An exonic splicing silencer in the testes-specific DNA ligase III β exon (original) (raw)
Abstract
Alternative pre-mRNA splicing of two terminal exons (α and β) regulates the expression of the human DNA ligase III gene. In most tissues, the α exon is expressed. In testes and during spermatogenesis, the β exon is used instead. The α exon encodes the interaction domain with a scaffold DNA repair protein, XRCC1, while the β exon-encoded C-terminal does not. Sequence elements regulating the alternative splicing pattern were mapped by in vitro splicing assays in HeLa nuclear extracts. Deletion of a region beginning in the β exon and extending into the downstream intron derepressed splicing to the β exon. Two silencing elements were found within this 101 nt region: a 16 nt exonic splicing silencer immediately upstream of the β exon polyadenylation signal and a 45 nt intronic splicing silencer. The exonic splicing silencer inhibited splicing, even when the polyadenylation signal was deleted or replaced by a 5′ splice site. This element also enhanced polyadenylation under conditions unfavourable to splicing. The splicing silencer partially inhibited assembly of spliceosomal complexes and functioned in an adenoviral pre-mRNA context. Silencing of splicing by the element was associated with cross-linking of a 37 kDa protein to the RNA substrate. The element exerts opposite functions in splicing and polyadenylation.
INTRODUCTION
The human DNA ligase III gene is alternatively spliced, an important mode of regulation of its expression and function. The α DNA ligase III cDNA was cloned from a HeLa library and is of low general abundance, with increased expression in testes, thymus, prostate and heart (1). The β DNA ligase III cDNA was cloned from a testes library (2). This species has low expression in all tissues, except in testes, where expression is high. In mouse testes, the β isoform mRNA is tightly regulated, with expression confined to late stages of spermatogenesis, especially pachytene spermatogenesis (2). With the recent cloning of the mouse DNA ligase III cDNA and genomic DNA, the difference between mouse DNA ligase III β and α was confirmed as being due to alternative use of two terminal exons (β or α). The functional difference between the isoforms is at the C-terminus of DNA ligase III; the α isoform contains a domain that interacts with XRCC1, while the β isoform does not (3,4).
Alternative splicing of pre-mRNA is an important mode of regulating gene expression in higher eukaryotes, and occurs in many pre-mRNAs (5,6). In Drosophila, genetic and biochemical evidence shows the important role of pre-mRNA processing mechanisms in sex determination (for a review see 5), sexual behaviour (7,8) and spermatogenesis (9). In higher eukaryotes, regulated alternative splicing is a feature of the expression of many pre-mRNAs during the process of spermatogenesis (10,11). The regulation of alternative splicing in general involves many pre-mRNA sequence elements, including the splice sites, branch site, polypyrimidine tract and exonic and intronic splicing enhancers and silencers (for reviews see 12,13). Calcitonin/CGRP and immunoglobulin µ (IgM) pre-mRNAs have alternative splicing patterns that, as for DNA ligase III, involve 3′-terminal exons, and some information is available on these systems (reviewed in 14). In calcitonin/CGRP pre-mRNA, the choice is between the calcitonin exon 4 (a terminal exon) or CGRP exon 5 (an internal exon, the subsequent exon 6 being a terminal exon). The non-canonical 3′ splice site of exon 4 results in an intrinsic block to the splicing of this intron (15), which appears to be overcome by actions of splicing enhancer elements in the exon (16,17) and in the downstream intron (reviewed in 18). The downstream intronic splicing enhancer also stimulates polyadenylation and cleavage of exon 4, raising the possibility that cooperation between the polyadenylation/cleavage and splicing machineries enhances splicing to exon 4 (19). In the case of IgM, a choice is offered in the composite IgM C4 exon between a 5′ splice site and the polyadenylation signal. If the 5′ splice site is used, the downstream M1 and M2 exons are expressed and a membrane-bound form of IgM is produced. Competition occurs between the splicing and polyadenylation machineries. In B cells, increasing CstF-64 promotes use of the IgM C4 exon polyadenylation site, thereby subverting splicing to the M1 exon (20).
Thus, there are a number of possible modes of regulation of human DNA ligase III, involving splicing, polyadenylation or transcriptional termination. If the intronic splicing signals for the β exon are inherently weak, then a spermatogenesis-specific enhancement of splicing or polyadenylation via sequence elements may be required for β exon expression. Increasing the termination of transcripts between the β and α exons may also allow more time for splicing to occur or may promote polyadenylation and cleavage (21). Alternatively, repression of splicing to the β exon may be due to the function of silencer sequence elements, except in pachytene spermatogenesis, when this repression is relieved. Again, any silencer elements may act directly on splicing or indirectly, via inhibition of polyadenylation/cleavage. In this paper, we report on a mechanism of repression of splicing to the DNA ligase III β exon in HeLa nuclear extracts.
