Coxsackievirus and Adenovirus Receptor Amino-Terminal Immunoglobulin V-Related Domain Binds Adenovirus Type 2 and Fiber Knob from Adenovirus Type 12 (original) (raw)
Abstract
The extracellular region of the coxsackievirus and adenovirus receptor (CAR) is predicted to consist of two immunoglobulin (Ig)-related structural domains. We expressed the isolated CAR amino-terminal domain (D1) and a CAR fragment containing both extracellular Ig domains (D1/D2) in Escherichia coli. Both D1 and D1/D2 formed complexes in vitro with the recombinant knob domain of adenovirus type 12 (Ad12) fiber, and D1 inhibited adenovirus type 2 (Ad2) infection of HeLa cells. These results indicate that the adenovirus-binding activity of CAR is localized in the amino-terminal IgV-related domain and confirm our earlier observation that Ad2 and Ad12 bind to the same cellular receptor. Preliminary crystallization studies suggest that complexes of Ad12 knob bound to D1 will be suitable for structure determination.
Characterization of the molecular basis for virus attachment to cells has importance both for understanding virus tropism and for developing agents that inhibit virus binding or alter the specificity of binding. Recently, a cellular receptor for adenovirus type 2 (Ad2) and other closely related serotypes was identified. This receptor, encoded by a single gene on human chromosome 21 (17), is a 46-kDa glycoprotein which also serves as a receptor for group B coxsackieviruses (CBV) (4, 13, 25) and thus was termed the coxsackievirus and adenovirus receptor (CAR). CAR mRNA is present in varying abundance in many human tissues (5, 25). A broad tissue distribution of CAR protein expression correlates with the broad tropism of CBV (19), but subgroup C adenoviruses that are known to bind CAR have a much more restricted tropism, limited primarily to the upper respiratory tract (18). Thus, other factors in addition to receptor availability clearly have important roles in determining adenovirus tropism. Although adenovirus binds to CAR with high affinity (17, 28), virus titers are significantly reduced on cells with down-regulated CAR expression (10). These results suggest that adenovirus infection in vivo may be restricted to cells which express CAR at levels above a minimum threshold concentration. CAR protein levels are relatively low on the apical surfaces of differentiated (ciliated) respiratory epithelial-cell cultures (32), which may account for the poor efficiency of adenoviral gene transfer to human lung tissue in vivo (8, 11, 14, 31, 33).
Adenovirus binding to CAR results from an interaction between viral fibers, rod-shaped proteins located at the capsid vertices, and the extracellular region of CAR. The distal, carboxy-terminal end of fiber consists of a globular domain, termed the knob, which has receptor-binding activity (23). The knob domain of Ad5 was expressed in Escherichia coli as a soluble, trimeric, biologically active protein (12), and its 3-dimensional structure was determined by X-ray crystallography (29). The predicted amino acid sequence of CAR (4) suggests a structure consisting of two extracellular domains related to the immunoglobulin V (IgV) and IgC2 domain folds (6), a single membrane-spanning region, and one carboxy-terminal cytoplasmic domain. Regions of CAR necessary for binding the fiber knob domain have not yet been determined.
We report here the expression in E. coli of the fiber knob from Ad12 and fragments of human CAR corresponding to the extracellular immunoglobulin-like domains. The isolated amino-terminal IgV-related CAR domain (D1) and the entire extracellular region (D1/D2) both formed complexes with Ad12 knob, and D1 inhibited infection of HeLa cells by Ad2, indicating that D1 alone is sufficient for the adenovirus-binding activity of intact CAR. Infection of HeLa cells by Ad2 also was inhibited by Ad12 knob, complementing our earlier report that native Ad2 fibers inhibit infection by Ad12 and supporting the conclusion that CAR also serves as the major attachment receptor for Ad12, a subgroup A adenovirus (1). The recombinant protein fragments we describe provide a means to determine the 3-dimensional structure of the fiber-CAR complex by X-ray crystallography and to screen for antiviral agents that may interfere with virus attachment to cells.
MATERIALS AND METHODS
Expression and purification of Ad12 knob.
A DNA fragment encoding the entire Ad12 fiber knob domain and several flanking amino acids from the fiber shaft (amino acids 401 to 587) was amplified from viral DNA by PCR (30 cycles at 94°C for 30 s, 55°C for 40 s, and 72°C for 60 s) using forward primer 1, CATATGAGCAACACTCCATACG, and reverse primer 2, GGATCCTTATTCTTGGGTAATGT (Fig. 1a). The PCR product was cloned between the _Nde_I and _Bam_HI sites of vector pET15b (Novagen) and transformed into strain BL21-DE3 (Novagen) for expression of the hexahistidine-tagged knob protein. Overnight cultures in Luria-Bertani (LB) broth containing 150 mg of penicillin G (Sigma)/liter were diluted 100-fold into fresh LB-penicillin broth and grown at 37°C until mid-log phase (optical density of 0.8 at 600 nm), at which time they were chilled to 24°C and adjusted to 50 μM isopropyl β-d-thiogalactopyranoside (IPTG) to induce knob expression. After shaking (250 rpm) overnight at 24°C, the cells were collected by centrifugation, resuspended in 10% of the original culture volume of STE (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) containing 100 μg of lysozyme/ml, and subjected to three cycles of freezing and thawing. The viscous cell lysate was then sonicated and cleared by centrifugation at 25,000 × g for 10 min. Knob was precipitated from the supernatant by the addition of solid ammonium sulfate to 35% saturation (25°C), dialyzed against several changes of 10 mM Tris-HCl (pH 7.5), and passed over a column of DEAE-cellulose (DE52; Whatman) equilibrated in the same buffer. Knob was recovered from the flowthrough fractions essentially free of contaminating E. coli proteins and nucleic acids, and was further purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography according to the manufacturer’s instructions (Qiagen). Typically, >100 mg of purified Ad12 knob is obtained from 1 liter of bacterial culture.
