Inhibition of Epstein-Barr Virus Replication by a Benzimidazole l-Riboside: Novel Antiviral Mechanism of 5,6-Dichloro-2-(Isopropylamino)-1-β-l-Ribofuranosyl-1H-Benzimidazole (original) (raw)
Abstract
Although a number of antiviral drugs inhibit replication of Epstein-Barr virus (EBV) in cell culture, and acyclovir (ACV) suppresses replication in vivo, currently available drugs have not proven effective for treatment of EBV-associated diseases other than oral hairy leukoplakia. Benzimidazole riboside compounds represent a new class of antiviral compounds that are potent inhibitors of human cytomegalovirus (HCMV) replication but not of other herpesviruses. Here we characterize the effects of two compounds in this class against lytic replication of EBV induced in a Burkitt lymphoma cell line latently infected with EBV. We analyzed linear forms of EBV genomes, indicative of lytic replication, and episomal forms present in latently infected cells by terminal probe analysis followed by Southern blot hybridization as well as the high-molecular-weight unprocessed viral DNA by pulsed-field gel electrophoresis. d-Ribofuranosyl benzimidazole compounds that act as inhibitors of HCMV DNA maturation, including BDCRB (5,6-dichloro-2-bromo-1-β-d-ribofuranosyl-1H-benzimidazole), did not affect the accumulation of high-molecular-weight or monomeric forms of EBV DNA in the induced cells. In contrast, the generation of linear EBV DNA as well as precursor viral DNA was sensitive to the l-riboside 1263W94 [5,6-dichloro-2-(isopropylamino)-1-β-l-ribofuranosyl-1H-benzimidazole]. The 50% inhibitory concentration range for 1263W94 was 0.15 to 1.1 μM, compared with 10 μM for ACV. Thus, 1263W94 is a potent inhibitor of EBV. In addition, 1263W94 inhibited the phosphorylation and the accumulation of the essential EBV replicative cofactor, early antigen D.
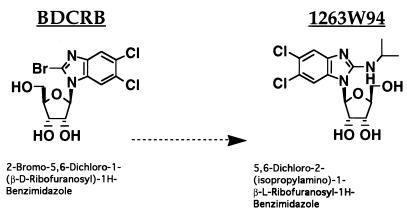
Benzimidazole riboside compounds with potent anti-human cytomegalovirus (HCMV) activity have recently been characterized (6, 22, 23, 36, 68, 69, 74). BDCRB (5,6-dichloro-2-bromo-1-β-d-ribofuranosyl-1H-benzimidazole [Fig. 1]) is a d-riboside compound that works by a unique mechanism, namely, inhibition of maturation of HCMV through potential targeting of a viral endonucleolytic activity necessary for processing of precursor DNA. Inhibition results in accumulation of unprocessed high-molecular-weight (HMW) forms of viral DNA which can be detected by pulsed-field gel electrophoresis (PFGE). The target of BDCRB has been identified as the HCMV UL89 gene product, a putative viral terminase (73). BDCRB is specific for HCMV; no other viruses tested thus far are inhibited by BDCRB (5, 22, 68, 69).
FIG. 1.
Structures of benzimidazole ribosides. BDCRB is the 2-substituted derivative of the compound DRB (5,6-dichloro-1-[β-d-ribofuranosyl) benzimidazole] (66, 67). The synthesis and activity of BDCRB have been described elsewhere (22, 68, 69). The compound 1263W94 differs from BDCRB in that it contains an l-sugar group and has a substitution of an isopropylamino group for bromine at the 2 position of the benzimidazole ring. Neither compound is phosphorylated in vitro or in vivo (5, 6).
l-Riboside benzimidazole analogs of BDCRB were synthesized to improve metabolic stability while retaining activity against HCMV. 1263W94 is a benzimidazole l-riboside with selective anti-HCMV activity. It is not active against herpes simplex virus types 1 and 2 (HSV-1 and HSV-2), varicella-zoster virus, or murine cytomegalovirus by plaque reduction assay, nor is it active against human immunodeficiency virus or hepatitis B virus (6). The compound has a 50% inhibitory concentration (IC50) of 0.04 to 0.1 μM against the AD169 laboratory strain of HCMV, with a similar range of 0.03 to 0.132 μM against HCMV clinical isolates (36). In addition, 1263W94 has >50% oral bioavailability in monkeys and rats and no toxic effects in monkeys dosed at 180 mg/kg for 30 days (74). 1263W94 inhibits HCMV replication by a novel mechanism: although viral DNA synthesis is inhibited, the viral DNA polymerase is not the target (6).
Compounds that inhibit replication of EBV in vitro include adenine arabinoside (3, 11), phosphonoformic acid, and phosphonoacetate (16, 60, 63, 77), a class of halogenated nucleoside analogs exemplified by FIAC [1-(2-fluoro-2-deoxy-β-d-arabinofuranosyl)-5-iodocytosine] (42, 46), BVDU [E-5-(2-bromovinyl)-2′-deoxyuridine) (43, 44), ganciclovir (39), acyclovir (ACV) (12, 24, 45), and the phosphonated nucleoside analogs HPMPC {(S)-1-[(3-hydroxy-2-phosphonylmethoxy)propyl]cytosine} and related compounds (40, 41). Nucleoside analogs and the pyrophosphate analogs phosphonoformic acid and phosphonoacetate inhibit replication of most or all of the herpesviruses. The mode of action of the nucleosides is via inhibition of the virally encoded herpesvirus DNA polymerases by the triphosphorylated metabolites. None of the compounds affects latent infection.
EBV infection of B cells is predominantly latent; only a subset of viral genes is expressed (38). In latently infected cells, the EBV genome is maintained as a closed circular episome (1, 51, 53) which is replicated by host DNA polymerase. In contrast, during productive infection, the viral DNA polymerase synthesizes large numbers of linear genomes (52). Latently infected B lymphocytes can be induced by cross-linking of surface immunoglobulin (64, 65) to trigger the switch to productive lytic infection. The d-riboside BDCRB and the l-riboside 1263W94 [5,6-dichloro-2-(isopropylamino)-1-β-l-ribofuranosyl-1H-benzimidazole (Fig. 1)] were tested for activity against EBV in a latently infected Burkitt lymphoma (BL) cell line in which viral reactivation and lytic replication can be efficiently induced (64, 65). Here we show that BDCRB was not effective in blocking either replication or maturation of EBV DNA; however, viral replication was highly sensitive to 1263W94. This compound inhibited both the accumulation and phosphorylation of the essential EBV replicative cofactor early antigen D (EA-D), the product of the BMRF1 gene, a DNA processivity factor which is essential for EBV lytic replication.