MATERIALS AND METHODS
In vitro transcription templates
Genomic clones for human DNA ligase III were isolated by PCR using primers 5′-GAAGCGGAAAGCTGCTGATGAGA and 5′-CTGGTGTGGAGGGTGGCAAGTA or by screening a human PAC library (clone ID 7f16 from the RPCI1 library made by the de Jong laboratory, Roswell Park Cancer Institute, Buffalo, NY). The introns surrounding exons β and α were sequenced on both strands, and the data matched those (accession no. AC004223) subsequently submitted to the databases by the Whitehead Institute/MIT Center for Genome Research as part of the human genome sequencing project. DNA templates for in vitro transcription were made by cloning DNA ligase III fragments into pGEMT (Promega, Madison, WI) and using unique restriction sites within the β exon (_Pvu_II), in the intron between the β and α exons (_Ava_II) or downstream of the α exon (_Nco_I, in the pGEMT polylinker) to run-off 32P-labelled pre-RNAs. A series of DNA templates were also made by taking aliquots every 0.5 min from an exonuclease III digestion (250 U; Promega) of 2.5 µg of an _Apa_I/_Nco_I-digested pGEM-DNA ligase III clone. After polishing with S1 nuclease and Klenow fragment, plasmids were religated and individual clones checked by restriction analysis and sequencing. 32P-labelled transcripts were run-off at _Fsp_I or _Xmn_I sites in the pGEM vector. For mutagenesis studies, templates were made by PCR, using the sense primer T7-DL3-2829S (5′-TAATACGACTC-ACTATAGGGAAGCGGAAAGCTGCTGATGAGA) with the following antisense primers: DL3-βA (5′-GGCCAGCAGCTGA-GTGAGT) for pre-mRNA substrate A; REP-DL3-βA (5′-GTAA-ATTTTGACTTGGGGCCAGCAGCTGAGTGAGT) for pre-mRNA B; CS-DL3bA (5′-GACTGGGCTTTTCCCGGCCAGCA-GCTGAGTGAGT) for pre-mRNA C; HHV-DL3bA (5′-CTG-CTTCCACCCTGACCACAAACTCAATGCTGCATTCCCA-CACCCAGGCCAGCAGCTGAGTGAGT) for pre- mRNA D; s5-DL3-366A (5′-ACTCACGTAAATTTTGACTTGGGG) for pre-mRNA E; s5-DL3bA (5′-ACTCACGGCCAGCAGCTGA-GTGAGT) for pre-mRNA F (all the above used in Fig. 2B); bpaΔCA (5′-TTTAATGTAAATTTTGACTTG) for REPpA (Figs 3 and 4); pA-DL3bA (5′-TTTAATGGCCAGCAGCTGA-GTGAGT) for ΔREPpA (Figs 3 and 4); paREP-C3 (5′-TT-TAATGTAGATTTTGACTTGGGGCCAGCAGCTGAGTG-AGT) for C3pA (Fig. 4). Transcription templates for heterologous pre-mRNAs (Figs 5 and 6) were made by PCR using pAdMLpar (22) as the PCR template. A T7 primer was used with the following primers: hetREPadmldelA (5′-GTAAATT-TTGACTTGGATCCAAGAGTACTGGAA) for AdML-REP; adml-L2-pA (5′-TTTAATATCCAAGAGTACTGGAA) for AdML-PARpA; hetREPadml (5′-TTTAATGTAAATTTTGAC-TTGGATCCAAGAGTACTGGAA) for AdML-REPpA; adml-L2-5c (5′-ACTCACATCCAAGAGTACTGGAA) for AdML-PAR5c; hetREPadml5c (5′-ACTCACGTAAATTTTGACTTGGATCCAAGAGTACTGGAA) for AdML-REP5c. The control AdML-PAR RNA was run-off from _Bam_HI-linearised pAdMLpar (22). All pre-mRNAs were capped and universally labelled with [α-32P]GTP.
Figure 2.

Sequence elements in the _Pvu_II–_Ava_II region silence splicing of the C-β intron in HeLa nuclear extracts. (A) A scheme of the _Pvu_II–_Ava_II region, including the polyadenylation signal (pA) and cleavage site (scissors), is shown. Constructs A–F are depicted. (B) Splicing reactions of pre-mRNAs A–F (indicated above lanes). Incubations were for 2 h. Pre-mRNAs, lariat intermediates and mRNA products are labelled on the right; an asterisk also shows the mRNA products.
Figure 3.

(A) The 16 nt exonic element silences in vitro splicing, but does not inhibit polyadenylation. Time courses (40, 80 and 120 min) under splicing conditions are shown. A bracket and the letter a on the right indicate polyadenylation smears above the pre-mRNAs. The positions of the pre-mRNAs, lariat intermediates and mRNAs (with expected sizes) are shown on the left. Lane 1, markers. (B) The 16 nt exonic sequence overlaps with an upstream polyadenylation element. Incubations were for 60 min under conditions unfavourable to splicing. The polyadenylation smear is marked with a bracket and the letter a. Lane 1, markers.
Figure 4.
Reduced spliceosomal complex assembly on DNA ligase III pre-mRNA containing the 16 nt exonic silencer. (A) Denaturing PAGE analysis of aliquots of pre-mRNA splicing reactions incubated for 60 min. The symbols on the left indicate the pre-mRNAs, lariat intermediates and mRNA products. (B) Non-denaturing PAGE of aliquots of splicing reactions after 30 or 60 min incubation. Control aliquots were kept on ice for 60 min (lanes 1, 4 and 7). Non-specific complexes, H, and spliceosomal complexes A and B are labelled on the right.
Figure 5.
Function of the 16 nt exonic element in the context of AdML pre-mRNAs. In both panels the RNAs are indicated by symbols on the left (triangles indicate a possible exonucleolytic degradation product previously seen with this substrate; 55). (A) Splicing reactions were for 120 min. Control pre-mRNAs (lanes 2 and 4) ended at the _Bam_HI run-off site (PAR) or a GUGAGU 5′ splice site (PAR5c), while heterologous pre-mRNAs (lanes 3 and 5) contained the 16 nt element alone or a 5′ splice site (REP and REP5c). Lane 1, markers. (B) Splicing is silenced by the 16 nt element, but polyadenylation is enhanced. Pre-mRNAs ending in AUUAAA were incubated under splicing conditions for 60 min (lanes 3 and 5) or kept on ice (lanes 2 and 4), in the presence of cordycepin. Parallel reactions, under conditions unfavourable for splicing (lanes 7 and 8), were without cordycepin. The polyadenylation smear is indicated by the bracket and the letter a. Lanes 1 and 6, markers.
Figure 6.
Inhibition of spliceosomal complex assembly and UV cross-linking of a 37 kDa protein by the 16 nt exonic silencer in AdML pre-mRNAs. (A) The upper panel shows denaturing PAGE of a time course (0, 20, 40 and 60 min) of splicing reactions. The identity of the RNAs is indicated on the left. The lower panel shows non-denaturing PAGE of aliquots of the splicing reactions. The non-specific complex, H, and spliceosomal complexes A, B and C are labelled. The additional complex (I) is indicated by an arrow. (B) The same experimental design as in (A) except that AdML-REPpA was compared to AdML-PAR. (C) The upper panel shows denaturing PAGE analysis of aliquots of splicing reactions with AdML pre-mRNAs incubated for 60 min under cross-linking conditions (lanes 2 and 4) or kept on ice (lanes 1 and 3). The identity of the RNAs is indicated on the left. The lower panel shows an autoradiograph of proteins from the splicing reactions after UV cross-linking and RNase treatment. An arrow indicates the protein cross-linked to the AdML-REP substrate. (D) The same experimental design as in (C) except that AdML-REPpA is compared to AdML-PAR.
