Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination (original) (raw)
Abstract
The development of an effective human immunodeficiency virus type 1 (HIV-1) vaccine is likely to depend on knowledge of circulating variants of genes other than the commonly sequenced gag and env genes. In addition, full-genome data are particularly limited for HIV-1 subtype C, currently the most commonly transmitted subtype in India and worldwide. Likewise, little is known about sequence variation of HIV-1 in India, the country facing the largest burden of HIV worldwide. Therefore, the objective of this study was to clone and characterize the complete genome of HIV-1 from seroconverters infected with subtype C variants in India. Cocultured HIV-1 isolates were obtained from six seroincident individuals from Pune, India, and virtually full-length HIV-1 genomes were amplified, cloned, and sequenced from each. Sequence analysis revealed that five of the six genomes were of subtype C, while one was a mosaic of subtypes A and C, with multiple breakpoints in env, nef, and the 3′ long terminal repeat as determined by both maximal χ2 analysis and phylogenetic bootstrapping. Sequences were compared for preservation of known cytotoxic T lymphocyte (CTL) epitopes. Compared with those of the HIV-1LAI sequence, 38% of well-defined CTL epitopes were identical. The proportion of nonconservative substitutions for Env, at 61%, was higher (P < 0.001) than those for Gag (24%), Pol (18%), and Nef (32%). Therefore, characterized CTL epitopes demonstrated substantial differences from subtype B laboratory strains, which were most pronounced in Env. Because these clones were obtained from Indian seroconverters, they are likely to facilitate vaccine-related efforts in India by providing potential antigens for vaccine candidates as well as for assays of vaccine responsiveness.
According to World Health Organization estimates, India will have the greatest number of human immunodeficiency virus (HIV)-infected individuals of any country by the end of this decade (1, 6). High rates of sexually transmitted diseases, rapidly increasing seroprevalence in female commercial sex workers, and inadequate facilities for HIV testing, counseling, and prevention are the major contributing factors in the recent explosive increases in the numbers of HIV infections (5, 6, 24, 29). While antiretroviral drugs have reduced mortality from AIDS in developed nations, their effect will be negligible elsewhere due to their cost. For most communicable diseases, vaccines offer the most cost-effective control strategy. It is likely that development of a vaccine for HIV will require knowledge of the viral variants being transmitted in the target population. Despite India’s impending predominance in the worldwide pandemic, little is known of the genetic diversity of HIV-1 in India.
The HIV-1 sequence database is growing exponentially, but the distribution of submitted sequences is not representative of the worldwide picture. Subtype C has been reported in nearly every region affected by HIV-1 (11, 23, 28) and predominates in India, and it also causes 74% of infections in southern Africa and 96% of infections in northern Africa (11, 18, 32). Given the combined population of India and the other regions affected, subtype C is likely to be the most commonly transmitted HIV-1 subtype worldwide. In contrast, 7% of the available HIV-1 sequence data is from subtype C-infected individuals (37), and of the 46 completely sequenced HIV-1 genomes (excluding multiple derivatives of HIV-1LAI), only two are of subtype C, one from a 1992 Brazilian sample and the other from a 1986 Ethiopian sample (37). In November 1997, an analysis of cross-clade epitope variation (9) excluded the C clade from evaluation of p24_gag_ epitopes because of a lack of sequence data, whereas there was sufficient data to analyze subtypes A, B, D, F, G, and H (no HIV-1 harboring a subtype E gag gene has been found). Further sequence data from subtype C is needed, but the past approach of generating data from small subgenomic amplicons is no longer sufficient.
Recent developments have made full-genome characterization of HIV-1 isolates both important and feasible. First, the recognition of intersubtype recombination in a significant proportion of HIV-1 sequences (44, 45) has led to detection of mosaic genomes in many regions of the world affected by multiple subtypes (14, 17, 31). Subtypes A, B, and C in India have been reported (4, 22, 30, 31, 59), but mosaic HIV-1 there has not been reported. The existence of such recombinants makes characterization of variants by analyzing subgenomic segments incomplete. Second, immune responses to vaccines based on single genes such as env have been limited (13), and attention is being shifted toward multivalent vaccines that incorporate other gene products. Third, interactions among discontinuous regions of the genome, such as between the long terminal repeat (LTR) and pol (26), can be detected only when such regions can be analyzed from the same template.
In an effort to characterize subtype C virus genomes being transmitted currently in India, viral isolates were obtained from individuals with seroincident infections in India. Three of the isolates (collected in 1994 and 1995) were known to be non-syncytium inducing (NSI) and therefore resembled viruses transmitted through unprotected sexual contact, which account for 75 to 85% of new infections (2, 15, 61). These isolates were cloned, and nearly full-length genomic sequences were determined. Detailed sequence analysis was performed, as was an analysis of variation in characterized cytotoxic T lymphocyte (CTL) epitopes.
MATERIALS AND METHODS
Study subjects and virus isolates.
As part of an ongoing prospective study of HIV seroconversion, HIV-1 was cultured from peripheral blood samples collected from six seroconverters from the city of Pune in western India (Table 1). Three subjects were identified in 1993 by indeterminate Western blotting result, which developed to a fully positive result over a period of months. The other three subjects were initially seronegative during follow-up in a sexually transmitted disease clinic but were found to be seropositive at a later visit. While none of the subjects had symptoms associated with HIV infection, all three subjects identified in 1994 and 1995 had syphilis and genital ulcer disease.
TABLE 1.