MATERIALS AND METHODS
Compounds.
BDCRB was synthesized at the University of Michigan and has been previously described (22, 68, 69). 1263W94 was synthesized at Glaxo Wellcome Co. Thymidine phosphorylase (TPase) and ACV were supplied by Glaxo Wellcome Co.
Virus and cells.
Akata is a type 1 BL cell line latently infected with EBV (64, 65). DG75 is an EBV-negative BL line (2). Lymphocyte lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Gibco BRL, Gaithersburg, Md.) and antibiotics, at 37°C in a 5% CO2 atmosphere. For viral reactivation studies, Akata cells were treated with anti-human immunoglobulin G (0.1 mg/ml; Sigma, St. Louis, Mo.) to cross-link surface receptor. For transient assays, 0.5 ml of DG75 cells (107 cells) were transfected by electroporation with a Bio-Rad (Hercules, Calif.) Gene Pulser at 320 V and 925 μF.
Plasmids.
Plasmid Xho1.9 contains EBV sequence adjacent to the terminal repeat region and has been previously described (57). EcoRI-C contains the _Eco_RI C fragment of EBV cloned into Mua3 vector (56). Both plasmids were generous gifts from Nancy Raab-Traub. pSG5 vector was purchased from Stratagene (La Jolla, Calif.). pSG5-BMRF1, a gift from David Dorsky, contains the _Bcl_I-_Bgl_II fragment from the EBV _Bam_H M fragment cloned into pSG5 (79). pCMVZ contains EBV Z cDNA in the pGEM2-based vector pHD1013 (79).
PFGE.
Cells were harvested following 72 h of treatment with compound and were washed with phosphate-buffered saline (PBS) solution. Akata cells (107) were resuspended in 45 μl of PBS, mixed with 45 μl of 1.6% low-melting-point agarose (FMC, Rockland, Maine), pipetted into preformed wells (Bio-Rad), and chilled. The agarose plug was incubated for 48 h at 48°C in lysis buffer (0.5 M EDTA [pH 9.0], 1% sodium sarcosyl, 0.5 mg of proteinase K per ml) and then transferred to storage buffer (10 mM Tris [pH 7.5], 50 mM EDTA) prior to PFGE. Electrophoresis was performed as described previously (73) for 20 h at 14°C with a linear ramping pulse of 60 to 90 s through 120°C (Bio-Rad CHEF Mapper). Gels were UV-irradiated (Stratalinker; Stratagene) and then incubated in 0.5 M NaOH–1.5 M NaCl to denature the DNA. DNA was transferred to Nytran membrane (Schleicher & Schuell, Keene, N.H.), and Southern blot analysis was performed. Hybridization was carried out with nick-translated DNA probe Eco-C (56) overnight at 42°C. The membranes were rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then washed with 0.1× SSC–0.1% sodium dodecyl sulfate (SDS) for 30 min at 42°C and twice for 30 min at 65°C. The dried filters were analyzed by autoradiography with a Molecular Dynamics PhosphorImager.
Terminal probe analysis.
Akata cells were harvested following 72 h of treatment with compound and were washed in PBS. The cells were resuspended in 300 μl of DNA digestion buffer (100 mM NaCl, 10 mM Tris [pH 7.9], 25 mM EDTA, 0.5% SDS, 0.1 mg of proteinase K per ml) and incubated for 20 h at 48°C. Next, 100 μg of RNase A was added, and incubation was continued for 2 h at 37°C. The DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and then precipitated with 1 ml of isopropyl alcohol. Terminal probe analysis was performed essentially as described previously (57). Briefly, DNA was digested overnight with _Bam_HI, and 10 μg of DNA was separated by gel electrophoresis (0.7% agarose, 1× Tris-borate-EDTA) for 20 h at 50 V. The gel was incubated in denaturing buffer (1.5 M NaCl, 0.5 N NaOH) for 30 min and then incubated in neutralization buffer (1.5 M NaCl, 1 M Tris [pH 7.4]) for 30 min. The DNA was blotted onto a nitrocellulose membrane, and hybridization was performed as described above. The filters were hybridized at 45°C with a 32P-labeled riboprobe prepared by transcription of _Hin_dIII-digested plasmid Xho1.9 with SP6 RNA polymerase (Promega, Madison, Wis.).
Northern blot analysis.
Total RNA was prepared from Akata cells with an RNeasy kit (Qiagen, Hilden, Germany); 25 μg of RNA was used for Northern blot analysis. Probes were prepared by random priming; 100 ng of EA-D, Z, or actin cDNA was labeled with [32P]dCTP with a Prime-It II kit as specified by the manufacturer (Stratagene). Actin cDNA was purchased from Clontech (Palo Alto, Calif.). EA-D was prepared by digestion of pSG5 EA-D with _Bam_HI and _Bgl_II (79). Z cDNA was prepared by digesting cytomegalovirus Z with _Eco_RI and _Xba_I (79). EA-D and Z cDNAs were gel purified (Qiaex II, Qiagen) prior to labeling.
Western blot analysis.