In vitro splicing reactions
Splicing reactions were performed by incubating 20 fmol of pre-mRNA at 30°C, in the presence of 0.5 mM ATP, 3.2 mM MgCl2, 2.6% polyvinyl alcohol, 60 mM KCl and 32% HeLa nuclear extract. Splicing reactions were reproducible in several batches of nuclear extracts prepared at the Cold Spring Harbor Cell Culture Facility or obtained from the Computer Cell Culture Centre (Mons, Belgium). Reaction products were extracted with phenol and precipitated in ethanol before electrophoresis in denaturing polyacrylamide gels (10% in Figs 1B and 4A; 5.5% in Figs 2B, 3, 5 and 6). Splicing reactions used to analyse protein–RNA cross-linking and spliceosome assembly were performed as described above, except in the presence of 20 ng/µl tRNA and with the omission of polyvinyl alcohol. For cross-linking, reactions were irradiated with 254 nm UV light twice at 900 mJ/cm2 for 60 s (XL-1000; Spectronics Corp.) and treated with 400 ng/µl RNase A (Sigma) and 4 U/µl RNase T1 (Gibco BRL) for 45 min at 37°C, before electrophoresis on 12% SDS–polyacrylamide gels. For analysis of spliceosomal complexes, heparin was added to 5 µg/µl and the reactions were analysed on 1.5 mm thick non-denaturing polyacrylamide gels at 4 W for 5 h in 50 mM Tris–glycine running buffer (23). Cordycepin was added to reactions at concentrations of 1 mM where indicated (Figs 3A, 4, 5B and 6B and D). To analyse polyadenylation in isolation from splicing, the splicing reaction was weakened by lowering the MgCl2 and KCl concentrations to 1.5 and 50 mM.
Figure 1.
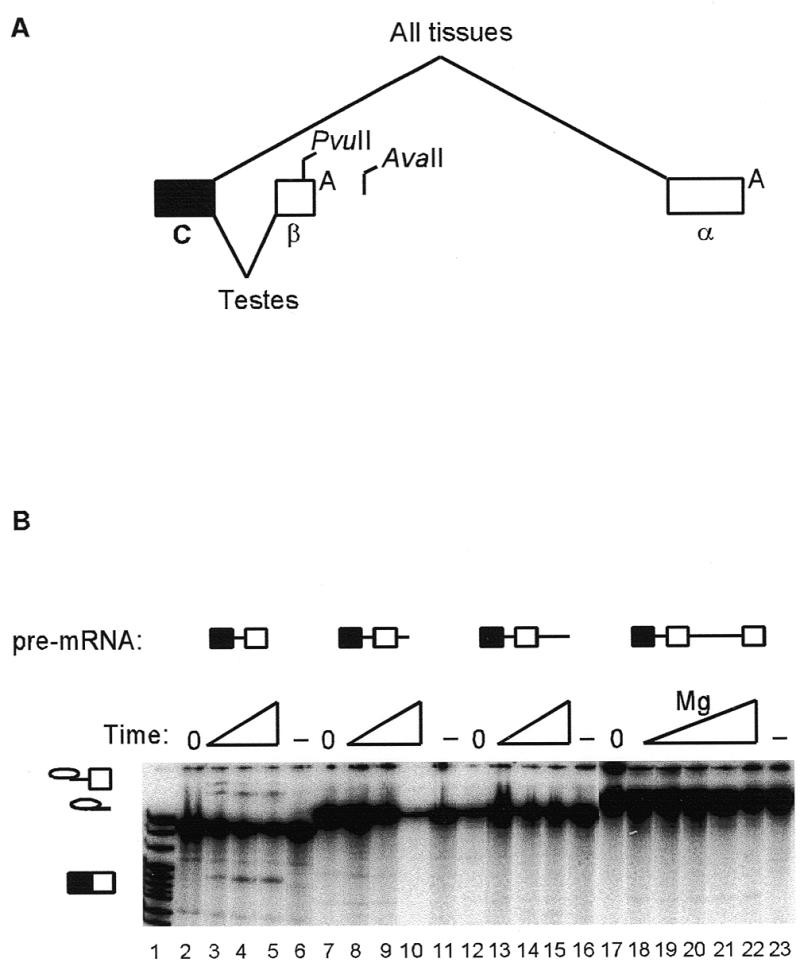
Genomic structure of the last three exons of the DNA ligase III gene and deletion mapping of splicing silencer elements by exonuclease III methods. (A) The genomic structure is described in the first paragraphs of Materials and Methods and Results. The filled box represents the last constitutive exon, C, open boxes the alternative terminal exons (β and α) and diagonal lines the alternative splicing patterns, with _Pvu_II and _Ava_II restriction sites and polyadenylation signals, A, also shown. (B) Pre-mRNA splicing substrates are identified above the lanes. A time course of in vitro splicing is shown (20, 40 and 60 min), except for lanes 18–22, where splicing reactions were for 60 min over a range of magnesium concentrations (1–3 mM). Control reactions omitted ATP (and were incubated under splicing conditions for 60 min; lanes 6, 11, 16 and 23) or were kept on ice (lanes 2, 7, 12 and 17). Splicing intermediates and mRNA products are indicated by symbols on the left. Lane 1, pBR322/_Msp_I markers.
RESULTS
The genomic structure of the last three exons of the human DNA ligase III gene indicates that the basis for variant α and β mRNA isoforms is a choice between alternative terminal exons (Fig. 1A). The intron between the last constitutive exon (C) and the β exon was 203 nt, and contained 5′ splice site, branch site, polypyrimidine tract and 3′ splice site sequences that were reasonable matches to consensuses for these elements. An intron of 1134 nt was found from the cleavage site of the β exon to the α exon. DNA ligase III transcription templates with different length truncations from the 3′-end were made by exonuclease III, restriction enzyme and PCR methods. In vitro splicing was detected in HeLa nuclear extracts when truncations reached 16 nt upstream of the β exon polyadenylation signal, close to a _Pvu_II site (Fig. 1B, lanes 3–5). However, no splicing was detected in pre-mRNAs with smaller 3′ truncations (including those at the β exon polyadenylation signal) or when the pre-mRNA was run-off at an _Ava_II site downstream of the β exon (data not shown). This suggested that the intron between the C and β exons contained adequate splicing sites, and that splicing was being silenced by elements between the _Pvu_II and _Ava_II sites.