Characteristics of study subjects
| Isolate | Age (yr), gendera | Risk factorb | Date HIV-negative or indeter-minate sample obtained (day-mo-yr) | Date HIV-positive sample obtained (day-mo-yr) | Date sample for coculture obtained (day-mo-yr) |
|---|---|---|---|---|---|
| 93IN904 | 28, F | Transfusion | 17-Sep-92 | 04-Nov-92 | 27-Mar-93 |
| 93IN905 | 23, F | Transfusion | 17-Jun-92 | 30-Sep-92 | 27-Mar-93 |
| 93IN999 | 52, M | Sex with men and CSW | 09-Nov-92 | 22-Dec-92 | 24-Apr-93 |
| 94IN11246 | 26, M | Sex with CSW | 23-May-94 | 15-Oct-94 | 25-Oct-94 |
| 95IN21068 | 21, M | GUD | 13-Apr-94 | 01-Aug-94 | 18-Feb-95 |
| 95IN21301 | 40, M | Sex with CSW | 24-Jun-94 | 01-Feb-95 | 03-Apr-95 |
Viruses were propagated by short-term cocultivation of the subjects’ peripheral blood mononuclear cells (PBMCs) with phytohemagglutinin-stimulated PBMCs from healthy donors (51). The samples obtained in 1994 and 1995 were tested for in vitro phenotype, and they were all NSI. High-molecular-weight DNA was extracted from infected cells using the Easy DNA kit as recommended by the manufacturer (Invitrogen, Carlsbad, Calif.).
Heteroduplex mobility analysis.
Subtype determinations by heteroduplex mobility analysis (HMA) were performed according to the WHO heteroduplex mobility analysis protocol (version 3), using supplied reagents (3, 16, 22). This kit was obtained from the NIH Research and Reference Reagent Program (catalog no. 2751).
Large template amplification and cloning.
Virtually full-length HIV-1 genome amplification was performed with the LTR primers MSF12 (5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′) and MSR5 (5′-GCACTCAAGGCAAGCTTTATTGAGGCT-3′) described previously (48), which amplify the entire HIV-1 genome except for 75 bp in the U5 region of the LTR. They are located at nucleotide positions 623 through 646 and 521 through 547 in HIV-1LAI, respectively. PCR was performed with the Expand long template polymerase preparation (Boehringer Mannheim, Indianapolis, Ind.) in a volume of 50 μl overlaid with mineral oil. The reaction mixtures were incubated in a PTC-100 thermal cycler (MJ Research, Watertown, Mass.) in thin-walled tubes. DNA templates from PBMC cocultures were titrated over a set of dilutions (500, 250, 50, and 10 ng) to obtain a 9-kb amplification product with a minimum number of nonspecific bands and to minimize the number of input templates. The thermal cycling consisted of 94°C for 2 min followed by 10 cycles of 94°C for 10 s, 62°C for 30 s, and 68°C for 10 min. This was followed by 20 cycles of 94°C for 10 s, 55°C for 30 s, and 68°C for 10 min. The final incubation was at 72°C for 10 min. Products of amplification reactions were analyzed on 0.6% agarose gels (SeaKem GTG; FMC Biolabs), and the 9-kb bands were gel purified by using a Qiaquick gel purification kit (Qiagen, Chatsworth, Calif.). The gel-purified PCR products were cloned by using the pCR 2.1 vector in the TA cloning kit (Invitrogen), and colonies were picked at random. Transformed bacteria were incubated at 25 to 30°C. The clones were screened for plasmids carrying a 9-kb insert on the basis of their electrophoretic migration and banding pattern after _Eco_RI digestion, and a group of clones with the same banding pattern was called a clonotype. One clone, representing the predominant clonotype for each isolate, was chosen for automated sequencing. Sequencing of both strands was performed by using cycle sequencing and dye termination on an automated sequencer (Applied Biosystems, Inc., Foster City, Calif.).
Sequence analysis.
Sequenced fragments were assembled into contiguous sequences, and a consensus of the two strands was formed by using the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.). A representative genome for each major subtype was selected from a current reference list (33). Sequences were aligned by using Clustal W (56), and the alignments were edited manually in Vised version 1.2 (40), in order to shift gaps to restore codons and perform translation.
For phylogenetic tree construction, sites containing a gap in any aligned sequence were removed (gap stripped), as were areas of ambiguous alignment, and each alignment was then used to generate a distance file by using DNADIST from the PHYLIP package (20) (maximum likelihood option; transition/transversion ratio of 1.5) and subjected to a bootstrap analysis (19) by using SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE. The original distance matrix was then used as input for NEIGHBOR to generate a final phylogenetic tree, which was visualized by using Tree View (39).
Analysis for intersubtype mosaicism.
SimPlot, an interactive 32-bit software program for Microsoft Windows computers, was created to plot similarity versus position (41) and is similar in purpose to the Recombination Inference Program (RIP) for UNIX computers (52). The results of SimPlot were equivalent to the graphical output from RIP (data not shown). Briefly, SimPlot calculates and plots the percent identity of the query sequence to a panel of reference sequences in a sliding window, which is moved across the alignment in steps. The window and step sizes are adjustable.
Evidence of mosaicism led to more extensive analysis, in which the env gene was aligned to consensus sequences for subtypes A and C (with subtype B as an outgroup) from a 0% threshold consensus env alignment (from http://hiv-web.lanl.gov/RIP/BACKGD_ALIGNMENTS/). The aligned sequence was then compared to the 50% consensus file, to exclude poorly conserved sites. Alignments were analyzed for recombination breakpoints by maximization of χ2 as previously described (44, 53). Briefly, the SimPlot program was used to identify informative sites as described by Robertson et al. (44). Phylogenetically informative sites in this context are those at which four taxa are divided equally into two groups, each of which has identity at that site. Each informative site supports one of three possible phylogenetic relationships among the four taxa, and a cluster analysis maximizing the value of χ2 is then used to select breakpoints among the clusters. P values for the resultant divisions of sites were calculated by using Fisher’s exact test. These breakpoints were used to divide the alignment into segments for phylogenetic tree construction as described above.