Cells were harvested, washed with PBS, and then resuspended in ELB+ buffer (0.25 M NaCl, 0.1% Nonidet P-40, 0.05 M HEPES [pH 7.0], 0.001 M phenylmethylsulfonyl fluoride, 0.005 EDTA, 0.5 mM dithiothreitol). The sample was lysed by freeze-thawing three times, and the supernatant was collected following centrifugation at 13,000 × g for 15 min at 4°C. The protein concentration of the supernatant was determined with the Bradford reagent (Bio-Rad). For Western analysis, equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to an Immobilon membrane (Millipore, Bedford, Mass.). The membrane was probed with antibody to the following proteins as indicated in the figure legends: EA-D (catalog no. C65026M; Biodesign, Kennebunk, Maine), Z (BZ-1; Dako, Glostrup, Denmark), R (8C12; Argene S.A. Varilhes, France), actin (clone AC-15; Sigma), or interferon response factor 1 (IRF-1) (sc497-X; Santa Cruz Biotechnology, Santa Cruz, Calif.). The membranes were washed for three times for 10 min each with TBST buffer (0.9% NaCl, 0.02 M Tris [pH 7.5], 0.05% Tween 20), then probed with the appropriate secondary antibody (horseradish peroxidase-conjugated anti-mouse or anti-rabbit; Amersham, Arlington Heights, Ill.), and washed again with TBST. The reaction was visualized by enhanced chemiluminescence (Amersham).
Phosphatase assay.
Cell lysate (20 μg) was digested with 2,000 U of lambda phosphatase in a volume of 50 μl for 3 h at 30°C as specified by the manufacturer (New England Biolabs, Beverly, Mass.). The samples were analyzed by SDS-PAGE followed by immunoblotting as described above.
RESULTS
1263W94, but not BDCRB, inhibits EBV replication.
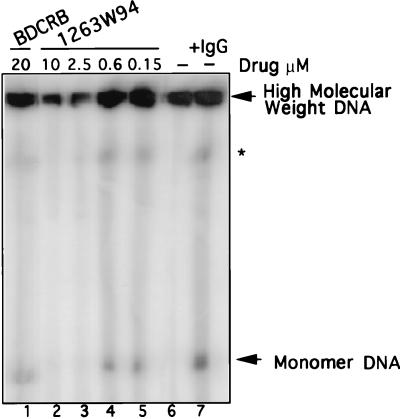
Akata cells were induced by cross-linking of surface immunoglobulin; then compounds were added, and incubation continued for 72 h. The cells were harvested, and DNA was analyzed by PFGE followed by Southern blot hybridization with an EBV-specific probe, Eco-C. Two forms of viral DNA were detected; monomeric DNA represents the unit-length (172-kbp) EBV genome, consisting of both linear and episomal DNA, and HMW DNA represents the unprocessed precursor viral DNA. The appearance of nicked DNA results from partial digestion of HMW DNA (73). An increase in monomeric DNA, representing newly synthesized linear viral genomes, was detected upon induction of the cells (Fig. 2; compare lanes 6 and 7). Increasing amounts of 1263W94 inhibited the appearance of both forms of EBV DNA (lanes 2 to 5). In contrast, BDCRB at a high dose (20 μM) (lane 1) inhibited the appearance of neither monomeric nor HMW DNA, unlike its effect on HCMV (73). The amount of hybridized monomer DNA was quantitated by PhosphorImager analysis. IC50 values calculated by PFGE analysis in several experiments were 0.15 to 1.1 μM for 1263W94, compared with 10 μM for ACV.
FIG. 2.
The l-riboside 1263W94 inhibits replication of EBV DNA. Induced Akata cells were incubated in the presence or absence of drug for 72 h and then harvested. Samples were prepared, and PFGE was performed to detect viral replication. EBV DNA was detected by hybridization with 32P-labeled _Eco_RI C fragment of EBV. HMW and monomer forms of DNA were quantitated following PhosphorImager analysis. A band representing nicked HMW DNA (∗) migrates to the resolution limit of the gel.
1263W94 is more active than ACV against EBV replication.
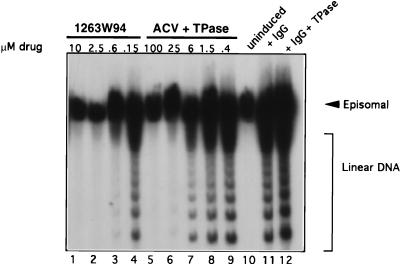
Terminal probe analysis was used to measure inhibition of EBV viral DNA replication, as it easily distinguishes between the circular, latent form of the viral genome and linear replicative forms (57). Increasing amounts of 1263W94 (Fig. 3, lanes 1 to 4) or ACV (lanes 5 to 9) were added to Akata cells immediately after induction and incubated for 72 h. The samples were harvested, and viral DNA was analyzed as described above. Ladder arrays of bands, which represent linear genomic forms of newly replicated EBV DNA, were detected in the IgG-induced cells (lanes 11 and 12) but not in the sample from uninduced cells (lane 10). Linear genomes were reduced by more than 50% with 0.6 μM 1263W94 (lane 3) or with 25 μM ACV (lane 6). This result confirms the PFGE results, namely, that 1263W94 is more effective than ACV in inhibiting EBV DNA replication. None of the EBV DNA replication forms was inhibited by 20 μM BDCRB (data not shown).
FIG. 3.
1263W94 inhibits replication of EBV. Akata cells were induced by IgG cross-linking and incubated in the presence or absence of drug for 3 days. Cells were harvested, and terminal probe analysis was performed as described in the text. DNA was digested with _Bam_HI, separated on a 0.7% agarose gel, then transferred to nitrocellulose, and probed with 32P-labeled riboprobe Xho1.9 containing sequences specific for the EBV terminal repeat region. Samples containing ACV were incubated in the presence of TPase.
The medium of samples receiving ACV was pretreated with TPase for 1 h at 37°C (2 U/ml), followed by filter sterilization. Thymidine, which is present in bovine serum, can compete with ACV for uptake into the cell, decreasing the effectiveness of the drug (15). TPase did not affect viral replication induced by anti-IgG treatment (Fig. 3, lane 12).
1263W94 inhibits the appearance of EBV EA-D.