The region between the _Pvu_II and _Ava_II sites was arbitrarily divided into three parts: from the _Pvu_II site to the polyadenylation signal; from the polyadenylation signal to the cleavage site; and from the cleavage site to the _Ava_II site (Fig. 2A). Each of these parts was added to the end of a truncated DNA ligase III pre-mRNA (A) that was competent for splicing (Fig. 2B, lane 1). The 16 nt exonic element, from near the _Pvu_II site to the polyadenylation signal, substantially silenced splicing of the intron (pre-mRNA B, lane 2), as did the intronic sequence (pre-mRNA D, lane 4). The element between the polyadenylation signal and the cleavage site was at least neutral (pre-mRNA C, lane 3). Additional pre-mRNAs, in which the 16 nt exonic silencer element was replaced with its antisense sequence, showed efficient splicing (data not shown), further ruling out a non-specific effect of exonic length on splicing efficiency. Note that the polyadenylation signal was not present in any of these pre-mRNAs. This suggested that the mechanism of in vitro silencing of DNA ligase III β exon splicing was independent of the polyadenylation signal. To test whether silencing by the exonic silencer element could be relieved by the presence of a 5′ splice site, a consensus 5′ splice site was positioned at the end of the β exon. Pre-mRNA E (lane 5) spliced significantly less efficiently than pre-mRNA F (lane 6) and with similar efficiency to pre-mRNA B (lane 2). Thus, silencing of splicing by the 16 nt element could also occur in the context of a juxtaposed 5′ splice site.
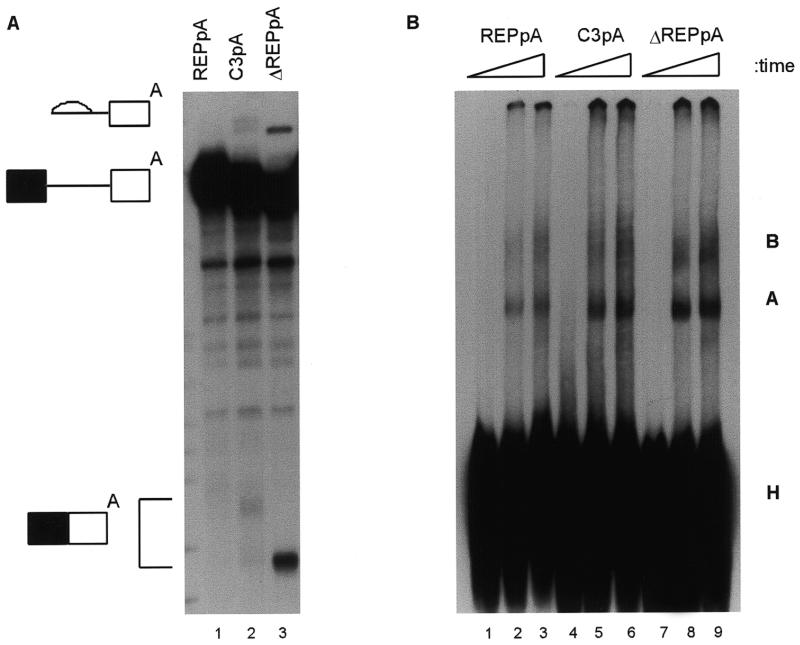
The next question was whether the 16 nt exonic splicing silencer suppressed polyadenylation. Although the element was able to silence splicing in the absence of the polyadenylation signal, another mode of repression might have been through an inhibition of polyadenylation and 3′-end formation (24–26). Therefore, the interaction between splicing and polyadenylation was tested by making pre-mRNAs with the β exon AUUAAA signal at the 3′-end of the exon, either with (REPpA) or without (ΔREPpA) the 16 nt exonic splicing silencer. No splicing was detected with the REPpA pre-mRNA (Fig. 3A, lanes 2–4), whereas efficient splicing was seen when the silencer was deleted (ΔREPpA, lanes 5–7). When cordycepin was omitted from parallel reactions, smears representing polyadenylation were seen above all the pre-mRNAs (lanes 8–12). Polyadenylation was somewhat reduced in the ΔREPpA pre-mRNA (lanes 11 and 12). This was possibly because of consumption of pre-mRNA substrates by the splicing apparatus or because the splicing silencer overlapped with an upstream element of the polyadenylation signal. Thus, reactions were also carried out under conditions where splicing of DNA ligase III was inhibited (Fig. 3B). While some polyadenylation was seen with pre-mRNA REPpA (lane 3), none was detected with ΔREPpA (lane 2). The 16 nt exonic splicing silencer did not inhibit polyadenylation and the primary mechanism of silencing of the β exon by this element did not appear to involve coordinate inhibition of polyadenylation and splicing mechanisms. Instead, the splicing silencer element appeared to overlap with an upstream element of the polyadenylation signal.
The mechanism by which the 16 nt exonic silencer interacts with the splicing apparatus was explored by analysis of spliceosomal complex assembly. Experiments were done with the REPpA and ΔREPpA pre-mRNAs used in Figure 3. A third substrate (C3pA) was also used. This substrate was identical to REPpA except that the third U in the silencer (CCAAGUCAA-AAUUUAC) was changed to a C (CCAAGUCAAAAUCUAC). It had been made as part of an initial systematic mutagenesis in which each U in the 16 nt element was changed to a C. C3pA showed partial relief of silencing, with weakly detectable splicing (Fig. 4A), whereas the other mutant pre-mRNAs spliced undetectably or very inefficiently (data not shown). Splicing reactions in the presence of cordycepin were analysed in parallel on denaturing or non-denaturing polyacrylamide gels. The same hierarchy of splicing activity (REPpA < C3pA < ΔREPpA) was seen for spliceosomal complex assembly (Fig. 4B). The amount of A complex was reduced at every time point for REPpA (lanes 2 and 3), intermediate for C3pA (lanes 5 and 6) and highest for ΔREPpA (lanes 8 and 9). Note that the pre-mRNAs contain only the polyadenylation signal and no downstream sequence, therefore weak binding of cleavage/polyadenylation specificity factor (CPSF) is not enhanced by cooperative interaction with cleavage stimulation factor (27). The weak binding of CPSF is not detectable as a separate complex (28) under these gel conditions.