Analysis of epitope sequence variation.
Predicted protein sequences were compared to the optimal epitope sequences for the best-defined HIV CTL epitopes (8), which are based on the subtype B isolate HIV-1LAI, also known as human T-cell leukemia virus type IIIB (HTLV-IIIB). The epitopes were compared to the corresponding sequences for the Indian isolates, and each was scored as identical, different due to conservative changes only, or different due to at least one nonconservative change. Conservative changes were changes within one of six physicochemical groups as described by George, Hunt, and Barker (27). For comparison, protein sequence distances were calculated for entire genes by using the program PROTDIST in the PHYLIP package. To count all differences, the Kimura distance option was used, whereas to count only physiochemical differences the Categories (George/Hunt/Barker) option was used.
Nucleotide sequence accession numbers.
The sequences described here have been submitted to GenBank and assigned accession no. AF067154 through AF067159.
RESULTS
Amplification of virtually full-length genomes from India.
PCR amplification of cocultured virus resulted in visible bands of the proper size in ethidium bromide-stained agarose gels for the isolates from all six of the HIV-1-infected subjects from India. After ligation and transformation, 7 to 14 clones for each of the six isolates were found to contain inserts of at least 9 kb. _Eco_RI digestion allowed selection of one representative of the predominant clonotype for each subject, and herein these six clones are referred to by the identifier of the source subject. A BLAST search did not reveal any evidence of sample contamination. For the entire genome, the mean intersubject sequence diversity among these six clones was 5.6% (range, 2.8 to 9.0%), while the mean difference between these isolates and the Ethiopian subtype C reference clone C2220 was 9.4% (range, 8.8 to 11.3%).
Evidence for mosaicism.
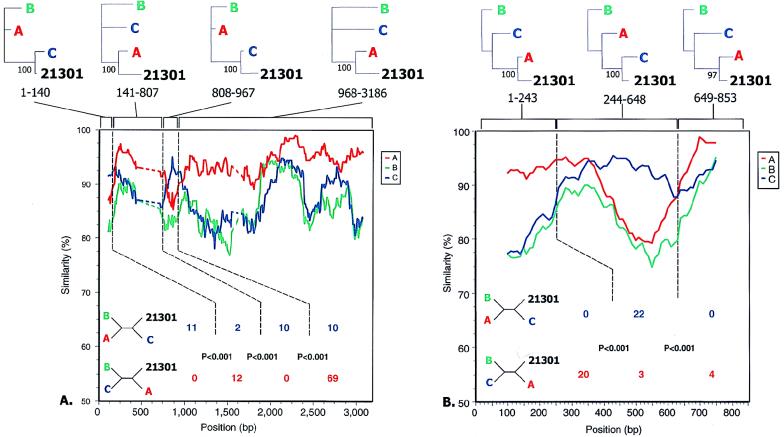
We performed genome-wide comparisons of all clones to available full-length reference sequences in order to determine the viral subtype, to detect evidence of intersubtype recombination, and to identify potential breakpoints for any such events (Fig. 1). The reference sequences chosen for subtypes A (92UG037), B (RF), C (C2220), D (NDK), F (93BR020), and H (90CF056) are ones for which there is no evidence of mosaicism. There is no nonmosaic sequence available for subtype E, and the mosaicism of the available sequence for subtype G is debated, so well-characterized sequences were selected (10, 25). Isolate 93TH253 is a prototypical A/E recombinant, composed primarily of subtype A sequence, with the areas assigned to subtype E limited to vif, vpr, env, and nef. The subtype G isolate 92NG083.2 was recently described as an A/G recombinant, with subtype A homology in the accessory gene (vif and vpr) regions (25, 43). Available sequences for subtypes I and J were too short to merit inclusion in Fig. 1. Five of the six clones were most similar to subtype C throughout the entire genome, a finding supported by subsequent analyses, described below. However, while clone 95IN21301 is predominantly subtype C, it shows greater similarity to the subtype A sequence in the 3′ half of the genome: in env and nef and in the U3-R region of the 3′ LTR.
FIG. 1.
Plots of similarity (generated by SimPlot) of a set of reference sequences to the 93IN999 (upper panel) and 95IN21301 (lower panel) genomes. Each curve is a comparison between the genome being analyzed and a reference genome. Each point plotted is the percent identity within a sliding window 600 bp wide centered on the position plotted, with a step size between points of 20 bp. Positions containing gaps were excluded from the comparison. The horizontal bars above the curves are a cartoon of the open reading frames of the HIV-1 genome, arranged as indicated in Fig. 3. The colors are consistent with those used for the similarity curves and indicate the subtype to which that part of the genome is most similar based on the adjacent similarity plot. Results for the remaining four genomes discussed in this report were consistent with those for 93IN999.
More detailed analysis of the env and nef and LTR regions of 95IN21301 was performed to confirm mosaicism and determine recombination breakpoints. Similarity plots of the 95IN21301 sequence with subtype consensus sequences suggested the presence of five points of crossover (Fig. 2A and B). By the method of maximization of χ2, the most likely breakpoints were located (44, 53). These positions, as well as the subtype assignments, were corroborated when phylogenetic trees with bootstrap analysis were constructed for the resulting regions (Fig. 2A and B), though the small number of informative sites at the 3′ end of nef makes the precise site of crossover difficult to determine. The results of this detailed analysis, which are depicted in Fig. 3, show that 95IN21301 is distinct from the only two previously reported A/C recombinant genomes, ZAM184 and 92RW009.6.
FIG. 2.