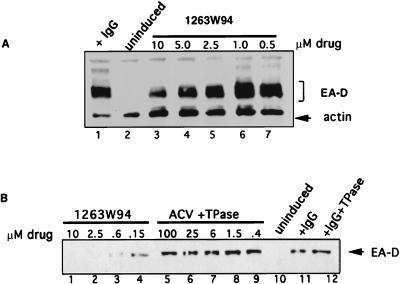
The EBV early gene BMRF1 encodes EA-D, a processivity factor for the viral DNA polymerase (71). Detection of EA-D is commonly used as a marker for EBV lytic reactivation. Total cellular lysate was prepared from induced Akata cells incubated for 72 h in the presence of increasing amounts of drug. As expected, EA-D was detected in the induced cells (Fig. 4A, lane 1) but not in uninduced cells (lane 2), indicating that the uninduced cells are tightly latent and efficiently induced by IgG antibody. Surprisingly, 1263W94 affected the appearance of EA-D in this and other assays in a dose-dependent manner (Fig. 4A [lanes 3 to 7] 4B [lanes 1 to 4]). This effect was specific for 1263W94 as the amount of EA-D protein was not decreased after the addition of ACV (Fig. 4B, lanes 5 to 9), even at levels of ACV which completely inhibited viral DNA replication (25 to 100 μM ACV [Fig. 3, lanes 5 and 6). The decrease in EA-D protein was probably not due to inhibition of viral DNA replication, as levels were similarly reduced 12 h following induction (data not shown), well before the peak of viral DNA replication in induced Akata cells (14). The addition of TPase to samples containing ACV (Fig. 3) did not affect EA-D protein levels.
FIG. 4.
1263W94 inhibits appearance of EBV EA-D. Akata cells were induced by cross-linking with IgG and incubated in the presence or absence of 1263W94 for 3 days. Cells were harvested, and total protein lysate was prepared for immunoblot analysis; 30 μg of lysate were separated on an SDS–9% polyacrylamide gel, transferred to Immobilon, and probed with an EA-D-specific antibody. Detection was by enhanced chemiluminescence.
1263W94 does not inhibit transcription of EA-D.
As EA-D protein levels were reduced in the presence of 1263W94, we analyzed BMRF1 (EA-D) RNA by Northern blot analysis (Fig. 5). BMRF1 RNA levels were not lower in samples incubated with 5 μM 1263W94 (Fig. 5, lanes 7 to 11) than in control samples (lanes 2 to 6). Therefore, the reduced levels of EA-D protein do not correlate with decreased RNA levels. In contrast, BMRF1 RNA levels increased in the presence of ACV (lanes 12 to 16).
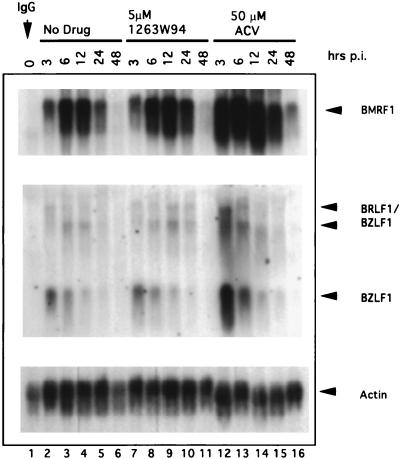
FIG. 5.
1263W94 does not inhibit the transcription of EA-D or the immediate-early genes R and Z. Akata cells were induced and then incubated in the absence of drug or with 5.0 μM 1263W94 or 50 μM ACV. Samples were harvested at times postinduction (p.i.), and total RNA was prepared. Northern blot analysis was performed with radiolabeled probes specific for EA-D, Z, or actin.
Since BMRF1 promoter activity is regulated by the viral immediate-early gene products R and Z (29, 32), these two proteins could be potential targets for 1263W94. Therefore we also measured levels of R and Z RNAs. R and Z are transcribed from two separate promoters located in the immediate-early region of the EBV genome. One, Rp, encodes two bicistronic transcripts of 4.0 and 3.3 kb, each of which contains the entire coding sequence for Z and R. A second promoter, Zp, encodes a 1.0-kb BZLF1 transcript (4, 47). There was no significant change in the levels of the 1.0-kb (BZLF1) or 3.3- or 4.0-kb (BRLF1/BZLF1) RNA in samples incubated with 1263W94 (Fig. 5, lanes 6 to 11) compared with samples which did not receive compound (lanes 2 to 6). Surprisingly, there was a significant increase in R and Z RNAs in the samples treated with ACV (lanes 12 to 16), and this effect may account for the increase in EA-D RNA.
1263W94 inhibits phosphorylation of EA-D.
Since EA-D RNA levels were unaffected by 1263W94, we investigated the stability of EA-D protein. Akata cells were induced and then incubated with compounds as indicated for 24 h. Next, cycloheximide was added to a final concentration of 50 μg/ml, and samples were harvested at 0, 2, 6, and 12 h after cycloheximide addition. Equivalent amounts of protein lysate were used for immunoblot analysis.
Overall, EA-D protein levels were lower in the presence of 1263W94 (Fig. 6A, lanes 7 to 10) than in the controls (lanes 2 to 5). However, the most striking observation is the virtual disappearance of the more slowly migrating form(s) of EA-D in the samples treated with 1263W94 (lanes 7 to 10). This form of EA-D appeared less stable in the presence of 1263W94. It has been previously shown, by 32P labeling, that multiple phosphorylated forms of EA-D exist in lytically infected cells (76); this result suggested that 1263W94 may inhibit the phosphorylation of EA-D and that this form of the protein may be less stable in the presence of 1263W94.
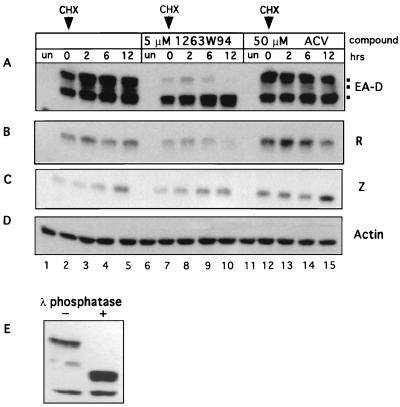
FIG. 6.
1263W94 inhibits the phosphorylation of EA-D. Akata cells were induced, and compound was immediately added. The samples were incubated for 24 h, then cycloheximide (CHX) was added to a final concentration of 50 μg/ml, and incubation was continued. Samples were harvested at the times indicated after addition of cycloheximide. Whole-cell lysates were prepared, and protein was quantitated by the Bradford method; 50 μg of each sample were separated on an SDS–10% polyacrylamide gel, and the protein was transferred to an Immobilon membrane. The membrane was probed with antibodies specific for EA-D (A), R (B), Z (C), or actin (D). (E) Akata cell lysates induced for 24 h (20 μg) were incubated with lambda phosphatase for 3 h at 30°C. The samples were separated by SDS-PAGE and then probed for EA-D as described above. un, untreated.