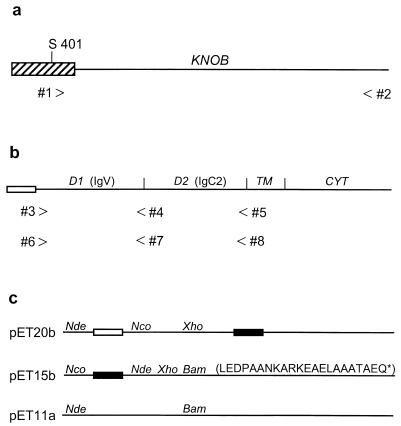
FIG. 1.
Cloning of the Ad12 fiber knob and the extracellular domains of human CAR. (a) The Ad12 knob domain (line) begins at a conserved Thr-Leu-Trp-Thr motif (amino acids 409 to 412) and extends to the fiber protein carboxy terminus (Glu587). A fragment of Ad12 DNA encoding the entire knob domain and several amino acids from the preceding fiber shaft region (striped box) beginning at Ser401 was amplified by PCR using forward primer 1 and reverse primer 2. The resulting PCR product was cloned between the _Nde_I and _Bam_HI sites of pET15b. (b) The human CAR protein consists of an amino-terminal signal peptide (open box), two extracellular Ig-related domains (D1 and D2), a membrane-spanning region (TM), and a cytoplasmic domain (CYT). cDNA fragments encoding D1 and D1/D2 were amplified by PCR using forward primer 3 and reverse primers 4 and 5. The resulting PCR products were cloned between the _Nco_I and _Xho_I sites of pET20b. Similar D1- and D1/D2-encoding cDNA fragments were amplified by PCR using forward primer 6 and reverse primers 7 and 8. The resulting PCR products were cloned between the _Nde_I and _Bam_HI sites of pET15b. (c) pET vectors for protein expression in E. coli. The open and filled boxes represent bacterial signal peptides and hexahistidine tags, respectively. The restriction sites used in this study are shown, and the sequence of the pET15b-encoded 22-amino-acid carboxy-terminal extension of sD1 is given in single-letter code.
Expression and purification of CAR protein fragments.
cDNA fragments encoding the human CAR extracellular domains (D1 and D1/D2 [Fig. 1b]) were amplified by reverse transcription-PCR of total RNA from a mouse A9 cell line transformed with multiple copies of the cloned human CAR gene (to be described elsewhere) and correspond exactly to the CAR cDNA sequence in GenBank file Y07593. First-strand cDNA synthesis was primed by oligo(dT). Primers 3 through 5 (CCATGGGTATCACTACTCCTGAAGAGA, CTCGAGCGCACCTGAAGGCTTA, and CTCGAGTGAAGGAGGGACAAC, respectively [Fig. 1b]) were designed for cloning D1- and D1/D2-encoding PCR products between the _Nco_I and _Xho_I sites of expression vector pET20b (Novagen). The PCR cycling program was identical to that used for Ad12 knob. These same PCR products also were cloned into pET15b as _Nco_I-_Xho_I restriction fragments and thus lacked the vector-encoded hexahistidine tag but had 22-amino-acid carboxy-terminal extensions that were encoded by vector sequences downstream of the _Xho_I site (Fig. 1c). Primers 6 through 8 (CATATGGGTATCACTACTC, GGATCCTACGCACCTGAAGGCT, and GGATCCTATCCAGCTTTATTTGAAG, respectively [Fig. 1b]) were designed to adapt the CAR PCR products for cloning between the pET15b _Nde_I and _Bam_HI restriction sites, which provides for attachment of the amino-terminal hexahistidine tag to the expressed proteins. Stop codons were built into the reverse primers in order to avoid synthesis of the CAR fragments with the vector-encoded carboxy-terminal extensions.
The procedure used for expression of the initial pET15b-D1 construct (PCR product from primers 3 and 4) was similar to that described above for Ad12 knob except that the culture was induced at 18°C. Soluble D1 (sD1) was precipitated from cleared cell lysates by ammonium sulfate precipitation (35 to 60% cut; 25°C) and was partially purified by anion-exchange chromatography (DE52) in 10 mM Tris-HCl buffer (pH 7.5). About 5 mg of partially purified sD1 was recovered from 1 liter of bacterial culture. The hexahistidine-tagged CAR fragments expressed from the second set of pET15b constructs (by using primers 6 through 8) were insoluble but were recovered from inclusion bodies. Cultures were induced at 37°C, and cleared lysates were prepared as described above. After centrifugation, the supernatant was discarded, and the pellet was washed several times in STE containing 0.1% Nonidet P-40, dissolved in 8 M urea–50 mM β-mercaptoethanol–50 mM Tris-HCl (pH 9.2) (20 ml per liter of initial culture), and then diluted with 15 volumes of 20 mM Tris-HCl (pH 8.0). The slightly hazy solution was passed through a 10-ml bed volume of DEAE-Sepharose Fast Flow (Pharmacia) equilibrated in 20 mM Tris-HCl (pH 8.0). Approximately half of the bound CAR fragments eluted with 50 mM NaCl and were essentially pure. The remaining bound CAR eluted with 300 mM NaCl along with contaminating E. coli proteins and was discarded.