(A) Similarity plot as in Fig. 1 for the env gene of isolate 95IN21301, with a window size of 200 bp and a step size of 10 bp. The subtype reference sequences were majority (50%) consensus sequences for each of the subtypes, obtained from the Los Alamos web site (http://hiv-web.lanl.gov). The dashed regions indicate areas in V1 and V2 in which less than 50% of the sites could be compared due to gaps or lack of subtype consensus. Dotted vertical lines indicate breakpoints identified by maximization of χ2 as described in Materials and Methods, with numbers of informative sites shared by 95IN21301 and the subtype in each bounded region indicated below in the color assigned to that subtype. P values were calculated by using Fisher’s exact test. Four-member trees consistent with these sites are shown at the left. Above are phylogenetic trees for each region bounded by the recombination breakpoints showing the proportion of 100 bootstrapped trees surrounding the indicated relationship. The predicted gp120/gp41 processing site is at base 2044 in this alignment. (B) Similarity plot as in panel A for the nef gene and the U3/R LTR region of isolate 95IN21301. The LTR begins at position 296 in this alignment, and the nef termination codon is at position 634. Subtype majority consensus sequences were determined by using SF170, U455, and UG037 (subtype A), RF, MN, and TH475 (subtype B), and C2220, BR025, 93IN904, 93IN905, 93IN999, 94IN11246, and 95IN21068 (subtype C).
FIG. 3.
Cartoon depicting the subtype assignment of each region of the HIV-1 genome for all characterized A/C recombinant genomes.
Initial analysis for mosaicism suggested the presence of subtype E and G sequences in a small portion of the 3′ half of the 95IN21301 env gene, but difficulty separating subtypes A, G, and E in this region has been reported previously and may be the result of a recent common ancestry of this region of the genome as well as diversity within the A subtype (52). Further analysis provided no statistical support for the presence of a subtype other than subtype A in this region (data not shown).
To exclude the possibility that the recombination event was an artifact that occurred during coculture or PCR amplification (7), uncultured PBMCs from subject 21301 were obtained. Genomic DNA was isolated, and PCR was performed with the ED5 and ED12 primers, which span the V1 to V5 region of env, provided in the HMA kit (16). Direct sequencing was performed. The resulting sequence was 97.2% similar to the 95IN21301 sequence, in contrast to the other five isolates, which showed 80.7 to 82.5% similarity to the uncultured 21301 sequence. Intersubtype recombination analysis revealed the same subtype A and C breakpoints (data not shown). Therefore, this intersubtype mosaic was not an artifact of in vitro manipulation.
The extreme 3′ end of the 95IN21301 sequence appeared to have a final subtype A to C breakpoint. Consistent with the data for the nef gene, the overlapping U3 region was highly similar to subtype A sequences and contained only two NF-κB binding motifs (data not shown), whereas the subtype C LTR characteristically contains three NF-κB sites (60). In contrast, the TAR region of 95IN21301 was most similar to subtype C and contained a three-base TAR bulge rather than the two-base bulge seen in subtype A (26). The location of a breakpoint between the U3 and R regions would be consistent with reverse transcription of a genome with a subtype C 5′ LTR and a subtype A 3′ LTR. The 5′ LTR serves as the template for the R region of the resultant 3′ LTR.
Comparison of sequence-based and HMA-based subtyping.
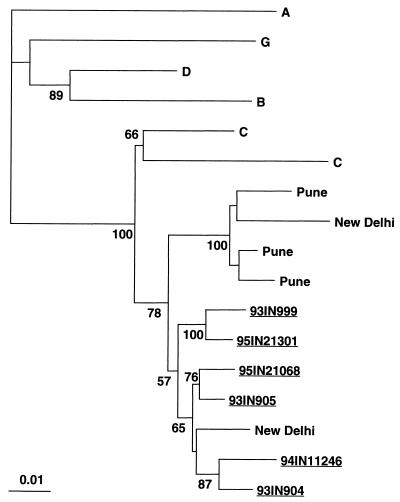
The presence of a subtype A env sequence in cloned 95IN21301 was not surprising, as preliminary HMA for the V3 to V5 region of env had shown that the isolate from which this clone was derived was most closely related to subtype A, while the isolates from IN21068 and IN11246 were most closely related to subtypes C2 (Zambia) and C3 (India), respectively (data not shown). Construction of a phylogenetic tree from the region assessed by the HMA revealed that all of the clones clustered with the subtype C3 (India) prototype except for clone 95IN21301, which clustered with subtype A (Fig. 4).
FIG. 4.
Phylogenetic tree based on the env gene sequences compared in the HMA reaction used to identify genotypes. The tree is based on 834 sites that remained after gap stripping of the alignment predicted for the ED5 to ED12 PCR product. Numbers at nodes indicate clades supported in more than 50 of 100 bootstrapped trees, and the scale for genetic distance is indicated below. The prototype sequences for the subtypes indicated were as follows: A1, RW020; A3, SF170; C1, MA959; C2, ZAM18; C3, IN868; and C4, BR025.
Detailed characterization of genomic sequences.
Having determined the genetic subtypes of the clones, we turned to an analysis of the sequence features of each of the clones. There were very few lethal mutations. Conceptual translation of the eight recognized genes of HIV-1 revealed that these reading frames were all open for clones 94IN11246, 95IN21301, 93IN904, 93IN905, and 93IN999. In contrast to the other clones, 95IN21068 had what is expected to be a lethal mutation, a premature termination codon in pol 57 bp downstream from the putative gag-pol frameshift site (37). The other potentially lethal mutation was in clone 95IN21301, an A-to-G transition in the 3′ splice acceptor site of the tat-rev intron.