To confirm that HMW EA-D bands were phosphorylated forms of the protein, we treated the control lysate sample (used in lane 2) with lambda phosphatase. The slowly migrating form of EA-D protein was resolved into a more rapidly migrating band in the presence of phosphatase (Fig. 6D), confirming that EA-D is differentially phosphorylated.
The amount of R protein, but not Z protein, was also slightly reduced in the presence of 1263W94 (Fig. 6B). Occasionally, a reduction in the levels of Z protein was also noted; however, these differences were not consistently seen at this level (5 μM) of drug (data not shown), and the decrease was not as dramatic as that noted for EA-D. In contrast, the amounts of Z and R proteins were increased by addition of ACV as expected, as increased levels of RNA were also observed (Fig. 5). To confirm that the cycloheximide block was effective, 0- and 2-h lysate samples were probed for IRF-1, a short-lived cellular protein (28, 75). IRF-1 was abundant at 0 h but virtually disappeared by 2 h following addition of cycloheximide (data not shown).
EA-D is poorly phosphorylated in an EBV-negative cell line.
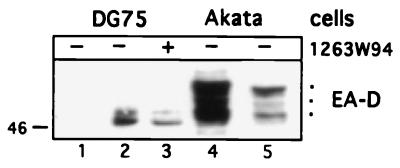
Next, we determined the phosphorylation status of EA-D in an EBV-negative cell line, DG75. DG75 cells were transfected with pSG5 vector (Fig. 7, lane 1) or an expression plasmid pSG5-EA-D and incubated without compound (lane 2) or with 5 μM 1263W94 (lane 3). Cells were harvested 24 h after transfection, and protein lysate was analyzed by Western blotting for EA-D along with control lysates from 24-h-induced Akata cells (lanes 4 and 5). Multiple forms of EA-D are detected in cells transfected with EA-D plasmid. However, the most highly phosphorylated form(s) of EA-D, which is readily detectable in induced Akata cells (54 kDa, lanes 4 and 5), is not present in the transfected cells (lanes 2 and 3). These data suggest that EA-D may be phosphorylated to some extent by a cellular kinase; however, further phosphorylation of EA-D involves a virally encoded kinase, a virally induced kinase, or possibly a cellular kinase which is induced by IgG cross-linking.
FIG. 7.
EA-D is underphosphorylated in DG75 cells. DG75 cells (107) were transfected with 5 μg of pSG5 vector (lane 1) or pSG5-BMRF1, with (lane 3) or without (lane 2) the addition of 5 μM 1263W94. Cells were harvested 24 h after transfection, and protein lysates were prepared. Akata cells (lanes 4 and 5) were induced by IgG cross-linking, and lysates were prepared from cells harvested 24 h postinduction. Following SDS-PAGE separation, the proteins were transferred and then probed with EA-D antibody as described in the text. Lane 5 is a shorter exposure of lane 4. Size at the left is indicated in kilodaltons.
EA-D phosphorylation is temporally regulated in Akata cells.
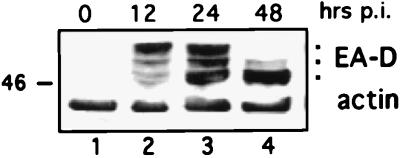
Since the data suggested that EA-D might be regulated in part by phosphorylation, we examined the kinetics of EA-D phosphorylation in induced Akata cells. EA-D phosphorylation appears to be temporally regulated following induction of the lytic cycle (Fig. 8). At 12 h postinduction, EA-D is predominantly hyperphosphorylated (lane 2). However, by 24 h approximately half of the total EA-D is unphosphorylated (lane 3), and by 48 h virtually all of the protein is unphosphorylated. This finding expands upon a previous report which shows that multiple forms of EA-D occur 9 and 12 h postinduction, concurrent with the initiation of viral DNA synthesis (14). These data suggest that EA-D function might be regulated in part by phosphorylation.
FIG. 8.
EA-D phosphorylation is temporally regulated during the lytic cycle. Akata cells were induced by IgG cross-linking, and samples were harvested at 0, 12, 24, and 48 h postinduction (p.i.). Lysate samples (50 μg) were analyzed for EA-D protein.
DISCUSSION
The l-riboside 1263W94 is a potent inhibitor of EBV DNA replication, with an IC50 of 0.15 to 1.1 μM, at least 10-fold more potent than ACV in identical assays. The toxicity (50% tissue culture inhibitory dose) of 1263W94 was determined for several lymphocyte lines, IM-9, MOLT4, and U937, and varied from 55 to 90 μM. Therefore, the in vitro therapeutic index for 1263W94 is 50 to 100 in this assay system. Phase I trials in asymptomatic human immunodeficiency virus-positive volunteers show that the drug is well tolerated at these levels, with no adverse side effects (74).
In contrast, another HCMV inhibitor, the d-riboside BDCRB, was ineffective against EBV replication. In contrast to its activity against HCMV, neither inhibition of processing of EBV precursor DNA nor inhibition of formation of linear genomes was detected. This difference in antiviral selectivity may be due to the lack of identity between HCMV UL89, a putative terminase which is the target for BDCRB, and its EBV homolog, BD/BGRF1, at a single critical residue (73).
EA-D is a component of the EBV DNA polymerase complex (9, 35, 72) and is essential for EBV lytic replication (25, 27). The protein functions as a DNA processivity factor (8, 71). In addition, EA-D is a nonspecific DNA-binding protein (70) and has transactivator function. EA-D transactivates the promoter for the BHLF1 gene through a GC-rich element which is also an essential element of the lytic origin of replication (_ori_-Lyt) (78, 79). As EA-D is essential for viral lytic replication, functional inactivation of this protein would inhibit viral DNA synthesis.