Assays for detection of knob-CAR complexes.
Metal affinity chromatography was used to detect the association of knob with His-tagged refolded CAR fragments or the association of sD1 with the His-tagged knob. The hexahistidine tag was cleaved from Ad12 knob by using biotinylated thrombin and was then passed through Ni-NTA and avidin columns in order to remove residual His-tagged proteins and thrombin. The resulting knob was mixed with purified Ad2 hexon protein and then divided into three equal samples. His-tagged D1 or D1/D2 was then added to two of the samples, and an equivalent volume of buffer was added to the third (control) sample. Each sample was then batch-adsorbed to Ni-NTA beads, washed, and eluted with 100 mM EDTA–25 mM Tris-HCl (pH 8.0). Samples were then electrophoresed in sodium dodecyl sulfate (SDS)-polyacrylamide gels and stained with Coomassie blue. Alternatively, His-tagged knob was added to a partially purified preparation of sD1, and the mixture was then chromatographed on a Ni-NTA column and processed as described above.
Inhibition of Ad2 infection of HeLa cells.
HeLa monolayer cultures were grown in 50% Dulbecco’s modified Eagle medium (DMEM; Gibco)–50% Ham’s F-12 Nutrient Mixture (Gibco) containing 10% calf serum. Monolayers were seeded in 24-well cluster plates 1 day before infection. Ad2 diluted in binding buffer (50% DMEM–50% phosphate-buffered saline [PBS]–0.4% bovine serum albumin) was divided into three equal samples and mixed with an equal volume of Ad12 knob, sD1 (both at approximately 2 mg/ml in PBS), or binding buffer. Each preparation was adsorbed in triplicate (0.2 ml/well) for 30 min at 4°C, and the wells were then washed twice with PBS and incubated for 2 days at 37°C in DMEM containing 2% calf serum. The number of infected cells in each culture was then determined by immunoassay for the viral hexon antigen as previously described (2). To control for possible cytotoxic effects of the recombinant proteins, additional sets of cultures were preincubated with Ad12 knob or sD1 (1 mg/ml) in binding buffer for 30 min, washed twice with PBS, and then infected with Ad2.
Analysis of Ad12 knob by STEM.
The mass of Ad12 knob (with the His tag removed) was measured by scanning transmission electron microscopy (STEM). Five microliters of the purified protein (∼10 μg/ml) was applied to an electron microscope holey grid covered with thin (∼2-nm) carbon, and after 1 min was wicked and washed 10 times with 20 mM ammonium acetate. The grid was blotted and rapidly frozen in liquid nitrogen slush, then freeze-dried overnight. Data were collected with the Brookhaven National Institutes of Health (NIH) Biotechnology Resource STEM (26) at a scan width of 0.512 μm, with a dose of 200 electrons/nm2. Protein particle masses were measured (27) off-line by using the PC-Mass program, and statistics and curve fitting were generated with SigmaPlot. Mass calibration was carried out by using tobacco mosaic virus particles that adhered to the grid before the sample was applied.
Gel filtration analysis of Ad12 knob, sD1, and knob-sD1 complexes.
The native molecular masses of His-tagged Ad12 knob, sD1, and knob-sD1 complexes were estimated by size exclusion chromatography using a Superose 6 gel permeation column. Aliquots (20 μl) of purified proteins or protein complexes were chromatographed at 0.3 ml/min in 20 mM Tris-HCl (pH 7.8)–200 mM NaCl–1 mM dithiothreitol–0.1 mM EDTA. Aliquots of the fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). These experiments were run over a range of protein (monomer) concentrations from 1 to 500 μM. Molar extinction coefficients of 2.61 × 104 and 1.34 × 104 were calculated for monomers of His-tagged Ad12 knob and sD1, respectively, according to a published method (22). His-tagged knob-sD1 complexes eluted from a Ni-NTA column were separated from excess uncomplexed His-tagged knob by anion-exchange chromatography at pH 7.5, a condition under which the complexes bind to the ion exchanger but the free knob does not. Complexes were eluted with 100 mM NaCl in 10 mM Tris (pH 7.5) and were then chromatographed on a Superose 6 column.
RESULTS
Expression and purification of CAR extracellular fragments.
To localize the adenovirus-binding activity of CAR, fragments corresponding the amino-terminal CAR IgV domain (D1) and the combined IgV plus IgC2 domains (D1/D2) were expressed in E. coli. A cDNA fragment coding for D1 (Fig. 1b) was cloned into pET20b, an expression vector designed to export expressed proteins into the E. coli periplasmic space (Fig. 1c), but synthesis of D1 (expected molecular size, about 16 kDa) was undetectable (data not shown). When the initial construct was enlarged to include the downstream IgC2 domain, however, the resulting D1/D2 polypeptide was overexpressed but was insoluble in E. coli cells grown at temperatures ranging from 18 to 37°C (data not shown). These results imply that the amino-terminal domain (D1) specified by the initial construct also entered the secretory pathway but probably was rapidly degraded in the periplasmic space.
To determine if D1 could be stabilized by restricting its synthesis to the cytoplasm, the D1-encoding PCR product was transferred as an _Nco_I-_Xho_I restriction fragment from pET20b into pET15b. Because of restriction site differences between these two expression vectors (Fig. 1c), the CAR protein fragment specified by the resulting construct (pET15b-sD1) had a vector-encoded 22-amino-acid carboxy-terminal extension, and it lacked the amino-terminal hexahistidine tag that is normally attached to proteins expressed from pET15b. The D1 fragment accumulated to moderate abundance at 37°C in BL21-DE3 cells transformed with pET15b-sD1 but was completely insoluble (data not shown). When the cultures were induced at 18°C, however, a significant amount of D1 was contained in the soluble fraction of cell lysates (Fig. 2). The larger CAR cDNA fragment encoding D1/D2 also was transferred from pET20b into pET15b, but none of the expressed protein was detected in the soluble fraction of cell lysates (Fig. 2).