In addition to the mutations expected to be lethal, the five nonmosaic subtype C clones contained mutations which have been noted in other isolates and are unlikely to be lethal. All of these clones have a premature termination codon at 251 bp downstream from the splice acceptor site. This termination appears to be a feature of the C subtype, present in all of the subtype C clones presented here as well as in previously sequenced subtype C genomes C2220 (47) and BR025 (25). It is unlikely to be lethal, since in vitro data have led some authors to state that this region of rev has no functional significance (34). In clone 93IN999 the vpu start codon is replaced by ATA, a feature shared by many previously reported isolates, which is thought to modulate the relative expression of Vpu and Env from the same spliced mRNA (55). Clone 94IN11246 has an eight-base deletion at nucleotide 572 of nef, resulting in a frameshift and an open reading frame that is 136 bp longer than usual. The impact of this is difficult to predict, but the deletion does not affect the SH3 binding motifs and alters only the last 19 amino acids of the usual open reading frame.
Another characteristic of subtype C genomes is the presence of three NF-κB binding motifs, one more than usual (36). All but two available subtype C LTR sequences have three NF-κB binding motifs (60). The two exceptions have two (92BR025.8) and four (C-Altr) such sites. It is noteworthy that clone 95IN21301 lacks both the rev truncation feature and the extra NF-κB site feature of subtype C, both of which fall in areas assigned to subtype A in the analysis above.
We investigated the known pol sequence markers for drug resistance, with the caveat that since reported resistance markers are based almost entirely on subtype B sequences their relevance to subtype C remains to be determined. Using the markers summarized by Mellors et al. (35), we searched the reverse transcriptase (RT) and protease (PR) genes for known resistance mutations. In this anti-retroviral drug-naïve cohort, none of the recognized RT resistance mutations was present, consistent with a previous report about subtype C isolates in Africa (50). Markers of PR inhibitor resistance (K20R, M36I, D60E, and L63P) were present, but these are felt to be minor and are commonly seen in drug-naïve individuals (12, 49).
Relationship to previously reported Indian HIV-1 sequences.
The degree to which these sequences represent variants circulating in India was studied through construction of a series of phylogenetic trees that included sequences previously obtained in India by other groups. A tree constructed with pol sequences obtained from New Delhi and Pune (54, 57) (Fig. 5) demonstrates strong clustering of the sequences discussed in this report with subtype C isolates from other cities in India but segregation of the sequences from subtype C isolates from Brazil and Ethiopia. Similar trees were obtained by using smaller gag sequences from samples from a group of Indian expatriates in Kuwait (58), env genes from samples collected in New Delhi and Pune (57), and unpublished nef sequences from samples collected in northern India (GenBank accession numbers Y15116 to Y15123). Based on these trees, the sequences reported here are more similar to previously reported sequences from strains circulating in India than to that of Ethiopian subtype C strain C2220. It is also notable that the (mosaic) 95IN21301 sequences for gag and pol are clearly related to the other Indian subtype C sequences, suggesting that the recombination event occurred in India. These results are consistent with the possible existence of a distinct Indian clade within subtype C as previously postulated (54).
FIG. 5.
Phylogenetic tree comparing the sequences reported here to Indian sequences reported previously for pol (692 bp) (54), with bootstrap values greater than 50% indicated. The representatives for each of the subtypes were as follows: A, U455; B, RF; C, C2220 and BR025; D, NDK; and G, 92NG083.
For gag, the mean genetic distance between the 1993 isolates and the 1994 and 1995 isolates was 3.2% (range, 2.1 to 4.4%), which is intermediate between the value of 4.1% (3.0 to 4.7%) among the 1994 and 1995 isolates and that of 3.0% (2.3 to 3.4%) among the 1993 isolates. These results suggest that the older isolates were more closely related to one another, that the more recent isolates have diverged from the earlier isolates, and that the more recent isolates are diverging from each other as well, consistent with general estimates of 1% divergence per year for HIV-1 (38).
Analysis of amino acid variation in known epitopes.
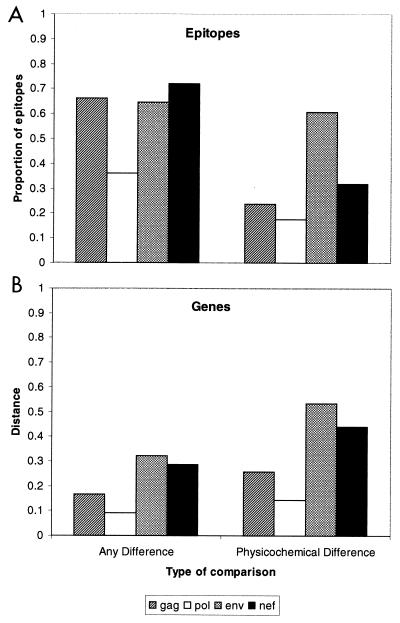
Genomic sequences offer an opportunity to assess conservation of known CTL epitopes that may have an impact on vaccine effectiveness. In order to provide an estimate of cross-clade CTL epitope conservation, the predicted protein sequences of these viruses were compared to the optimal epitope sequences for the best-defined HIV CTL epitopes, which are based on the subtype B isolate HIV-1LAI (8). Of 100 HIV-1 CTL epitopes, 38, 18, 23, and 21 are located in Gag, Pol, Env, and Nef, respectively. The results of this analysis are summarized in Table 2. Because nonconservative changes are most likely to abrogate major histocompatibility complex or T-cell receptor binding, we compared the proportion of epitopes containing nonconservative changes with that of epitopes that were identical or differed by only conservative changes. These data suggest that HIV isolates from India have strong similarity in 77, 78, and 70% of known CTL epitopes in Gag, Pol, and Nef, respectively, while in Env they share strong similarity in only 48% of these epitopes (P < 0.001 for each pairwise comparison). This difference was not entirely due to a greater degree of variation in Env, because the proportions of epitopes which were identical were 34, 64, 28, and 36% for Gag, Pol, Nef, and Env, respectively. These findings suggest that when a mutation appears in an epitope, it is more likely to be nonconservative if it is located in Env.
