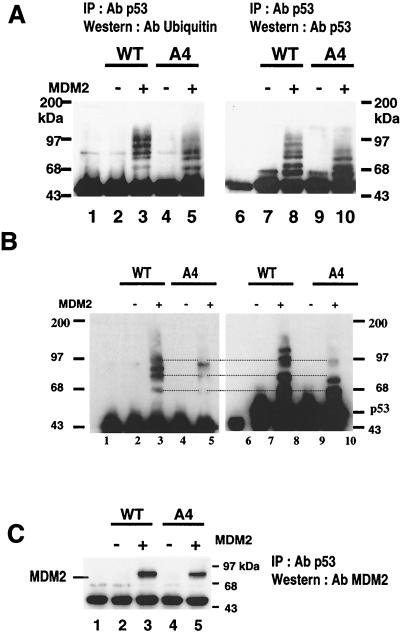
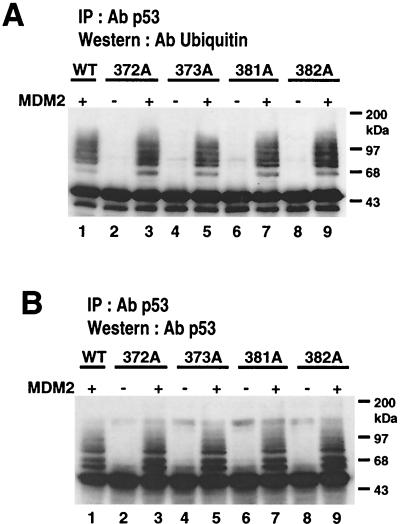
Next, p53 was immunoprecipitated with agarose-conjugated p53 antibody from the extract of transfected cells and then examined by immunoblotting with the ubiquitin antibody P1A6 (Fig. 4A, lanes 1 to 5). A ladder of protein bands with molecular weights comparable to those of p53 ubiquitin complexes was clearly detected in the lane containing protein from the cells transfected with wild-type p53 and MDM2. On the other hand, A4 mutant cotransfected with MDM2 showed fewer protein bands (Fig. 4A, lanes 3 and 5). When the same blot was stripped and reprobed with the p53 antibody, the same ladder of bands was recognized (Fig. 4A, lane 8). p53 and the immunoglobulin G (IgG) heavy chain have similar molecular weights and were therefore difficult to resolve on the Western blots. We used untransfected control cell extracts as negative controls (lanes 1 and 6). In Fig. 4A, lane 6 shows a band comprised of IgG heavy chain from an extract of control untransfected cells, while lanes 7 to 10 show major bands of immunoprecipitated p53 that migrated close to IgG. Using the same immunoprecipitation strategy and a ubiquitin-specific antibody, a longer gel was run to separate the ubiquitinated p53 protein complexes for better clarity (Fig. 4B). Three ubiquitinated forms of p53 protein were clearly detected, whereas A4 protein bands were less intense when probed with ubiquitin antibody (Fig. 4B, lane 3) or with p53 antibody (Fig. 4, lane 8). Together, these results indicated that the ladder of bands contained ubiquitinated p53 and that A4 mutant protein was inefficiently ubiquitinated upon MDM2 coexpression.
FIG. 4.
MDM2-mediated ubiquitination of wild-type p53 and A4 mutant. (A) H1299 cells were transfected with 5 μg of MDM2 plasmid and 5 μg of either wild-type p53- or A4 mutant-expressing plasmid. At 24 h after transfection, cell extracts were prepared as described in Materials and Methods and then immunoprecipitated with DO-1 anti-p53 antibody. The immunoprecipitates were subjected to Western blot analysis. A blot was probed first with ubiquitin monoclonal antibody P1A6 (lanes 1 to 5), then stripped, and then reprobed with Bp53-12 (lanes 6 to 10). Lanes 1 and 6, immunoprecipitates from untransfected cells serving as negative controls and showing the position of the IgG heavy chain immediately below the p53 protein band. (B) To separate the p53-ubiquitinated bands more clearly, a longer gel was run using the same strategy as above. Ubiquitin p53 complexes were marked. (C) A blot was probed first with anti-MDM2 antibody (upper), then stripped, and then reprobed with anti-p53 antibody (lower panel). Lane 1, immunoprecipitates from untransfected cells serving as negative controls.