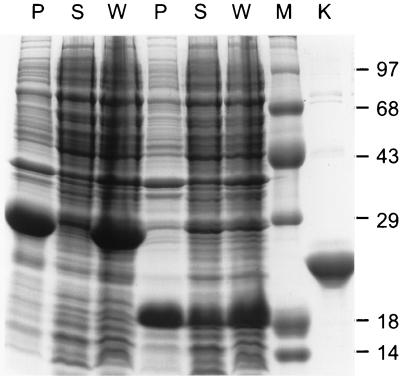
FIG. 2.
D1 and D1/D2 expression and solubility in the E. coli cytoplasm. BL21-DE3 cells transformed with pET15b-D1 (lanes 4 to 6) and pET15b-D1/D2 (lanes 1 to 3) (PCR products from reactions with primers 3 through 5 [Fig. 1b]) were induced with IPTG at 18°C. Protein contents of whole-cell lysates (W) and of the soluble (S) and insoluble (P) fractions of cell sonicates were analyzed by SDS-PAGE. The molecular sizes (in kilodaltons) of protein standards loaded in lane M are indicated. A sample of purified Ad12 knob was loaded in lane K.
To determine if removal of the vector-encoded carboxy-terminal extension would increase the yields of soluble CAR fragments produced in the E. coli cytoplasm, cDNA fragments encoding D1 and D1/D2 were amplified with new primer sets (primers 6 through 8 [Fig. 1b]) that introduced downstream stop codons and also fused the proteins to the pET15b vector-encoded amino-terminal hexahistidine tag. Both CAR fragments were overexpressed but were insoluble at culture growth temperatures between 18 and 37°C (data not shown), suggesting that the carboxy-terminal extension specified by the initial pET15b-sD1 construct may enable the IgV domain to fold into a soluble structure within E. coli cells. To further investigate whether D1 solubility within intact E. coli cells depends on the presence of the 22-amino-acid C-terminal extension, the D1-encoding insert was transferred from pET15b-sD1 into pET11a as an _Nde_I-_Bam_HI fragment (Fig. 1c), removing both the N-terminal hexahistidine tag and the 22-residue C-terminal extension. BL21-DE3 cells transformed with the resulting pET11a-D1 construct overexpressed D1, but the D1 fragment again was completely insoluble (Fig. 3). Taken together, these data indicate that D1 solubility in E. coli cells is enhanced by the pET15b-encoded 22-amino-acid carboxy-terminal extension.
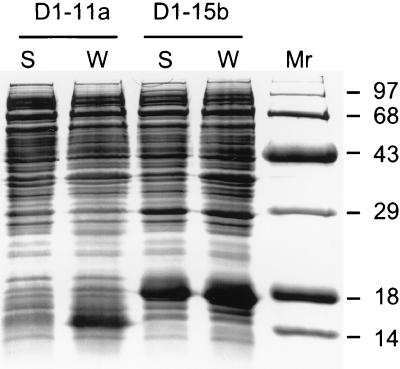
FIG. 3.
D1 solubility in the E. coli cytoplasm. BL21-DE3 cells transformed with pET11a-D1 (D1-11a) or pET15b-sD1 (sD1-15b) were induced with IPTG at 18°C. The protein content of whole-cell lysates (W) and that of the soluble fraction of cell sonicates (S) were analyzed by SDS-PAGE. The molecular weights (in thousands) of protein standards loaded in lane _M_r are indicated.
sD1 was partially purified from lysates of pET15b-sD1-transformed BL21-DE3 cells by ammonium sulfate precipitation and anion-exchange chromatography. The insoluble, hexahistidine-tagged D1 and D1/D2 CAR fragments were both refolded from urea-solubilized inclusion bodies and were purified to apparent homogeneity by Ni-NTA affinity chromatography and anion-exchange chromatography.
Biological activity of CAR extracellular fragments.
To examine the activity of the refolded D1 and D1/D2 CAR fragments, we determined whether they could form specific complexes with recombinant fiber knob from Ad12. We previously reported that infection of HeLa cells by Ad12 is inhibited by purified native fiber protein from Ad2 (1), suggesting that CAR serves as the major attachment receptor for both Ad2 and Ad12. A fragment of Ad12 DNA coding for the fiber knob domain (Fig. 1a) was cloned in pET15b. Ad12 knob was abundantly expressed following IPTG induction of cultures at 37°C but accumulated entirely within the insoluble fraction of cell lysates. When cultures were induced at 24°C, however, the majority of knob was in the soluble fraction. The knob was purified by ammonium sulfate precipitation and anion-exchange chromatography (see Fig. 2, lane K), and the His tag was removed by digestion with thrombin. Ad12 knob was then incubated with the refolded His-tagged D1 or D1/D2 in the presence of purified Ad2 hexon protein (included as a specificity control). The mixtures were then adsorbed to Ni-NTA beads in order to capture the His-tagged CAR fragments. In control incubations lacking the CAR fragments, Ad12 knob and Ad2 hexon both failed to bind to Ni-NTA beads, but in the presence of His-tagged D1 or D1/D2, Ad12 knob was retained on the Ni-NTA beads whereas Ad2 hexon was not (Fig. 4A). Although these results were qualitatively reproducible, the relative amounts of knob and CAR components that eluted from Ni-NTA beads varied in different experiments, possibly resulting from structural heterogeneity in different preparations of the refolded D1 and D1/D2 proteins.