TABLE 2.
Epitope sequence comparisons between defined optimal HIV-1 epitopes and Indian clones
| Resulta | Gag | Pol | Env | Nef | Total (%) |
|---|---|---|---|---|---|
| Identical | 34 | 64 | 36 | 28 | 230 (38) |
| Conservative | 43 | 19 | 4 | 40 | 173 (29) |
| Nonconservative | 24 | 18 | 61 | 32 | 197 (33) |
DISCUSSION
We report the first virtually full-length genomic HIV-1 sequence data from India, with evidence that mosaic virus is present and that A and C intersubtype recombination has occurred. These clones were obtained from replicative virus of known HIV seroconverters and are therefore more likely to reflect HIV transmitted in India than previously available genomic sequences. Moreover, the addition of the sequences presented here more than doubles the number of published subtype C genomes.
That mosaic HIV-1 is present in India is important but not surprising. Subtypes A and C have been present in India since at least 1992 (11). HMA analysis has been used to detect the presence of HIV-1 subtypes A, B, and C as well as HIV-2 in this cohort in Pune (16). We are unaware of other A and C mosaic genomes with a breakpoint pattern similar to the one presented here. In the Thailand epidemic, an early balance between a subtype B strain and an A-E recombinant was quickly shifted to dominance of the recombinant variant (58). A similar A-E mosaic variant, apparently a descendant of the same recombination event, has been found in an infected individual in the Central African Republic, but to date a purely subtype E genome has not been found (26). This leads to speculation that the recombinant variant had some significant advantage over the parental E strain and that the nonrecombinant virus was eliminated by selection. Such advantages may include alterations in tropism, replication efficiency, or immune recognition. As with influenza virus, recombination may allow more efficient transmission in an exposed population.
Initial characterization of the A-C recombinant by HMA suggested that it was a subtype A isolate, illustrating the limitations of characterization of isolates on the basis of a single genomic region. In addition, the implications and accuracy of HMA for intrasubtype assignments are unknown. The phylogenetic results differed from the HMA results for subject 95IN21068; therefore, distinctions finer than subtype may not always be reliable when this technique is used.
While subtype assignment is useful for epidemiologic and virologic investigation, its relevance to vaccine development is unclear. Recent reports have demonstrated strong cross-subtype CTL recognition but have not addressed the degree to which such cross-clade responses vary from gene to gene or among large numbers of subjects (9, 21, 46). Our data suggest that there are significant differences in the degree of epitope variation among the genes, especially when the ratio of nonconservative changes to conservative changes in epitopes is considered. The present study was not designed to determine the impact of such variation.
To assess variation in relevant CTL epitopes, we utilized a database of well-characterized epitopes (8). This database is a valuable resource, but it is important to note its inherent biases. It is not generally feasible to perform CTL assays of PBMCs from infected individuals by using their HIV-1 quasispecies as the target antigen, as we have done previously (42). Instead, standard assays use laboratory strains like HIV-1LAI (also known as HTLV-IIIB), resulting in detection of T-cell clones which recognize more conserved epitopes. This is demonstrated by the use, with only two exceptions, of HIV-1LAI as the prototype sequence of the 100 best-characterized HIV-1 epitopes in the database (8). As a result, this epitope analysis is likely to be biased toward conserved epitopes.
Based on the epitope analysis, CTL-mediated immunological pressure does not appear to alter the relative rate of fixation of nonconservative substitutions compared with such substitutions in the overall genes, whether the method of counting substitutions assesses every change or only nonconservative substitutions (Fig. 6). This may be due to constraints on nonconservative changes due to their impact on protein function. In contrast, conservative changes in epitopes were distributed among the genes in a pattern that was quite different from changes in the overall gene sequences (Fig. 6). One explanation for these differences is that immunological pressure has a tendency to select for variants with conservative changes in epitope sequences. Conservative changes in epitopes may have a greater impact on CTL recognition than is appreciated from in vitro studies.
FIG. 6.
Distributions of amino acid sequence differences in epitopes (A) and overall gene sequences (B). All differences in sequences are shown on the left, while only nonconservative differences identified based on physicochemical properties are shown on the right.
We report the first HIV-1 full-length genomes from infected individuals in India. Five of them are of subtype C, and one is a new subtype A-C mosaic. Although two of the genomes appear to be defective, and they come from a single city in India, these sequences are genetically representative of the breadth of subtype C sequences previously reported from various regions of the country. These clones, particularly since they come from newly infected individuals, may be important in the development of an effective vaccine for use in India.
ACKNOWLEDGMENTS
We thank the patients and staff of the health clinics in Pune for providing the clinical samples and Dale Dondero, Michel Lubaki, and Melchior Mwandagalinwa Kashamuka. Critical review of the manuscript by Beatrice Hahn is gratefully acknowledged.
This investigation was supported by the Fogarty International Center, National Institutes of Health (NIH), Program of International Training Grants in Epidemiology Related to AIDS, D43 TW0000, and by the HIVNET contract with Family Health International (FHI) with funds from the National Institute of Allergy and Infectious Diseases (NIAID), NIH grant N01-AI-35173-113 and NIAID grant 1 R01 AI41369-01A1.