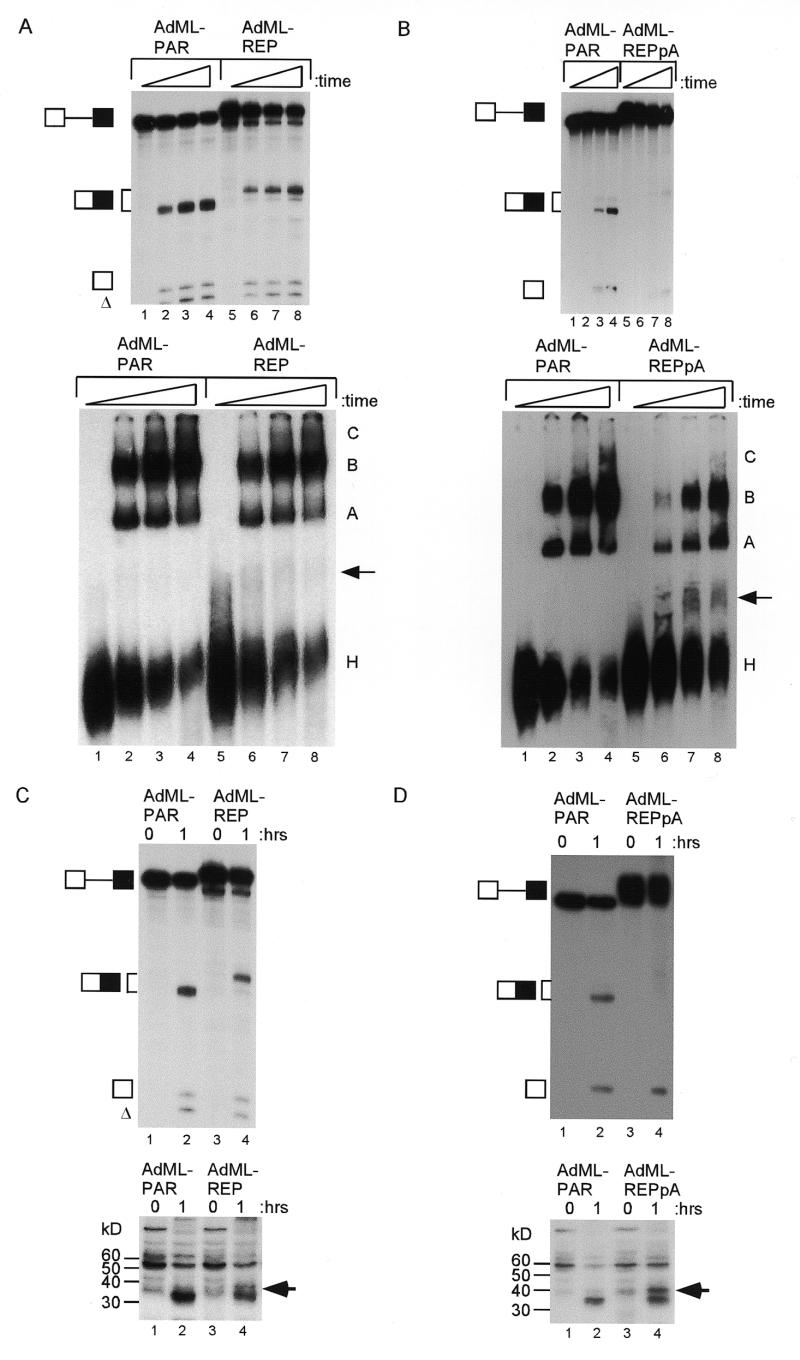
The efficiency of complex assembly for the DNA ligase III substrate was low in general. In addition, it was not clear whether the exonic splicing silencer could function in a context other than that of a DNA ligase III pre-mRNA. Therefore, heterologous pre-mRNAs based on the adenovirus major late (AdML) first and second leader exons were used in splicing experiments. The parental substrate (AdML-PAR) has been optimised to allow purification of spliceosomal complex components (22). The 16 nt exonic silencer was positioned at the end of the second leader (L2) exon, either alone (AdML-REP) or in the context of a downstream AUUAAA (AdML-REPpA) or consensus 5′ splice site GUGAGU (AdML-REP5c). In cognate control pre-mRNAs, the downstream AUUAAA or 5′ splice site were positioned at the end of AdML-PAR (AdML-PARpA and AdML-PAR5c). The efficiency of splicing was reduced when the silencer element was present (Fig. 5A, compare AdML-PAR and AdML-REP, lanes 2 and 3). However, in the context of a downstream consensus 5′ splice site, silencing of splicing was minimal (compare AdML-PAR5c and AdML-REP5c, lanes 4 and 5). In order to assess the effects of the 16 nt element on splicing or polyadenylation, reactions uncoupling the two processes were performed in the context of a downstream AUUAAA (AdML-PARpA and AdML-REPpA, Fig. 5B). Under splicing-only conditions (in the presence of cordycepin), the AdML-REPpA pre-mRNA spliced less efficiently than the control AdML-PARpA pre-mRNA (lanes 3 and 5). Under conditions unfavourable for splicing and in the absence of cordycepin, polyadenylation was detected with the AdML-REPpA pre-mRNA (lane 8), but not AdML-PARpA (lane 7). Note that some level of splicing remained detectable in the polyadenylation reactions, but was still substantially less than parallel splicing reactions (compare lane 3 with 7). An additional, consistent observation was that the accumulation of mRNA product was most affected by the presence of the silencer, with reduction of first step intermediates being less apparent.
To analyse the possible mechanisms of silencing, splicing complex assembly experiments were performed. As before, the splicing efficiency of the AdML-REP pre-mRNA was reduced (Fig. 6A, upper panel, compare lanes 1–4 to lanes 5–8). Parallel non-denaturing gel analysis of spliceosome assembly consistently showed a reduction of A, B and C complexes at all time points with the AdML-REP pre-mRNA (lower panel). In three independent experiments, the average reduction in A and B complexes (quantified relative to the H complex) was 30%. A faint additional complex was identified using the AdML-REP substrate (lanes 6–8, arrow). A substantial silencing of splicing was seen when AdML-REPpA was compared to AdML-PAR (Fig. 6B, upper panel, compare lanes 1–4 with 5–8). Again, complex assembly was reduced (lower panel) and the additional complex was clearly seen with the AdML-REPpA substrate (arrow). Similar results were obtained when spliceosome assembly was compared between AdML-PARpA and AdML-REPpA (data not shown).