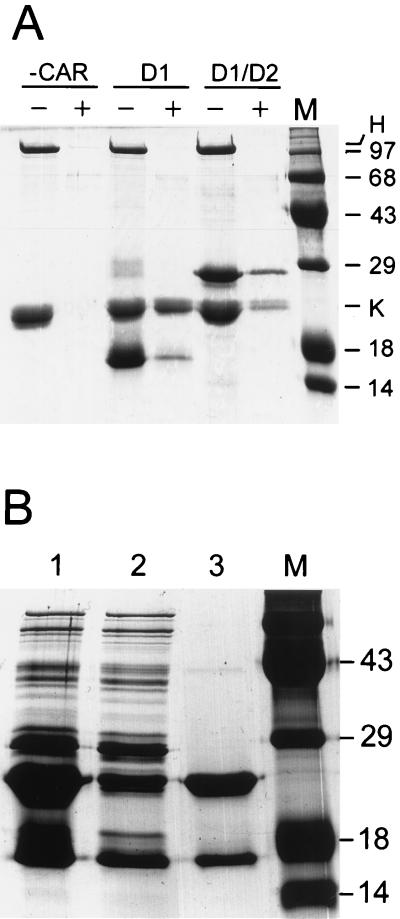
FIG. 4.
Formation of complexes between CAR fragments and Ad12 knob. (A) Insoluble His-tagged D1 and D1/D2 fragments were refolded, purified, and incubated with a mixture of Ad12 knob (His tag removed by thrombin cleavage) and native Ad2 hexon. CAR fragments were omitted from a control incubation (−CAR). A sample of each mixture was loaded in lanes marked −, and the remainder was then incubated with Ni beads. The beads were washed to remove unbound proteins, and bound material was eluted by boiling in SDS-PAGE sample buffer and loaded into lanes marked +. The positions in the gel of hexon and Ad12 knob (H and K) and the molecular sizes (in kilodaltons) of standards loaded in lane M are indicated. (B) sD1 was partially purified and mixed with purified His-tagged Ad12 knob (lane 1). The mixture was then adsorbed to Ni beads, and a sample of the unbound proteins was loaded in lane 2. Bound proteins were eluted from the beads by boiling in SDS-PAGE sample buffer and were loaded in lane 3. Molecular size standards were loaded in lane M.
To avoid complications that might result from heterogeneity of refolded CAR fragments, we focused our efforts on characterization of sD1, which is synthesized in E. coli as a soluble protein fragment. Purified, His-tagged Ad12 knob was mixed with a partially purified preparation of sD1 and incubated briefly to allow protein complexes to form. The mixture was then applied to a column of Ni-NTA beads, unbound proteins were washed from the column, and the bound fraction was eluted with EDTA. As shown in lane 3 of Fig. 4B, sD1 and His-tagged knob were the major protein species that eluted from the Ni-NTA column. The relative amounts of sD1 and knob shown in Fig. 4, lane 3, do not represent the actual stoichiometry of the knob-sD1 complex because excess knob was added to the crude sD1 preparation (Fig. 4, lane 1). Taken together, these experiments support the conclusion that the D1 domain alone is sufficient for the knob-binding activity of CAR.
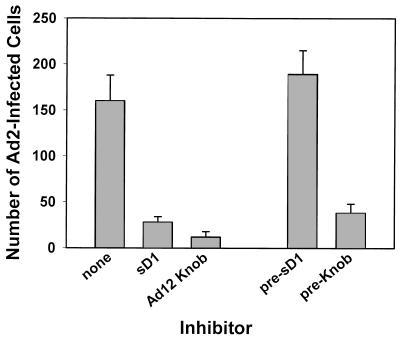
To determine if the in vitro interaction of Ad12 knob with the D1 domain of CAR represents physiologically relevant binding, the ability of the recombinant proteins to inhibit Ad2 infection of HeLa cells was tested. As shown in Fig. 5, Ad2 infectivity was significantly inhibited when either sD1 or Ad12 knob was included in the virus inoculum during virus adsorption. No inhibition of infection was observed in cell cultures that were pretreated with sD1 and then washed prior to virus adsorption, indicating that the inhibitory activity of sD1 does not result from a cytotoxic effect on cells. Cells similarly pretreated with Ad12 knob, however, were still partially refractory to infection by Ad2 virus, most likely resulting from incomplete dissociation of knob-CAR complexes on cells rather than from a cytotoxic effect of knob. The approximately equal inhibition by sD1 and Ad12 knob indicates that interaction of viral fibers with CAR is the predominant route to infection of HeLa cells and that alternate, CAR-independent infections (e.g., resulting from direct binding of virus to integrins) are rare in this system. Thus, the binding specificity of native fiber and CAR is reconstituted in the recombinant Ad12 knob and sD1 proteins.
FIG. 5.
Binding activity of sD1 and Ad12 knob. HeLa cell monolayers were infected with about 200 focus-forming units of Ad2 per well in the presence or absence of sD1 or Ad12 knob. After incubation of the infected cultures for 2 days at 37°C, monolayers were fixed, stained with rabbit anti-hexon serum, counterstained with horseradish peroxidase-goat anti-rabbit IgG, and developed with diaminobenzidine. The number of infected cells in triplicate wells was then counted and plotted (mean ± standard deviation). Control cultures were pretreated with sD1 or knob (pre-sD1 or pre-Knob) and then washed prior to infection.