REFERENCES
- 1.Anonymous. Global AIDS surveillance. WHO Weekly Epidemiological Record. 1993;68(27):195–196. [Google Scholar]
- 2.Anonymous. World Health Report. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 3.Bachmann M H, Delwart E L, Shpaer E G, Lingenfelter P, Singal R, Mullins J I. Rapid genetic characterization of HIV type 1 strains from four World Health Organization-sponsored vaccine evaluation sites using a heteroduplex mobility assay. WHO Network for HIV isolation and characterization. AIDS Res Hum Retroviruses. 1994;10:1345–1353. doi: 10.1089/aid.1994.10.1345. [DOI] [PubMed] [Google Scholar]
- 4.Baskar P V, Ray S C, Rao R, Quinn T C, Hildreth J E, Bollinger R C. Presence in India of HIV type 1 similar to North American strains. AIDS Res Hum Retroviruses. 1994;10:1039–1041. doi: 10.1089/aid.1994.10.1039. [DOI] [PubMed] [Google Scholar]
- 5.Bollinger R C, Brookmeyer R S, Mehendale S M, Paranjape R S, Shepherd M E, Gadkari D A, Quinn T C. Risk factors and clinical presentation of acute primary HIV infection in India. JAMA. 1997;278:2085–2089. [PubMed] [Google Scholar]
- 6.Bollinger R C, Tripathy S P, Quinn T C. The human immunodeficiency virus epidemic in India. Current magnitude and future projections. Medicine (Baltimore) 1995;74:97–106. doi: 10.1097/00005792-199503000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Bradley R D, Hillis D M. Recombinant DNA sequences generated by PCR amplification. Mol Biol Evol. 1997;14:592–593. doi: 10.1093/oxfordjournals.molbev.a025797. [DOI] [PubMed] [Google Scholar]
- 8.Brander C, Walker B D. Systematic identification of optimal HIV-1 CTL epitopes. In: Korber B T, Moore J P, Brander C, Walker B D, Haynes B, Koup R, editors. HIV molecular immunology database. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1997. pp. 1–11. [Google Scholar]
- 9.Cao H, Kanki P, Sankale J L, Dieng-Sarr A, Mazzara G P, Kalams S A, Korber B, Mboup S, Walker B D. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 11.Cassol S, Weniger B G, Babu P G, Salminen M O, Zheng X, Htoon M T, Delaney A, O’Shaughnessy M, Ou C Y. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res Hum Retroviruses. 1996;12:1435–1441. doi: 10.1089/aid.1996.12.1435. [DOI] [PubMed] [Google Scholar]
- 12.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 13.Connor R I, Korber B T, Graham B S, Hahn B H, Ho D D, Walker B D, Neumann A U, Vermund S H, Mestecky J, Jackson S, Fenamore E, Cao Y, Gao F, Kalams S, Kunstman K J, McDonald D, McWilliams N, Trkola A, Moore J P, Wolinsky S M. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp 120 subunit vaccines. J Virol. 1998;72:1552–1576. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J the UNAIDS Network for HIV Isolation and Characterization. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:8209–8212. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelissen M, Mulder-Kampinga G, Veenstra J, Zorgdrager F, Kuiken C, Hartman S, Dekker J, van der Hoek L, Sol C, Coutinho R, Goudsmit J. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J Virol. 1995;69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 17.Diaz R S, Sabino E C, Mayer A, Mosley J W, Busch M P the Transfusion Safety Study Group. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J Virol. 1995;69:3273–3281. doi: 10.1128/jvi.69.6.3273-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietrich U, Grez M, von Briesen H, Panhans B, Geissendorfer M, Kuhnel H, Maniar J, Mahambre G, Becker W B, Becker M L. HIV-1 strains from India are highly divergent from prototypic African and US/European strains, but are linked to a South African isolate. AIDS. 1993;7:23–27. doi: 10.1097/00002030-199301000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP (Phylogeny Inference Package, version 3.572c). Seattle, Wash: Department of Genetics, University of Washington; 1995. [Google Scholar]
- 21.Ferrari G, Humphrey W, McElrath M J, Excler J L, Duliege A M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadkari D, Moore D, Sheppard H W, Kulkarni S S, Mehendale S M, Bollinger R C. Transmission of genetically diverse strains of HIV-1 in Pune, India. Indian J Med Res. 1998;107:1–9. [PubMed] [Google Scholar]
- 23.Galai N, Kalinkovich A, Burstein R, Vlahov D, Bentwich Z. African HIV-1 subtype C and rate of progression among Ethiopian immigrants in Israel. Lancet. 1997;349:180–181. doi: 10.1016/S0140-6736(05)60982-8. [DOI] [PubMed] [Google Scholar]
- 24.Gangakhedkar R R, Bentley M E, Divekar A D, Gadkari D, Mehendale S M, Shepherd M E, Bollinger R C, Quinn T C. Spread of HIV infection in married monogamous women in India. JAMA. 1997;278:2090–2092. [PubMed] [Google Scholar]
- 25.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barré-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George D G, Hunt L T, Barker W C. Current methods in sequence comparison and analysis. In: Schlesinger D H, editor. Macromolecular sequencing and synthesis. New York, N.Y: Alan R. Liss; 1988. pp. 127–149. [Google Scholar]
- 28.Hu D J, Dondero T J, Rayfield M A, George J R, Schochetman G, Jaffe H W, Luo C C, Kalish M L, Weniger B G, Pau C P, Schable C A, Curran J W. The emerging genetic diversity of HIV. The importance of global surveillance for diagnostics, research, and prevention. JAMA. 1996;275:210–216. [PubMed] [Google Scholar]
- 29.Jain, M. K., T. J. John, and G. T. Keusch. 1994. Epidemiology of HIV and AIDS in India. AIDS 8(Suppl. 2):S61–S75. [PubMed]
- 30.Jameel S, Zafrullah M, Ahmad M, Kapoor G S, Sehgal S. A genetic analysis of HIV-1 from Punjab, India, reveals the presence of multiple variants. AIDS. 1995;9:685–690. doi: 10.1097/00002030-199507000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Kampinga G A, Simonon A, Van de Perre P, Karita E, Msellati P, Goudsmit J. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology. 1997;227:63–76. doi: 10.1006/viro.1996.8318. [DOI] [PubMed] [Google Scholar]