Candidate proteins involved in the silencing mechanism were sought by UV cross-linking to universally labelled AdML-PAR and AdML-REPpA pre-mRNAs under splicing or control conditions. Non-specific RNA–protein interactions were then blocked by addition of tRNA, before UV treatment. An aliquot of the reaction was analysed by denaturing gel electrophoresis after removing the protein. Splicing was detected with AdML-PAR pre-RNA and less efficiently with AdML-REP (Fig. 6C, upper panel, lanes 2 and 4). The remaining reactions were digested with RNase A and T1 and proteins were separated by 12% SDS–PAGE, Coomassie stained (not shown) and exposed to film. For both AdML-PAR and AdML-REP pre-mRNAs, the cross-linking pattern was different under splicing conditions (Fig. 6C, lower panel, lanes 2 and 4) compared to control reactions that had been kept on ice (lanes 1 and 3). A protein at ~37 kDa cross-linked more to the AdML-REP pre-mRNA (lane 4, see arrow). As before, when AdML-REPpA was compared to AdML-PAR, silencing of splicing occurred (Fig. 6D, upper panel), with cross-linking of the ~37 kDa protein (lower panel, lane 4, see arrow). This pattern was observed in two additional, separate experiments (data not shown) and also comparing AdML-PARpA and AdML-REPpA substrates (data not shown). Note that silencing of splicing and complex assembly and cross-linking to the ~37 kDa protein consistently appeared stronger in the context of a downstream AUUAAA.
DISCUSSION
The mechanisms of silencing of the human DNA ligase III β exon in HeLa nuclear extracts were first investigated by determining the genomic structure of the terminal exons. The intron between exons C and β appeared to have no theoretical splice site constraint on splicing to the β exon. Indeed, when sequences in the distal part of the β exon were deleted, splicing was detected, showing that there was no intrinsic barrier to splicing of the intron. Instead, two extrinsic silencing elements were detected: a 16 nt exonic sequence immediately upstream and adjacent to the β exon AUUAAA signal; an intronic element downstream of the β exon. The exonic silencer was sufficient to silence splicing of a DNA ligase III pre-mRNA, did not require the polyadenylation signal and could function in the context of a downstream adjacent 5′ splice site. Assembly of early spliceosomal complexes was reduced in both DNA ligase III and AdML pre-mRNA substrates containing the exonic splicing silencer. Increased cross-linking of a 37 kDa protein, under splicing conditions, was seen to AdML pre-mRNAs containing the silencer compared to those without the silencer.
Only a few exonic sequence elements that silence splicing of higher eukaryotic non-viral pre-mRNAs have been identified, although viral pre-mRNAs contain several well-characterised silencers (Table 1). Exonic splicing silencers are a diverse collection of sequences. The intensity of mapping the elements, and their dependence on the context of surrounding sequences, may explain the wide range of their sizes (from 4 to 74 nt in length). However, the sequences themselves vary. Comparison of the DNA ligase III exonic splicing silencer to other exonic silencers reveals little similarity. The sequence CAAG has been mapped in the fibronectin EDA exon (29) and is present in the DNA ligase III exonic element. Also, the sequence AAAUU occurs once in DNA ligase III and in the CD44 v5 (30) and fibronectin EDA (31) exonic silencers. However, it is unknown what importance these sequences may have to silencing function and more detailed mapping and analysis of this and other exonic silencers will be needed to determine this. Our preliminary point mutation data suggest that the third U in the element had some role in silencing, while mutation of the other U nucleotides had little effect. The intronic splicing silencer element downstream of the DNA ligase III β exon was not further defined, and although it is GU-rich, this is often found in this context (just downstream of the cleavage site). This, too, will require more detailed mapping.
Table 1. Exonic splicing silencer sequences.
| Sequence | Gene | Reference |
|---|---|---|
| Non-viral | ||
| UGUGGG | β-TM | (35) |
| UAAGUGUUCUGAGCU | α-TM | (51) |
| CAAG | Fibronectin | (29) |
| UAAG | FGF-2R | (52) |
| AGACAGAAUCAGCACCAGUGCUCAUGGAGAAAAUUGGACC | CD44 | (30) |
| CUAGUAAACUUAUUCUUACGUCUUUCCUGUGUUGCCCUCCAGCUUUUAUCUCUGAGAUGGUCUUCUUUCUAG | IgM | (33) |
| CACUGAUGUGGAUGUCGAUUCCAUCAAAAUUGCCUUGGGAAAGCCCACAGGGGCAAGUUUCCAGGUACAGGGUG | Fibronectin | (31) |
| CCAAGUCAAAAUUUAC | DNA ligase III | This work |
| Viral | ||
| AGAUCCUAGACUAGAGCCCU | HIV-1 tat2 | (53) |
| AGAUCCAUUCGAUUAGUGAA | HIV-1 tat3 | (32,53) |
| GGCUCCCCC | BPV | (38) |
| CCCGAAGGAAUAGUGGAGAGAGAGACAGAGACAGAUCCAUUCGA | HIV-1 tat-rev | (54) |
Despite the disparate nature of the exonic splicing silencers, some similarities in their mechanisms of action may be identified. The DNA ligase III exonic splicing silencer partially inhibits the early steps of spliceosome assembly, including the A complex. This is similar to an effect seen with the HIV-1 tat exon 3 silencer element (32). Partial inhibition of spliceosome assembly was also seen in a heterologous context. However, the reduction in complex formation may not be the only mechanism for splicing silencing, as some complexes were still observed in substrates containing the silencer. Another possible mechanism of silencing may involve later stages of the splicing reaction, for example the second step. Thus, with some substrates, quite high amounts of first step intermediates were seen, despite low levels of second step mRNA products. Indeed, we have recently reported that the IgM M2 exon silencer (33) appears to function in a second step _trans_-splicing assay (34). However, the function of the DNA ligase III β exon silencer has not yet been tested in the second step _trans_-splicing assay. When the exonic splicing silencer was placed in an AdML context, a faint additional complex was seen. This additional complex was evident when the silencer was in the context of a downstream AUUAAA. This extra complex may be similar to the I complex seen with the IgM exonic splicing silencer (33). However, we did not see this complex with a DNA ligase III pre-mRNA. This may be because the AdML pre-mRNA has been optimized for analysis of spliceosome assembly (22), making this a particularly efficient substrate for detection of complexes. We have no evidence that such an I complex was silencing splicing in this context, but if so, it may contain silencer proteins that have been identified by methods such as UV cross-linking (35).