1263W94 appeared to inhibit both the accumulation and the phosphorylation of EA-D, but it is not clear at this point if these two observations are related or are due to different effects of the drug. EA-D protein, but not RNA, levels were consistently decreased. As this decrease was noted at early times after induction, the reduction in EA-D protein levels are unlikely to be due to the effects of a replication block. It is possible that translation initiation is inhibited or that newly synthesized EA-D is rapidly degraded in the presence of the compound. In addition, the accumulation of a hyperphosphorylated form of EA-D was selectively inhibited, and this form of the protein specifically appeared to be less stable in the presence of the compound. This may contribute to the overall reduced levels of EA-D. These possibilities are now being investigated to determine the mechanism underlying these observations. No decrease in EA-D was detected in cells treated with ACV at levels that completely blocked replication. We frequently also noticed a decrease in the levels of R and Z proteins in the presence of 1263W94, although not as dramatic as that observed for EA-D. In addition, the decreased levels of R and Z did not result in noticeably decreased levels of EA-D mRNA. At this point it is unclear to what extent the decrease in R, Z, and EA-D proteins contributes to the antiviral mechanism of the compound. In contrast, incubation of cells with 50 μM ACV caused a dramatic increase in the transcription of R, Z, and EA-D RNAs. The mechanism underlying this increase remains to be addressed.
As mentioned previously, 1263W94 consistently inhibited the appearance of hyperphosphorylated forms of EA-D. This effect implies that a direct or indirect target of the compound may be a kinase that phosphorylates EA-D. EA-D was phosphorylated to low levels in the EBV-negative cell line DG75, presumably due to a cellular kinase. However, additional hyperphosphorylated forms of EA-D appear in lytically induced Akata cells, suggesting that a viral kinase, or possibly a virally induced cellular kinase, may be responsible for hyperphosphorylation of EA-D. In addition, we show that EA-D hyperphosphorylation is temporally regulated in induced Akata cells, being readily detected 12 h after induction and decreasing by 48 h. Taken together, these data suggest that phosphorylation may contribute to the regulation of EA-D activity. Since EA-D is essential for viral replication, the inhibition of EA-D phosphorylation and protein accumulation by 1263W94 may account for the antiviral effect of the compound.
A putative EBV kinase has been identified based on motifs present in catalytic domains of protein kinases (62). This gene, BGLF4, has recently been cloned (26) and homologs have been identified in the other human herpesviruses (7, 49, 50, 61, 62). These related kinases are in some instances nonessential for replication in vitro (13, 19, 25, 30, 54) but may be required for pathogenesis. The HSV homolog, UL13, phosphorylates an HSV early protein, ICP22 (55), as well as the cellular elongation factor EF-1δ (34) and has been implicated in pathogenesis in a mouse model (13). Another HSV kinase, US3, was shown to be required for replication in mice but not in tissue culture (37). The varicella-zoster virus homolog, open reading frame 47, has recently been shown to be essential for replication in thymocytes and skin cells (48). Therefore, it is possible that a virally encoded kinase is responsible for the phosphorylation of EA-D. BGLF4 has recently been cloned in this laboratory, and preliminary data suggest that EA-D may be a potential target for the kinase (26).
The essential role of EA-D in EBV DNA replication has been appreciated for years. In fact, its homolog in HSV, UL42, has been proposed as a target for inhibition of viral replication because of its specificity (10, 20, 21). Until recently, it was thought that most EBV-associated malignancies were not good candidates for antiviral therapy, as they are largely due to latent EBV infection. However, recent evidence suggests that in several of these diseases a period of viral replication may precede and perhaps trigger onset of disease, which raises the possibility of prophylactic therapy. Expression of viral replicative genes is detected in a subset of cells in B-cell lymphomas, including posttransplant lymphomas and AIDS-associated lymphoma (33, 58, 59). Prior to the onset of nasopharyngeal carcinoma, serum IgA titers to EBV replicative antigens, including EA-D, increase in response to viral reactivation at unknown sites (18, 31).
Currently, prospects for anti-EBV therapy are limited to a few drugs, all of which target the viral polymerase. Since drug-resistant mutants to the polymerase gene arise in other herpesviruses, and probably EBV, it is possible that cross-resistant viral mutants will arise during prolonged treatment with such drugs. Therefore, novel antiviral compounds that target genes other than herpesvirus polymerases, yet retain antiviral specificity, are likely to be needed.
In this work, we show that the antiviral compound 1263W94 inhibits EBV replication more effectively than ACV. In addition, we present evidence that the essential replication factor, EA-D, may be an ultimate target of the antiviral compound 1263W94, as both the accumulation and phosphorylation of EA-D are decreased in the presence of the compound. We also confirm that the phosphorylation of EA-D is temporally regulated, and propose that a EA-D may be a substrate for a viral kinase encoded by the BGLF4 ORF. Recently, Davis et al. mapped HCMV resistance to 1263W94 to the viral kinase, UL97 (17). Since 1263W94 is effective against both HCMV and EBV, the effects of the compound on both the HCMV replication cofactor, UL44, and the viral kinase, UL97, are now being investigated.
ACKNOWLEDGMENTS
We thank Shannon Kenney for helpful observations and discussions and for critical review of the manuscript. We thank David Dorsky for the BMRF1 plasmid.
This work was supported by NCI grant CA-19014 and by a grant from Glaxo Wellcome Co.
REFERENCES
- 1.Adams A, Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc Natl Acad Sci USA. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a ‘Burkitt like’ malignant lymphoma (line DG-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 3.Benz W C, Siegel P J, Baer J. Effects of adenine arabinoside on lymphocytes infected with Epstein-Barr virus. J Virol. 1978;27:475–482. doi: 10.1128/jvi.27.3.475-482.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggen M, Bodescot M, Perricaudet M, Farrell P. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J Virol. 1987;61:3120–3122. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biron, K. K. Unpublished data.