Physical characteristics of Ad12 knob and CAR domains.
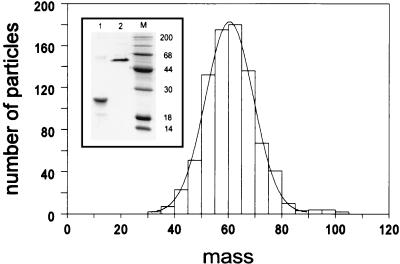
Analysis of heat-denatured and unheated samples of Ad12 knob by SDS-PAGE showed bands of 20 and 60 kDa, respectively (Fig. 6 inset, lanes 1 and 2), suggesting that, like the Ad5 fiber knob (12), the Ad12 knob is trimeric. To confirm this result, a sample of Ad12 knob was examined in the Brookhaven STEM, which measures the mass per unit length of macromolecules. In good agreement with the PAGE results, STEM analysis showed that the Ad12 knob has a mass of 60.6 kDa (Fig. 6).
FIG. 6.
Mass measurement of Ad12 knob by STEM. A sample of purified Ad12 knob protein (with the hexahistidine tag removed) was examined by STEM, and a mean mass of 60.6 kDa (n = 839) for individual molecules was determined as described in Materials and Methods. (Inset) SDS-PAGE analysis of Ad12 knob samples that were heat denatured (lane 1) or unheated (lane 2) before electrophoresis under reducing conditions.
Size exclusion chromatography was performed to estimate the sizes of sD1, Ad12 knob, and complexes of sD1 and Ad12 knob under nondenaturing conditions (Fig. 7). Ad12 knob eluted at a position consistent with a 60-kDa globular protein, in good agreement with the STEM and PAGE analyses. Surprisingly, sD1 also eluted at nearly the same position as the 60-kDa knob, suggesting either that sD1 is a multimeric globular protein or that it has an elongated shape. sD1 is unlikely to have an elongated shape given the predicted Ig-related (globular) fold of CAR. sD1 multimers could take the form of dimers, as was recently shown for the amino-terminal IgV-related domain of intercellular adhesion molecule 1 (ICAM-1) (7), or possibly higher oligomers such as dimers of dimers, which would better fit the observed molecular size of about 60 kDa (4 × 16 kDa). Both Ad12 knob and sD1 eluted as sharp, symmetrical peaks, demonstrating the stability and homogeneity of their folded conformations. An approximately equimolar mixture of Ad12 knob and sD1 proteins (based on monomers) eluted as a major peak at a position indicative of a globular protein of about 120 kDa and a minor peak at the position of the uncomplexed knob and sD1 components (60 kDa). Knob-sD1 complexes that were first purified by anion-exchange chromatography to remove excess uncomplexed knob eluted from the size exclusion column as a single, symmetrical peak centered at about 120 kDa (Fig. 7 inset), indicating that the trailing 60-kDa peak in the chromatogram of the knob-sD1 mixture shown in Fig. 7 corresponds to excess uncomplexed knob or sD1 component and does not result from partial dissociation of the complex during chromatography. Although the resolution of this method is not sufficient to unambiguously determine the stoichiometry of knob and sD1 components in the complex, it clearly shows that the complex is a homogeneous molecular species and does not contain high-molecular-weight aggregates, forms that might be expected if one or both multimeric components exhibited multivalent binding.
FIG. 7.
Size exclusion chromatography of His-tagged Ad12 knob, sD1, and knob-sD1 complexes. Aliquots (20 μl) of His-tagged Ad12 knob (dashed line), purified sD1 (dotted-and-dashed line), or an approximately equimolar mixture of His-tagged Ad12 knob and sD1 (solid line) were chromatographed on a Superose 6 gel filtration column at a flow rate of 0.3 ml/min. The elution positions of size markers are shown (ferritin, 440 kDa; bovine serum albumin dimer, 130 kDa; human hemoglobin, 68 kDa; RNase A, 13 kDa). (Inset) Size exclusion chromatogram of His-tagged Ad12 knob-sD1 complexes that were purified by anion-exchange chromatography to remove excess uncomplexed knob before chromatography on the Superose 6 column.
DISCUSSION
Amino acid sequence alignments show that CAR is an Ig superfamily (IgSF) member (4, 25) with an extracellular aspect consisting of an amino-terminal IgV-related domain (D1) and an adjacent IgC2-related domain (D2) (6). We expressed D1 and D1/D2 CAR fragments in E. coli and found that adenovirus-binding activity was associated with D1. The D1 fragment bound to recombinant Ad12 fiber knob and inhibited infection of HeLa cells by Ad2. Other IgSF members also serve as virus receptors, including ICAM-1, the receptor for rhinovirus (24); PVR, the receptor for poliovirus (20); and CD4, the receptor for human immunodeficiency virus type 1 (16). In each of these cases, the receptor amino-terminal domain also is related to the IgV domain fold and has virus-binding activity. Based on these results, it is expected that CBV also will bind to CAR through the D1 domain, thus explaining the earlier observation that adenovirus and CBV compete for cell binding sites (15). Confirmation of the putative interaction of CBV with D1 will exclude the alternative model where CBV binding to the D2 domain of CAR sterically hinders the binding of adenovirus to D1. However, the IgC2-like D2 domain most likely functions to support the D1 domain in a membrane-distal location, in which case D2 may be relatively inaccessible to viruses. Determination of the structure of the D1/D2 fragment would reveal how the interaction between domains relates to the adenovirus- and CBV-binding activity of CAR.