The regulation of splicing of several pre-mRNAs has been shown to involve exonic enhancer and silencer elements, including constitutively spliced exons, for example β-globin (36). How these sequence elements influence the splicing apparatus is not completely understood. Enhancers may bind a family of splicing regulators, the SR proteins, which, in turn, promote the interaction with early spliceosomal components, e.g. U2AF65, with intronic splicing signals such as the polypyrimidine tract (13,37). No such clear model is available for the function of exonic silencers. Although several proteins [including SR proteins (38), PTB (38), hnRNP H (35) and A1 (39)] and snRNPs (33,38) bind to or function with exonic silencers, it is not known how they inhibit splicing or interact with the splicing machinery. We have identified a protein of ~37 kDa by cross-linking to a pre-mRNA containing an exonic silencer. Several RNA-binding proteins lie in the range 34–40 kDa, including hnRNPs A1 (40), A1B (41), A2/B1 (42) and 2H9 (43) and SR proteins hTra2β (44) and SRp40 (45). Some of these proteins are candidates for the cross-linked protein in this study. Studies to identify this protein and its binding site on the pre-mRNA are underway.
Splicing to some terminal exons is linked to polyadenylation, such that an inhibition of polyadenylation also reduces splicing (24–26). However, there was no evidence that the 16 nt exonic silencer element inhibited DNA ligase III splicing indirectly, through interference with polyadenylation. On the contrary, the splicing silencer element appears to overlap with an upstream element, increasing the efficiency of polyadenylation of DNA ligase III or AdML pre-mRNA substrates. We also noted that silencing of splicing of the AdML pre-mRNA was more efficient in the context of the downstream AUUAAA. Other upstream polyadenylation promoting elements may have similar dual or overlapping roles in splicing. The 53 nt upstream element for polyadenylation of the C2 complement pre-mRNA binds PTB (46), a factor implicated in pre-mRNA splicing regulation and silencing (38). A 101 nt element of the mouse histone H2a coding region inhibits cryptic splicing and promotes polyadenylation of intronless β-globin transcripts (47). In the case of the DNA ligase III silencer, there was no evidence of changes in cross-linking to a protein matching the size of PTB. We also do not know if the element binds one factor with opposing functions in splicing and polyadenylation. Two or more factors, each with separate functions in splicing and polyadenylation, may interact with the element.
The identification of exonic and intronic elements that silence splicing to the β exon in HeLa nuclear extracts suggests that the control of DNA ligase III expression and function may involve repression of the β exon in tissues other than the testes, with a relief of such repression during pachytene spermatogenesis. The demonstration of an activity in testes or spermatogenesis capable of relieving this silencing mechanism will be a major step forward in understanding the control of gene expression in the testes and in the process of spermatogenesis in mammals. However, the possibility of an additional enhancer function, elsewhere in the pre-mRNA, during spermatogenesis has not been tested or excluded by this work. Understanding the mechanisms of increased β exon expression in pachytene spermatogenesis would require the analysis of splicing in functional testes- and spermatogenesis-specific tissue extracts. Such reagents are not available, due to technical difficulties (48). Nevertheless, several RNA-binding proteins have been implicated in spermatogenesis, although their function in the process is unknown (10). Some of these are nuclear (49,50), and one speculation is that they may have some role in the control of alternative splicing of pre-mRNAs such as DNA ligase III.
CONCLUSIONS
Two splicing silencer elements have been identified that repress splicing to the DNA ligase III β exon in HeLa nuclear extracts. The exonic element can silence splicing independently of a polyadenylation signal, but appears to overlap with an upstream polyadenylation element. The exonic splicing silencer functions in several sequence contexts, including a heterologous pre-mRNA. Its mechanism of function may include inhibition of assembly of early spliceosomal complexes, and is associated with cross-linking of the pre-mRNA to a protein of ~37 kDa.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to the de Jong laboratory and the UK HGMP Resource Centre for providing the human PAC library, J. Wiggins for technical help, Robin Reed for the gift of pAdMLpar, Adrian Krainer for use of his laboratory and reagents for some experiments, and Adrian Krainer, Hongxiang Liu, Akila Mayeda and Mike Murray for helpful discussions. We are also grateful to the Wellcome Trust for Fellowship support (045401) and to the Joint Research Board of St Bartholomew’s Hospital for additional funding (XMJA).
REFERENCES
- 1.Wei Y.-F., Robins,P., Carter,K., Caldecott,K., Pappin,D.J.C., Yu,G.-L., Wang,R.-P., Shell,B.K., Nash,R.A., Schar,P., Barnes,D.E., Haseltine,W.A. and Lindahl,T. (1995) Mol. Cell. Biol., 15, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., Tomkinson,A.E., Ramos,W., Mackey,Z.B., Danehower,S., Walter,C.I., Schultz,R.A., Besterman,J.M. and Husain,I. (1995) Mol. Cell. Biol., 15, 5412–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackey Z.B., Ramos,W., Levin,D.S., Walter,C.A., McCarrey,J.R. and Tomkinson,A.E. (1997) Mol. Cell. Biol., 17, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 5.Moore M.J., Query,C.C. and Sharp,P.A. (1993) In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, pp. 303–357.
- 6.Wang Y.-C., Selvakumar,M. and Helfman,D.M. (1997) In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. IRL Press, Oxford, UK, pp. 242–279.
- 7.Ryner L.C., Goodwin,S.F., Castrillon,D.H., Anand,A., Villella,A., Baker,B.S., Hall,J.C., Taylor,B.J. and Wasserman,S.A. (1996) Cell, 87, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 8.Finley K.D., Taylor,B.J., Milstein,M. and McKeown,M. (1997) Proc. Natl Acad. Sci. USA, 94, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazelrigg T. and Tu,C. (1994) Proc. Natl Acad. Sci. USA, 91, 10752–10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venables J. and Eperon,I. (1999) Curr. Opin. Genet. Dev., 9, 346–354. [DOI] [PubMed] [Google Scholar]
- 11.Walker W.H., Delfino,F.J. and Habener,J.F. (1999) In Chew,S.L. (ed.), Post-transcriptional Processing and the Endocrine System. Karger, Basel, Switzerland, pp. 34–58.