We examined the association between MDM2 and A4 mutant p53. The MDM2 protein has been shown to possess a ubiquitin ligase activity for p53 in vitro (7). Thus, in the present study, it was important to know whether inefficient ubiquitination of the A4 mutant was caused by impaired association of MDM2 or by inhibition of ubiquitination itself despite the binding of MDM2. To determine the physical association of MDM2 with p53, we performed coimmunoprecipitation experiments. Extracts from cells transiently transfected with the indicated combinations of plasmids were immunoprecipitated using the p53 antibody-agarose conjugates, and MDM2 was detected by immunoblotting with the MDM2 antibody (Fig. 4C). MDM2 was clearly detected in immunoprecipitates from the extracts of cells transfected with wild-type p53 and MDM2. In addition, the combination of the A4 mutant and MDM2 also produced an obvious but slightly weaker MDM2 band. Thus, the difference in p53 level cannot be explained by a difference in MDM2 binding.
Unimportance of single-lysine mutations for p53 ubiquitination.
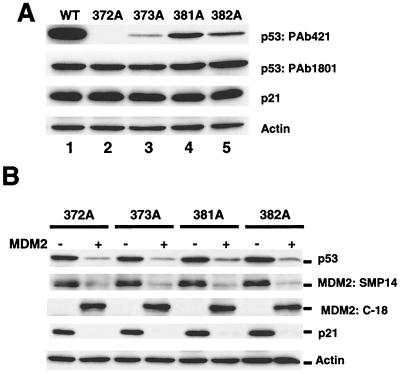
Because ubiquitin ligase transfers ubiquitin molecules to one or more lysine residues in the substrate (1), it is possible that critical lysine residues were removed from the A4 mutant. To determine whether any one of the altered lysine residues in A4 was critical for ubiquitination, point-mutated p53s carrying a single-lysine mutant at residue 372, 373, 381, or 382 were generated (Fig. 1). Individual plasmids containing the mutated genes were transfected into H1299 cells, and p53 expression in the transfected cells was analyzed. Immunoblotting with PAb1801 demonstrated that all four plasmids expressed p53 protein at levels comparable to that expressed by the wild-type p53 plasmid (Fig. 5A). On the other hand, the single-lysine mutants reacted differently to PAb421: 372A was not detected, while the others were weakly detected. Because the epitope of PAb421 comprises amino acids 371 to 380 of p53 (35), it is possible that some of the single-lysine mutants were weakly reactive to PAb421. In addition, all of the single-lysine mutants could induce endogenous p21 as effectively as wild-type p53 could.
FIG. 5.
Expression of single-lysine mutants and their degradation in the presence of MDM2 overexpression. H1299 cells were transfected with either 5 μg of each indicated plasmid or a combination of plasmids as indicated. At 24 h after transfection, cell extracts were prepared, and expression of p53, expression of MDM2, and induction of p21 were analyzed by Western blot analysis. MDM2 protein was detected by mouse monoclonal antibody SMP14 and rabbit polyclonal antibody C-18 (Santa Cruz). WAF1 monoclonal antibody EA10 was used for detecting p21. (A) Expression of wild-type p53 and single-lysine mutants. (B) Single-lysine mutant protein expression in the presence or absence of MDM2.
The protein levels of all the single-lysine mutant p53s decreased upon cotransfection with MDM2-expressing plasmid (Fig. 5B), indicating that the mutants were sensitive to MDM2-mediated degradation. As Fig. 6A depicts, immunoprecipitates with p53 antibody from the extracts of transfected cells contained ubiquitin-conjugated p53. The single-lysine mutants and wild-type p53 produced similar ladders that were recognized by both the ubiquitin antibody and the p53 antibody (Fig. 6B). Together, these results suggested that multiple mutations of lysine residues are required for inhibiting ubiquitination of p53.