The finding that CAR D1 forms a complex in vitro with the recombinant knob domain of Ad12 fiber protein and that the Ad12 knob inhibits infection of HeLa cells by Ad2 confirms our earlier conclusion that Ad2 and Ad12 (representatives of viral subgroups C and A, respectively) bind to the same receptor. We previously showed that saturation of receptors on HeLa cells with native fiber protein from Ad2 blocked the subsequent binding of radiolabeled Ad12 virions and also inhibited infection of the cells by Ad12 (1). Presumably, the knob amino acids that form the interface with CAR are conserved in all adenovirus serotypes that bind to CAR and are different in serotypes which bind to a different receptor. The crystal structure of the Ad5 knob revealed two different molecular features that were proposed to be potential receptor-binding sites, a central threefold symmetric cavity formed by the trimer interface and three identical cavities, one associated with each polypeptide subunit, located around the knob perimeter (30). However, it was not possible to conclude which of these two alternate sites corresponded to the receptor-binding site based on alignment of knob amino acid sequences from different adenovirus serotypes (30). Determination of the structure of the Ad12 knob complexed to CAR D1 would directly identify the amino acids of each of the proteins that form the receptor-knob interface. The importance of these amino acids to knob-CAR binding could readily be tested through site-directed mutagenesis of the recombinant proteins we describe here.
The subgroup B adenoviruses (e.g., types 3 and 7) do not cross-compete with subgroup C viruses for cell binding sites (9), and the subgroup B and C knob sequences differ significantly in both regions that could define receptor-binding specificity (30). It will be of interest to determine whether the cellular receptor for the subgroup B adenoviruses also is an IgSF member or whether the structural framework of the fiber knob, which probably is similar in all serotypes, can support the evolution of binding sites that are complementary to cell surface molecules with physical characteristics that are distinct from those of Ig domains. Binding of the receptor to the threefold symmetric central cavity formed by the knob trimer interface might restrict the range of potential receptors to a particular class of molecules, for example, those with corresponding threefold symmetry or those possessing pseudo-threefold symmetry. In this regard, the recent identification of CAR, an IgSF protein without apparent threefold symmetry, as the adenovirus receptor suggests that the knob central cavity may not correspond to the receptor-binding site. Although the knob central cavity is an attractive receptor-binding site based on its topological similarity to the canyons on picornavirus capsids, the canyons differ significantly in that they are formed from the interface of nonidentical protein subunits and thus do not have threefold symmetry (21).
We found evidence that sD1 exits in solution as a multimeric protein, but there is no evidence we are aware of indicating that native CAR forms multimers on intact human cells or that CAR multimers are required for adenovirus infection. It should be noted that all the CAR fragments described here were produced in E. coli and therefore are not glycosylated. Carbohydrate groups could mask regions that, when exposed, might cause the proteins to aggregate, possibly through hydrophobic interactions. Thus, D1 or D1/D2 multimers could be artifacts of a recombinant protein system. Evidence has been reported that the recombinant amino-terminal domain of ICAM-1 forms dimers (3, 7), but it is not clear whether ICAM-1 dimers have physiological relevance. Interestingly, the rhinovirus binding site on the amino-terminal domain of ICAM-1 is not occluded in the dimer; therefore, rhinovirus may be able to bind to a dimeric ICAM-1 receptor (7).
The solubility of D1 within E. coli also depended on the presence of a 22-amino-acid carboxy-terminal extension encoded by the expression vector. Curiously, the D1 protein lacking this extension precipitated within cells but could be refolded from urea-solubilized inclusion bodies as a soluble active protein, suggesting either that the conformations of the refolded and soluble D1 proteins are different or that the 22-amino-acid extension aids folding of the nascent D1 polypeptide chain into a soluble conformation rather than enhancing the solubility of the folded molecule. Recent data indicate that partial removal of the carboxy-terminal extension by trypsin digestion does not alter the solubility of sD1 (data not shown).
There were significant differences between expression of the Ad12 knob in our system and expression of the Ad5 knob reported by Deisenhofer and colleagues (12). The yield of soluble Ad12 knob (>100 mg/liter of culture) was about 20-fold greater than the yield reported for Ad5 knob. This does not likely result from differences in the expression systems that were used because we found that the Ad2 knob, which is closely related in amino acid sequence to the Ad5 knob, also was recovered in low yields from the soluble fraction of E. coli lysates (the majority of the Ad2 knob protein was in the insoluble fraction) when expressed from the same pET vector systems and under the same conditions as those reported here for the Ad12 knob (data not shown). In addition, whereas the Ad5 knob was soluble in bacteria grown at 37°C, the Ad12 knob was completely insoluble at this temperature but was predominantly soluble when expressed at 24°C. Both Ad2 and Ad12 knob polypeptides were overexpressed to approximately the same extent in our system; therefore, differences in the yields of the soluble knob proteins must reflect differences in knob folding and trimerization in bacterial cells. Optimization of Ad2 knob solubility through mutagenesis approaches could provide insights into intrinsic factors that regulate protein folding and multimerization in vivo.
The soluble forms of CAR and knob described here have potential applications as antiviral agents or as means to screen for novel drugs that interfere with virus-CAR binding. In addition, D1-based reagents, including D1-antibody fusion proteins or complexes, might be used to retarget the binding of adenovirus gene delivery vectors to novel cellular receptors.
ACKNOWLEDGMENTS
We thank M. Simon, B. Lin, and F. Kito for assistance with the STEM experiments.