- 6.Biron K K, Davis M G, Stanat S C, Walton L M, Smith A, Koszalka G W, Drach J C, Townsend L B, Harvey R J. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. Antiviral activity and mechanism of action of 1263W94, a benzimidazole riboside inhibitor of human cytomegalovirus, abstr. H85; p. 178. [Google Scholar]
- 7.Chee M S, Lawrence G L, Barrell B G. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- 8.Chen L-W, Lin L-S, Chang Y S, Liu S T. Functional analysis of EA-D of Epstein-Barr virus. Virology. 1995;211:593–597. doi: 10.1006/viro.1995.1443. [DOI] [PubMed] [Google Scholar]
- 9.Chiou J F, Li J K, Cheng Y C. Demonstration of a stimulatory protein for virus-specified DNA polymerase in phorbol ester-treated Epstein-Barr virus-carrying cells. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.17.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow C S, Coen D M. Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J Virol. 1995;69:6965–6971. doi: 10.1128/jvi.69.11.6965-6971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coker-Vann J, Dolin R. Effect of adenine arabinoside on Epstein-Barr virus in vitro. J Infect Dis. 1977;135:447–453. doi: 10.1093/infdis/135.3.447. [DOI] [PubMed] [Google Scholar]
- 12.Colby B M, Shaw J E, Elion G B, Pagano J S. Effect of acyclovir [9-(2-hydroxyethoxymethyl)guanine] on Epstein-Barr virus DNA replication. J Virol. 1980;34:560–568. doi: 10.1128/jvi.34.2.560-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulter L J, Moss H W, Lang J, McGeoch D J. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 14.Daibata M, Sairenji T. Epstein-Barr virus (EBV) replication and expressions of EA-D (BMRF1 gene product), virus-specific deoxyribonuclease, and DNA polymerase in EBV-induced Akata cells. Virology. 1993;196:900–904. doi: 10.1006/viro.1993.1555. [DOI] [PubMed] [Google Scholar]
- 15.Daluge S M, Purifoy D J M, Savina P M, St. Clair M H, Parry N R, Dev I K, Novak P, Ayers K M, Reardon J E, Roberts G B, Fyfe J A, Blum M R, Averett D R, Dornsife R E, Domin B A, Ferone R, Lewis D A, Krenitsky T A. 5-Chloro-2′,3′-dideoxy-3′-fluorouridine (935U83), a selective anti-human immunodeficiency virus agent with an improved metabolic and toxicological profile. Antimicrob Agents Chemother. 1994;38:1590–1603. doi: 10.1128/aac.38.7.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta A K, Hoop R E. Mechanism of inhibition of Epstein-Barr virus replication by phosphonoformic acid. Virology. 1981;114:52–59. doi: 10.1016/0042-6822(81)90251-8. [DOI] [PubMed] [Google Scholar]
- 17.Davis M G, Talarico C L, Underwood M R, Baldanti F, Biron K K. Mapping of a mutation in human cytolomegalovirus resistant to 1263W94. Presented at the 23rd International Herpesvirus Workshop, 1–7 Aug. 1998, York, United Kingdom. 1998. [Google Scholar]
- 18.de The G. Epidemiology of Epstein-Barr virus and associated diseases in man. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Press; 1982. pp. 25–103. [Google Scholar]
- 19.de Wind N, Domen J, Berns A. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J Virol. 1992;66:5200–5209. doi: 10.1128/jvi.66.9.5200-5209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Digard P, Chow C S, Pirrit L, Coen D M. Functional analysis of the herpes simplex virus UL42 protein. J Virol. 1993;67:1159–1168. doi: 10.1128/jvi.67.3.1159-1168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digard P, Williams K P, Hensley P, Brooks I S, Dahl C E, Coen D M. Specific inhibition of herpes simplex virus DNA polymerase by helical peptides corresponding to the subunit interface. Proc Natl Acad Sci USA. 1995;92:1456–1460. doi: 10.1073/pnas.92.5.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drach J C, Townsend L B, Nassiri M R, Turk S R, Coleman L A, Devivar R V, Genzlinger G, Kreske E D, Renau T R, Westerman A C, Shipman C, Jr, Biron K K, Dornsife R, Kern E R. Benzimidazole ribonucleosides: a new class of antivirals with potent and selective activity against human cytomegalovirus. Antiviral Res. 1992;21:49. [Google Scholar]
- 23.Drew W L, Lalezari J P, Wang L H, Miner R C, Aberg J A, Wire M B, Jacobson M A. Program and Abstracts of the 38th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. In vivo anti-CMV activity and safety of oral 1263W94 in HIV-infected subjects with asymptomatic CMV shedding, abstr. H-25; p. 322. [Google Scholar]
- 24.Elion G B, Furman P A, Fyfe J A, de Miranda P, Beauchamp L, Schaeffer H J. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl)guanine. Proc Natl Acad Sci USA. 1977;74:5716–5720. doi: 10.1073/pnas.74.12.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gershburg, E. Unpublished data.