FIG. 6.
Efficient ubiquitination of single-lysine mutants and their degradation by MDM2. H1299 cells were transfected with 5 μg of MDM2 plasmid and 5 μg of plasmid expressing either wild-type p53 or a single-lysine mutant. At 24 h after transfection, cell extracts were prepared and immunoprecipitated with DO-1 anti-p53 monoclonal antibody as described above. The immunoprecipitates were subjected to Western blot analysis. (A) Blot probed with ubiquitin monoclonal antibody P1A6. (B) Same blot stripped and reprobed with anti-p53 antibody.
p53 mutation and subcellular distribution.
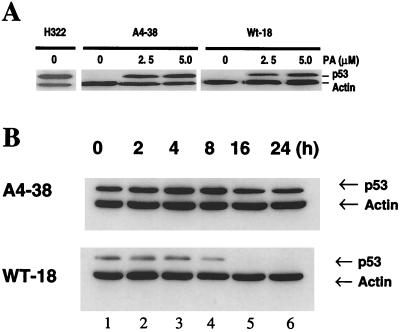
To further analyze the biological effect of the A4 mutant, we established H1299 cell lines expressing either wild-type or A4 mutant p53 under the control of an ecdysone-inducible system. One of these lines, a pVgRXR-transfected H1299 cell line called clone R-11, served as a negative parental control cell line. Several clones were screened for inducible p53 expression using immunohistochemical staining. Clones WT-18 and A4-38 expressing inducible wild-type p53 and A4 mutant genes, respectively, were selected. In both clones, the expression of p53 became detectable by immunohistochemistry at 4 to 6 h after incubation with 5 μM PA and reached a maximum at 16 h (data not shown). Clones WT-18 and A4-38 were both found to express comparable and significantly high levels of p53 as determined by Western blot analysis and so were used for further studies. Figure 7A shows the induction of p53 expression by different concentrations of PA. Total protein extract from a H322, another lung cancer cell line harboring a spontaneous mutant p53, was used as a positive control. Thirty micrograms of total cell extract from each cell line was analyzed by Western blotting using anti-p53 and anti-actin monoclonal antibodies as described in Materials and Methods. Results indicated that the stable clones did not leak p53 and that maximum expression could be obtained at a dose of 2 to 5 μM PA. The level and kinetics of p53 expression correlated with the induction of p21 expression (data not shown). Wild-type p53 protein turnover in the WT-18 and A4-38 cell lines was compared by treating the cells with 10 μg of cycloheximide per ml for different time intervals (Fig. 7B). As a result, the wild-type p53 protein level decreased after 8 h of cycloheximide treatment, while the A4 protein level remained unchanged even after 24 h of such treatment. Thus, it appeared that A4 proteins were stable and perhaps unable to be degraded by the MDM2-mediated pathway.
FIG. 7.
PA-induced expression of p53 protein in H1299 cells stably transfected with WT-18 and A4-38 as shown by immunoblot analysis of cell lysates. (A) p53 protein in WT-18 and A4-38 cells incubated for 24 h in the presence of as indicated PA. Actin protein served as a loading control. An H322 lung cancer cell line carrying a spontaneously mutated p53 gene was used as a positive control. (B) The stability of wild-type and A4 p53 protein in these clones was examined. The p53 protein in these cells was induced by 5 μM PA for 24 h. Induced cells were then incubated with 10 μg of cycloheximide per ml for different time intervals and then processed to prepare total cell extracts. p53 and actin proteins were detected by Western blot analysis using monoclonal antibodies as described earlier. (Upper panel) A4-38 cells. (Lower panel) WT-18 cells. Lanes 1 to 6 show p53 and actin proteins after 0, 2, 4, 8, 16, and 24 h of cycloheximide treatment, respectively.