- 32.Korber B T, Hoelscher M. HIV-1 subtypes: implications for epidemiology, pathogenicity, vaccines and diagnostics. Workshop report from the European Commission (DG XII, INCO-DC) and the Joint United Nations Programme on HIV/AIDS. AIDS. 1997;11:UNAIDS17–UNAIDS36. [PubMed] [Google Scholar]
- 33.Leitner T, Korber B, Robertson D L, Gao F, Hahn B H. Updated proposal of reference sequences of HIV-1 genetic subtypes. In: Korber B T, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F E, Kuiken C L, editors. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. pp. 1–6. [Google Scholar]
- 34.Luciw P A, Shacklett B L. Molecular biology of human and simian immunodeficiency viruses. In: Morrow W J W, Haigwood N L, editors. HIV molecular organization, pathogenicity, and treatment. Amsterdam, The Netherlands: Elsevier; 1993. pp. 123–219. [Google Scholar]
- 35.Mellors J W, Schinazi R F, Larder B A. Mutations in retroviral genes associated with drug resistance. In: Myers G, Korber B T, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S, editors. Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. pp. III-206–III-241. [Google Scholar]
- 36.Montano M A, Novitsky V A, Blackard J T, Cho N L, Katzenstein D A, Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71:8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers G, Korber B T, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S. Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 38.Myers G, Korber B T, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis G N. Human retroviruses and AIDS 1994: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 39.Page R D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 40.Peters K. Vised (Version 1.2). 1997. Parksville, British Columbia, Canada. Distributed by author. [Google Scholar]
- 41.Ray, S. C. 1998. SimPlot for Windows (version 1.6). Baltimore, Md. Distributed by author (http://www.welch.jhu.edu/∼sray/download).
- 42.Ray S C, Lubaki N M, Dhruva B, Siliciano R F, Bollinger R C. Autologous strain-specific cytolytic T lymphocyte responses directed against human immunodeficiency virus type 1 env. AIDS Res Hum Retroviruses. 1998;14:3–13. doi: 10.1089/aid.1998.14.3. [DOI] [PubMed] [Google Scholar]
- 43.Robertson D L, Gao F, Hahn B H, Sharp P M. Intersubtype recombinant HIV-1 sequences. In: Korber B T, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F E, Kuiken C L, editors. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. pp. III-25–III-30. [Google Scholar]
- 44.Robertson D L, Hahn B H, Sharp P M. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 45.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 46.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 47.Salminen M O, Johansson B, Sonnerborg A, Ayehunie S, Gotte D, Leinikki P, Burke D S, McCutchan F E. Full-length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res Hum Retroviruses. 1996;12:1329–1339. doi: 10.1089/aid.1996.12.1329. [DOI] [PubMed] [Google Scholar]
- 48.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 49.Schmit J C, Ruiz L, Clotet B, Raventos A, Tor J, Leonard J, Desmyter J, De Clercq E, Vandamme A M. Resistance-related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538) AIDS. 1996;10:995–999. doi: 10.1097/00002030-199610090-00010. [DOI] [PubMed] [Google Scholar]
- 50.Shafer R W, Eisen J A, Merigan T C, Katzenstein D A. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J Virol. 1997;71:5441–5448. doi: 10.1128/jvi.71.7.5441-5448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheppard H W, Lang W, Ascher M S, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS. 1993;7:1159–1166. [PubMed] [Google Scholar]
- 52.Siepel A C, Korber B T. Scanning the database for recombinant HIV-1 genomes. In: Myers G, Korber B T, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. pp. III-35–III-60. [Google Scholar]
- 53.Smith J M. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 54.Soto-Ramirez L E, Tripathy S, Renjifo B, Essex M. HIV-1 pol sequences from India fit distinct subtype pattern. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:299–307. doi: 10.1097/00042560-199612010-00001. [DOI] [PubMed] [Google Scholar]
- 55.Strebel K. Structure and function of HIV-1 Vpu. In: Myers G, Korber B T, Foley B, Jeong K-T, Mellors J W, Wain-Hobson S, editors. Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. pp. III-19–III-27. [Google Scholar]
- 56.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tripathy S, Renjifo B, Wang W K, McLane M F, Bollinger R, Rodrigues J, Osterman J, Essex M. Envelope glycoprotein 120 sequences of primary HIV type 1 isolates from Pune and New Delhi, India. AIDS Res Hum Retroviruses. 1996;12:1199–1202. doi: 10.1089/aid.1996.12.1199. [DOI] [PubMed] [Google Scholar]
- 58.Voevodin A, Crandall K A, Seth P, al Mufti S. HIV type 1 subtypes B and C from new regions of India and Indian and Ethiopian expatriates in Kuwait. AIDS Res Hum Retroviruses. 1996;12:641–643. doi: 10.1089/aid.1996.12.641. [DOI] [PubMed] [Google Scholar]
- 59.Weniger, B. G., Y. Takebe, C. Y. Ou, and S. Yamazaki. 1994. The molecular epidemiology of HIV in Asia. AIDS 8(Suppl. 2):S13–S28. [PubMed]
- 60.Zacharova V, Becker M L, Zacher V, Ebbesen P, Goustin A S. DNA sequence analysis of the long terminal repeat of the C subtype of human immunodeficiency virus type 1 from southern Africa reveals a dichotomy between B subtype and African subtypes on the basis of upstream NF-IL6 motif. AIDS Res Hum Retroviruses. 1997;13:719–724. doi: 10.1089/aid.1997.13.719. [DOI] [PubMed] [Google Scholar]
- 61.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]