- 12.Berget S.M. (1995) J. Biol. Chem., 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- 13.Black D.L. (1995) RNA, 1, 763–771. [PMC free article] [PubMed] [Google Scholar]
- 14.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emeson R.B., Hedjran,F., Yeakley,J.M., Guise,J.W. and Rosenfeld,M.G. (1989) Nature, 341, 76–80. [DOI] [PubMed] [Google Scholar]
- 16.Yeakley J.M., Hedjran,F., Morfin,J.-P., Merillat,N., Rosenfeld,M.G. and Emeson,R.B. (1993) Mol. Cell. Biol., 13, 5999–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zandberg H., Moen,T.C. and Baas,P.D. (1995) Nucleic Acids Res., 23, 248–255. [PMC free article] [PubMed] [Google Scholar]
- 18.Lou H. and Gagel,R.F. (1999) In Chew,S.L. (ed.), Post-transcriptional Processing and the Endocrine System. Karger, Basel, Switzerland, pp. 18–33.
- 19.Lou H., Gagel,R.F. and Berget,S.M. (1996) Genes Dev., 10, 208–219. [DOI] [PubMed] [Google Scholar]
- 20.Takagaki Y., Seipelt,R.L., Peterson,M.L. and Manley,J.L. (1996) Cell, 87, 941–952. [DOI] [PubMed] [Google Scholar]
- 21.Yonaha M. and Proudfoot,N.J. (1999) Mol. Cell, 3, 593–600. [DOI] [PubMed] [Google Scholar]
- 22.Gozani O., Patton,J.G. and Reed,R. (1994) EMBO J., 13, 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konarska M.M. (1989) Methods Enzymol., 180, 442–453. [DOI] [PubMed] [Google Scholar]
- 24.Niwa M. and Berget,S.M. (1991) Genes Dev., 5, 2086–2095. [DOI] [PubMed] [Google Scholar]
- 25.Wassarman K.M. and Steitz,J.A. (1993) Genes Dev., 7, 647–659. [DOI] [PubMed] [Google Scholar]
- 26.Gunderson S.I., Vagner,S., Polycarpou-Schwarz,M. and Mattaj,I.W. (1997) Genes Dev., 11, 761–773. [DOI] [PubMed] [Google Scholar]
- 27.Colgan D.F. and Manley,J.L. (1997) Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 28.Zarkower D. and Wickens,M. (1988) J. Biol. Chem., 263, 5780–5788. [PubMed] [Google Scholar]
- 29.Caputi M., Casari,G., Guenzi,S., Tagliabue,R., Sidoli,A., Melo,C.A. and Baralle,F.E. (1994) Nucleic Acids Res., 22, 1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konig H., Ponta,H. and Herrlich,P. (1998) EMBO J., 17, 2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staffa A., Acheson,N.H. and Cochrane,A. (1997) J. Biol. Chem., 272, 33394–33401. [DOI] [PubMed] [Google Scholar]
- 32.Si Z.-H., Rauch,D. and Stoltzfus,C.M. (1998) Mol. Cell. Biol., 18, 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan J.L. and Green,M.R. (1999) Genes Dev., 13, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chew S.L., Liu,H.-X., Mayeda,A. and Krainer,A.R. (1999) Proc. Natl Acad. Sci. USA, 96, 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C.D., Kobayashi,R. and Helfman,D.M. (1999) Genes Dev., 13, 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaal T.D. and Maniatis,T. (1999) Mol. Cell. Biol., 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J.Y. and Maniatis,T. (1993) Cell, 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Z.-M., Huynen,M. and Baker,C.C. (1998) Proc. Natl Acad. Sci. USA, 95, 14088–14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Gatto-Konczak F., Olive,M., Gesnel,M.C. and Breathnach,R. (1999) Mol. Cell. Biol., 19, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riva S., Morandi,C., Tsoulfas,P., Pandolfo,M., Biamonti,G., Merrill,B., Williams,K.R., Multhaup,G., Beyreuther,K. and Werr,H. (1986) EMBO J., 5, 2267–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buvoli M., Cobianchi,F., Bestagno,M.G., Mangiarotti,A., Bassi,M.T., Biamonti,G. and Riva,S. (1990) EMBO J., 9, 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burd C.G., Swanson,M.S., Gorlach,M. and Dreyfuss,G. (1989) Proc. Natl Acad. Sci. USA, 86, 9788–9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahe D., Mahl,P., Gattoni,R., Fischer,N., Mattei,M.-G., Stevenin,J. and Fuchs,J.-P. (1997) J. Biol. Chem., 272, 1827–1836. [DOI] [PubMed] [Google Scholar]
- 44.Tacke R., Tohyama,M., Ogawa,S. and Manley,J.L. (1998) Cell, 93, 139–148. [DOI] [PubMed] [Google Scholar]
- 45.Screaton G.R., Caceres,J.F., Mayeda,A., Bell,M.V., Plebanski,M., Jackson,D.G., Bell,J.I. and Krainer,A.R. (1995) EMBO J., 14, 4336–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreira A., Takagaki,Y., Brackenridge,S., Wollerton,M., Manley,J.L. and Proudfoot,N.J. (1998) Genes Dev., 12, 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y., Wimler,K.M. and Carmichael,G.G. (1999) EMBO J., 18, 1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Branche H., Frappier,D. and Chabot,B. (1991) Nucleic Acids Res., 19, 4509–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elliott D.J., Oghene,K., Makarov,G., Makarova,O., Hargreave,T.B., Chandley,A.C., Eperon,I.C. and Cooke,H.J. (1998) J. Cell Sci., 111, 1255–1265. [DOI] [PubMed] [Google Scholar]
- 50.Venables J., Vernet,C., Chew,S.L., Cowmeadow,R.B., Wu,J., Cooke,H.J., Artzt,K. and Eperon,I.C. (1999) Hum. Mol. Genet., 8, 959–969. [DOI] [PubMed] [Google Scholar]
- 51.Graham I.R., Hamshere,M. and Eperon,I.C. (1992) Mol. Cell. Biol., 12, 3872–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Del Gatto F. and Breathnach,R. (1995) Mol. Cell. Biol., 15, 4825–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amendt B.A., Si,Z.-H. and Stoltzfus,C.M. (1995) Mol. Cell. Biol., 15, 4606–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staffa A. and Cochrane,A. (1995) Mol. Cell. Biol., 15, 4597–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson K. and Moore,M.J. (1997) Science, 276, 1712–1716. [DOI] [PubMed] [Google Scholar]