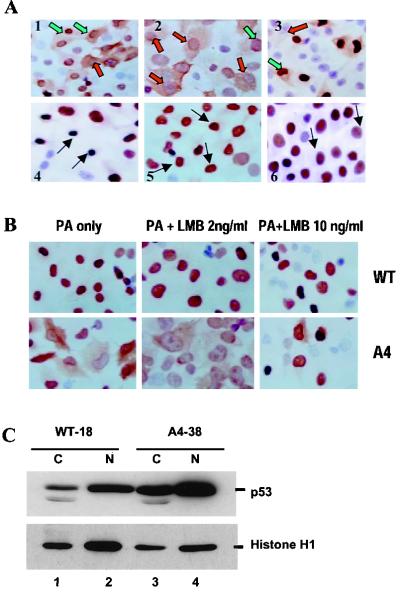
Immunohistochemical staining using p53 antibody demonstrated that a population of A4 mutant cells displayed p53 protein in both nucleus and cytoplasm, while WT-18 clones displayed wild-type p53 protein mostly in the nucleus (Fig. 8A). The cytoplasmic distribution of mutated protein in A4 clone, however, was not truly due to overexpression of the p53 protein since a significant number of cells with a low level of nuclear staining for p53 showed intense cytoplasmic distribution of the p53 (Fig. 8A). When we used the H322 lung cancer cell line expressing spontaneously mutated p53 protein as a positive control, we found no cytoplasmic distribution despite a high level of nuclear p53 accumulation. In these cell clones, about 80% of cells were induced to express fair amounts of p53 protein, although the level of p53 expression varied among the cells. We also found that, in a great majority of A4 cells expressing p53 protein, the p53 that was localized both in the cytoplasm and nucleus. Another stably transfected clone (A4-41), in which mutant p53 protein was expressed in fewer cells following PA administration, showed a similar cytoplasmic distribution (Fig. 8A).
FIG. 8.
View of immunohistochemically stained wild-type and mutant p53 protein in PA-induced cells. (A) Cells were grown on a culture slide and treated with 5 μM PA for 24 h. They were then stained with PAb1801 anti-p53 antibody. In the majority of cells, mutant p53 protein was distributed in both the nucleus and the cytoplasm of A4-38 clones (panels 1 and 2), while wild-type p53 was predominantly confined to the nucleus despite intense nuclear accumulation (panels 4 and 5). A4-41, another clone of A4, showed a similar cytoplasmic distribution (panel 5). A lung cancer cell line with a spontaneous mutation at codon 248 was used as a positive control; in this line, mutant p53 was restricted to the nucleus (panel 6). The red-filled arrow shows a strong cytoplasmic distribution of A4 mutant protein and a much lower nuclear accumulation. The green-filled arrow indicates the strong nuclear accumulation of p53 but cytoplasmic distribution. The black-filled arrow indicates strong nuclear localization of p53 with no cytoplasmic distribution. (B) Nuclear localization of leptomycin B-induced p53 protein. WT-18 cells (a, b, and c) and A4-38 cells (d, e, and f) were left untreated (a and d) or were treated with 2 ng of leptomycin B (b and e) or 10 ng of leptomycin B per ml for 18 h prior to fixation. p53 protein was detected in cells using anti-p53 monoclonal antibody as primary antibody and goat anti-mouse antibody as secondary antibody. Cells were also processed for immunohistochemical staining using an ABC kit. In both cases, the induced p53 protein was localized to the nucleus of the cells following leptomycin B treatment. (C) WT-18 and A4-38 cells were induced with PA for 24 h and fractionated into cytosolic (C) and nuclear (N) extracts as described in Materials and Methods. The extracts from an equivalent number of cells for each cell line were loaded onto a SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was then probed with anti-p53 antibody and reprobed with histone HI monoclonal antibody.