Research was supported by the Office of Biological and Environmental Research of the U.S. Department of Energy under Prime Contract DE-AC02-98CH10886 with Brookhaven National Laboratory and by NIH grant AI36251 (to P.F.). STEM analysis was supported by grants from the NIH and the U.S. Department of Energy Office of Health and Environmental Research.
REFERENCES
- 1.Bai M, Campisi L, Freimuth P. Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2. J Virol. 1994;68:5925–5932. doi: 10.1128/jvi.68.9.5925-5932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bella J, Kolatkar P R, Marlor C W, Greve J M, Rossmann M G. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson J M, Krithivas A, Celi L, Droguett G, Horwitz M S, Wickham T, Crowell R L, Finberg R W. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J Virol. 1998;72:415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 7.Casasnovas J M, Stehle T, Liu J H, Wang J H, Springer T A. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crystal R G, McElvaney N G, Rosenfeld M A, Chu C S, Mastrangeli A, Hay J G, Brody S L, Jaffe H A, Eissa N T, Danel C. Administration of an adenovirus containing the human CFTR cDNA to the respiratory tract of individuals with cystic fibrosis. Nat Genet. 1994;8:42–51. doi: 10.1038/ng0994-42. [DOI] [PubMed] [Google Scholar]
- 9.Defer C, Belin M T, Caillet-Boudin M L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freimuth P. A human cell line selected for resistance to adenovirus infection has reduced levels of the virus receptor. J Virol. 1996;70:4081–4085. doi: 10.1128/jvi.70.6.4081-4085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay J G, McElvaney N G, Herena J, Crystal R G. Modification of nasal epithelial potential differences of individuals with cystic fibrosis consequent to local administration of a normal CFTR cDNA adenovirus gene transfer vector. Hum Gene Ther. 1995;6:1487–1496. doi: 10.1089/hum.1995.6.11-1487. [DOI] [PubMed] [Google Scholar]
- 12.Henry L J, Xia D, Wilke M E, Deisenhofer J, Gerard R D. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu K H, Lonberg-Holm K, Alstein B, Crowell R L. A monoclonal antibody specific for the cellular receptor for the group B coxsackieviruses. J Virol. 1988;62:1647–1652. doi: 10.1128/jvi.62.5.1647-1652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles M R, Hohneker K W, Zhou Z, Olsen J C, Noah T L, Hu P C, Leigh M W, Engelhardt J F, Edwards L J, Jones K R, et al. A controlled study of adenoviral-vector-mediated gene transfer in the nasal epithelium of patients with cystic fibrosis. N Engl J Med. 1995;333:823–831. doi: 10.1056/NEJM199509283331302. [DOI] [PubMed] [Google Scholar]
- 15.Lonberg-Holm K, Crowell R L, Philipson L. Unrelated animal viruses share receptors. Nature. 1976;259:679–681. doi: 10.1038/259679a0. [DOI] [PubMed] [Google Scholar]
- 16.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 17.Mayr G A, Freimuth P. A single locus on human chromosome 21 directs the expression of a receptor for adenovirus type 2 in mouse A9 cells. J Virol. 1997;71:412–418. doi: 10.1128/jvi.71.1.412-418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei Y F, Wadell G. Molecular determinants of adenovirus tropism. Curr Top Microbiol Immunol. 1995;199:213–228. doi: 10.1007/978-3-642-79586-2_11. [DOI] [PubMed] [Google Scholar]
- 19.Melnick J L. My role in the discovery and classification of the enteroviruses. Annu Rev Microbiol. 1996;50:1–24. doi: 10.1146/annurev.micro.50.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 21.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philipson L, Lonberg-Holm K, Pettersson U. Virus-receptor interaction in an adenovirus system. J Virol. 1968;2:1064–1075. doi: 10.1128/jvi.2.10.1064-1075.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 25.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall J S. Biological scanning transmission electron microscopy. In: Hren J J, Goldstein J I, Joy D C, editors. Introduction to analytical electron microscopy. New York, N.Y: Plenum; 1979. pp. 333–342. [Google Scholar]
- 27.Wall J S, Hainfeld J F. Mass mapping with the scanning electron microscope. Annu Rev Biophys Biophys Chem. 1986;15:355. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 28.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 29.Xia D, Henry L, Gerard R D, Deisenhofer J. Structure of the receptor binding domain of adenovirus type 5 fiber protein. Curr Top Microbiol Immunol. 1995;199:39–46. doi: 10.1007/978-3-642-79496-4_3. [DOI] [PubMed] [Google Scholar]
- 30.Xia D, Henry L J, Gerard R D, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]
- 31.Zabner J, Couture L A, Gregory R J, Graham S M, Smith A E, Welsh M J. Adenovirus-mediated gene transfer transiently corrects the chloride transport defect in nasal epithelia of patients with cystic fibrosis. Cell. 1993;75:207–216. doi: 10.1016/0092-8674(93)80063-k. [DOI] [PubMed] [Google Scholar]
- 32.Zabner J, Freimuth P, Puga A, Fabrega A, Welsh M J. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J Clin Investig. 1997;100:1144–1149. doi: 10.1172/JCI119625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabner J, Ramsey B W, Meeker D P, Aitken M L, Balfour R P, Gibson R L, Launspach J, Moscicki R A, Richards S M, Standaert T A, et al. Repeat administration of an adenovirus vector encoding cystic fibrosis transmembrane conductance regulator to the nasal epithelium of patients with cystic fibrosis. J Clin Investig. 1996;97:1504–1511. doi: 10.1172/JCI118573. [DOI] [PMC free article] [PubMed] [Google Scholar]