To determine further if the cytoplasmic distribution of the A4 mutant p53 was due to interference with the nuclear localization signal, WT-18 and A4-38 cells were treated with leptomycin B, a drug that blocks the nuclear export of p53. As shown by immunohistochemistry, the p53 protein in WT-18 cells was mostly localized in the cell nucleus (Fig. 8A). However, 20 h after the addition of 10 ng of leptomycin B per ml in culture, p53 was present in the nucleus only (Fig. 8B, upper panel). In the case of A4 cells, which contain cytoplasmic p53 (Fig. 8B, lower panel), p53 protein 20 h after the addition of leptomycin B was observed mainly in the nucleus. Furthermore, the difference in localization was also examined in nuclear and cytoplasmic extracts. Immunoblotting for p53 protein showed that only 10% of the wild-type p53 protein expressed in WT-18 cells was cytoplasmic versus 40% of the A4 mutant protein expressed in A4-38 cells (Fig. 8C). Given the postulated role of MDM2 in p53 export, the increase in cytoplasmic staining of a mutant that binds MDM2 less efficiently was rather surprising, though it was presumably due to p53 binding to and being shuttled by MDM2 not being degraded as a result.
DISCUSSION
In this study, we demonstrated that p53 proteins point mutated at the putative acetylation sites in their C-terminal domains have unique properties. These mutations enhanced p53's sequence-specific DNA binding, although upregulation of the p53-responsive promoter produced comparable levels of mutant p53 and wild-type p53. As shown in a previous study (20), enhancement of mutant p53 protein's DNA-binding activity does not further promote the transactivation of target genes in transient-transfection experiments. At the least, our findings from the transient- and stable-transfection experiments described here demonstrate that these mutants retain a transactivation activity.
The above-mentioned mutations of p53 impair ubiquitination and render p53 resistant to MDM2-mediated degradation. This is consistent with the results of a recent study showing that C-terminal deletion mutants are less sensitive to MDM2-mediated degradation (15). Our study demonstrates that the stability of the A4 mutant is due to inefficient ubiquitination, possibly because substitution of the four lysine residues may remove lysine residues critical for ubiquitination. To date, the ubiquitination sites on p53 have not been verified; yet it is known that some lysine residues are targets for ubiquitination. For example, ubiquitin ligase (E3) transfers the ubiquitin molecule to one or more lysine residues in the substrate (1). According to our results, no single mutation of any of these four lysine residues affected its ubiquitination or sensitivity to MDM2-mediated degradation, suggesting that none of the mutated lysine residues is a ubiquitination site. In addition, Maki (18) demonstrated that the truncated p53(1–363) is ubiquitinated efficiently by MDM2, suggesting that ubiquitination sites are located somewhere other than in the extreme C-terminal domain (amino acids 364 to 393). Therefore, the high stability of the A4 mutant protein is not apparently due to loss of its ubiquitination sites.
Interestingly, however, MDM2 appears to bind to the A4 mutant protein more weakly than it does to wild-type p53 protein. It has been demonstrated that MDM2 binds to p53 near its acidic activation domain, resulting in inhibition of p53-mediated transcriptional activation (23, 25, 36). Our own cotransfection experiment revealed that MDM2 inhibits the induction of p21 by the A4 mutant protein. Thus, MDM2 could bind to the A4 mutant protein and inactivate its function. In a previous study, the C-terminal deletion mutant p53(1–369) was shown to bind MDM2 only slightly more weakly than did wild-type p53 but to be ubiquitinated much less than expected (8). Taken together, these data suggest that binding of p53 to MDM2 is not likely to be the only determinant of ubiquitination followed by p53 degradation. The A4 mutant and C-terminal truncation proteins might have different conformations than wild-type p53, resulting in altered interactions between p53 and either MDM2 or some other important component of ubiquitination. In our study, more A4 mutant protein than wild-type p53 was localized in the cytoplasm of a stable clone. Moreover, the cytoplasmic A4 mutant protein was isolated from the nucleus, in which MDM2 binds to p53 and mediates ubiquitination (3). Therefore, this cytoplasmic compartmentalization of mutant p53 may contribute to inefficient ubiquitination. On the other hand, MDM2 can reportedly shuttle p53 from nucleus to cytoplasm in order to degrade it (3). Presumably, MDM2 binds to and shuttles, but does not degrade, p53 in the cells expressing A4 mutant. More recently, p53 has been demonstrated to be exported from nucleus to cytoplasm by a nuclear export signal located in p53's tetramerization domain, with no help from MDM2 (31). This nuclear export signal is thought to be silenced when p53 forms a tetramer and to be exposed when oligomerization is impaired. An exposed nuclear export signal can be accessed by the export receptor CRM1, resulting in nuclear exportation of p53 without MDM2. Interestingly, the basic amino acids in the region between residues 363 and 386 of p53 have been demonstrated to be critical for tetramerization (32), and it is possible that the A4 mutant cannot form a tetramer because of multiple amino acid substitutions in its C-terminal basic region. Further investigations are therefore necessary to determine whether nuclear exportation of A4 mutant protein is mediated by MDM2 or a nuclear export signal of p53.
ACKNOWLEDGMENTS
We thank Marjorie Johnson for her excellent technical assistance, Jude Richard (Department of Scientific Publications) for his editing assistance, and Monica Contreras for preparation of the manuscript. We thank Minoru Yoshida (University of Tokyo, Tokyo, Japan) for the leptomycin B.
This study was partially supported by an NIH project grant (PO1-CA78778-01) (J.A.R.), an NCI Specialized Program of Research Excellence (SPORE) in Lung Cancer grant (P50-CA70907) (J.A.R.), a Development Grant from the NCI for The University of Texas M. D. Anderson Cancer Center SPORE (P50-Ca70907) in Lung Cancer (T.M.), the W. M. Keck Center for Cancer Gene Therapy, a grant from the Tobacco Settlement Funds as appropriated by the Texas State Legislature (Project 8), gifts to the Division of Surgery from Tenneco and Exxon for the Core Laboratory Facility, and a sponsored research agreement with Introgen Therapeutics, Inc.
REFERENCES
- 1.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 3.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 6.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 7.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 8.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hupp T R, Lane D P. Regulation of the cryptic sequence-specific DNA-binding function of p53 by protein kinases. Cold Spring Harb Symp Quant Biol. 1994;59:195–206. doi: 10.1101/sqb.1994.059.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 11.Jayaraman J, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 12.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 13.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 14.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 15.Kubbutat M H, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 17.Lu H, Levine A J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maki C G. Oligomerization is required for p53 to be efficiently ubiquitinated by MDM2. J Biol Chem. 1999;274:16531–16535. doi: 10.1074/jbc.274.23.16531. [DOI] [PubMed] [Google Scholar]
- 19.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 20.Marston N J, Ludwig R L, Vousden K H. Activation of p53 DNA binding activity by point mutation. Oncogene. 1998;16:3123–3131. doi: 10.1038/sj.onc.1201856. [DOI] [PubMed] [Google Scholar]
- 21.Meek D W. Multisite phosphorylation and the integration of stress signals at p53. Cell Signal. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 22.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 23.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay T, Roth J A. A codon 248 p53 mutation retains tumor suppressor function as shown by enhancement of tumor growth by antisense p53. Cancer Res. 1993;53:4362–4366. [PubMed] [Google Scholar]
- 25.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 26.Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 27.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 29.Shieh S Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 30.Shieh S Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturzbecher H W, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins J R. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 33.Tang H Y, Zhao K, Pizzolato J F, Fonarev M, Langer J C, Manfredi J J. Constitutive expression of the cyclin-dependent kinase inhibitor p21 is transcriptionally regulated by the tumor suppressor protein p53. J Biol Chem. 1998;273:29156–29163. doi: 10.1074/jbc.273.44.29156. [DOI] [PubMed] [Google Scholar]
- 34.Thut C J, Chen J-L, Klemm R, Tijan R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Sci. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 35.Wade-Evans A, Jenkins J R. Precise epitope mapping of the murine transformation associated protein, p53. EMBO J. 1985;4:699–706. doi: 10.1002/j.1460-2075.1985.tb03686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 37.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]