- 27.Hammerschmidt W, Sugden B. Identification and characterization of orilyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 28.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 29.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heineman T C, Cohen J I. The varicella-zoster virus (VZV) open reading frame 47 (ORF47) protein kinase is dispensable for viral replication and is not required for phosphorylation of ORF63 protein, the VZV homolog of herpes simplex virus ICP22. J Virol. 1995;69:7367–7370. doi: 10.1128/jvi.69.11.7367-7370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henle G, Henle W. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int J Cancer. 1976;17:1–7. doi: 10.1002/ijc.2910170102. [DOI] [PubMed] [Google Scholar]
- 32.Holley-Guthrie E A, Quinlivan E B, Mar E-C, S K. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen D (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz B Z, Raab-Traub N, Miller G. Latent and replicating forms of Epstein-Barr virus DNA in lymphomas and lymphoproliferative diseases. J Infect Dis. 1989;160:589–598. doi: 10.1093/infdis/160.4.589. [DOI] [PubMed] [Google Scholar]
- 34.Kawaguchi Y, Van Sant C, Roizman B. Eukaryotic elongation factor 1δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiehl A, Dorsky D. Cooperation of DNA polymerase and EA-D (BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 36.Koszalka G W, Chamberlain S D, Harvey R J, Drach J C, Townsend L B, Biron K K. Benzimidazoles for the treatment of human cytomegalovirus. Presented at the Ninth International Conference on Antiviral Research, 19–24 May 1996, Fukushima, Japan. 1996. [Google Scholar]
- 37.Kurachi R, Daikoku T, Tsurumi T, Maeno K, Nishiyama Y, Kurata T. The pathogenicity of a US3 protein kinase-deficient mutant of herpes simplex virus type 2 in mice. Arch Virol. 1993;133:259–273. doi: 10.1007/BF01313767. [DOI] [PubMed] [Google Scholar]
- 38.Liebowitz D, Kieff E. Epstein-Barr Virus. In: Roizman B, Whitely R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 107–172. [Google Scholar]
- 39.Lin J-C, Nelson D J, Lambe C U, Cho E I. Metabolic activation of 9([2-hydroxy-1-hydroxymethyl)ethoxy]methyl)guanine in human lymphoblastoid cell lines infected with Epstein-Barr virus. J Virol. 1986;60:569–573. doi: 10.1128/jvi.60.2.569-573.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J C, De Clerq E, Pagano J S. Inhibitory effects of acyclic nucleoside phosphonate analogs, including (S)-1-(3-hydroxy-2-phosphonomethoxypropyl)cytosine, on Epstein-Barr virus replication. Antimicrob Agents Chemother. 1991;35:2440–2443. doi: 10.1128/aac.35.11.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J C, De Clerq E, Pagano J S. Novel acyclic adenosine analogs inhibit Epstein-Barr virus replication. Antimicrob Agents Chemother. 1987;31:1431–1433. doi: 10.1128/aac.31.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J C, Smith M, Cheng Y C, Pagano J S. Epstein-Barr virus: inhibition of replication by three new drugs. Science. 1983;221:578–579. doi: 10.1126/science.6306771. [DOI] [PubMed] [Google Scholar]
- 43.Lin J C, Smith M C, Choi E I, De Clercq E, Verbruggen A, Pagano J S. Effect of (E)-5-(2-bromovinyl)-2′-deoxyuridine on replication of Epstein-Barr virus in human lymphoblastoid cell lines. Antiviral Res. 1985;1985(Suppl. 1):121–126. doi: 10.1016/s0166-3542(85)80018-8. [DOI] [PubMed] [Google Scholar]
- 44.Lin J C, Smith M C, Pagano J S. Comparative efficacy and selectivity of some nucleoside analogs against Epstein-Barr virus. Antimicrob Agents Chemother. 1985;27:971–973. doi: 10.1128/aac.27.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J C, Smith M C, Pagano J S. Prolonged inhibitory effect of 9-(1,3-dihydroxy-2-propoxymethyl)guanine against replication of Epstein-Barr virus. J Virol. 1984;50:50–55. doi: 10.1128/jvi.50.1.50-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez C, Watanabe K A, Fox J J. 2′-Fluoro-5-iodo-aracytosine, a potent and selective anti-herpesvirus agent. Antimicrob Agents Chemother. 1980;42:963–972. doi: 10.1128/aac.17.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manet E, Gruffat H, Trescol-Biemont M C, Moreno N, Chamvard P, Giot J F, Sergeant A. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional transactivators. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat J F, Zerboni L, Sommer M H, Heineman T C, Cohen J I, Kaneshima H, Arvin A M. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc Natl Acad Sci USA. 1998;95:11969–11974. doi: 10.1073/pnas.95.20.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng T I, Grose C. Serine protein kinase associated with varicella-zoster virus ORF 47. Virology. 1992;191:9–18. doi: 10.1016/0042-6822(92)90161-h. [DOI] [PubMed] [Google Scholar]
- 50.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagano J S. Extrachromosomal DNA. New York, N.Y: Academic Press; 1979. pp. 234–248. [Google Scholar]
- 52.Pagano J S. Epstein-Barr virus: therapy of active and latent infection. In: Jeffries D J, De Clercq E, editors. Antiviral chemotherapy. New York, N.Y: John Wiley & Sons Ltd.; 1995. pp. 155–195. [Google Scholar]
- 53.Pagano J S, Nonoyama M, Huang C-H. Possible episomes in eukaryotes: Proceedings of the Fourth Lepetit Colloquium. Amsterdam, The Netherlands: North-Holland; 1973. Epstein-Barr virus in human cells; pp. 218–228. [Google Scholar]
- 54.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purves F C, Roizman B. The UL13 gene of herpes simplex virus encodes the functions for post-translational processing associated with phosphorylation of the regulatory protein alpha 22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raab-Traub N, Dambaugh T, Kieff E. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype is an unusual deletion derivative. Cell. 1980;22:257–267. doi: 10.1016/0092-8674(80)90173-7. [DOI] [PubMed] [Google Scholar]
- 57.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–9. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 58.Rea D, Delecluse H J, Hamilton-Dutoit S J, Marelle L, Joab I, Edelman L, Finet J F, Raphael M. Epstein-Barr virus latent and replicative gene expression in post-transplant lymphoproliferative disorders and AIDS-related non-Hodgkin’s lymphomas. French Study Group of Pathology for HIV-Associated Tumors. Ann Oncol Suppl. 1994;5:113–116. doi: 10.1093/annonc/5.suppl_1.s113. [DOI] [PubMed] [Google Scholar]
- 59.Rea D, Fourcade C, Leblond V, Rowe M, Joab I, Edelman L, Bitker M O, Gandjbakhch I, Suberbielle C, Farcet J P, et al. Patterns of Epstein-Barr virus latent and replicative gene expression in Epstein-Barr virus B cell lymphoproliferative disorders after organ transplantation. Transplantation. 1994;58:317–324. [PubMed] [Google Scholar]
- 60.Rickinson A B, Epstein M A. Sensitivity of the transforming and replicative function of Epstein-Barr virus to inhibition by phosphonoacetate. J Gen Virol. 1978;40:409–420. doi: 10.1099/0022-1317-40-2-409. [DOI] [PubMed] [Google Scholar]
- 61.Russo J J, Bohenzky R A, Chien M C, Chen J, Maddalena Y M D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith R F, Smith T F. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J Virol. 1989;61:450–455. doi: 10.1128/jvi.63.1.450-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Summers W C, Klein G. Inhibition of Epstein-Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J Virol. 1976;18:151–155. doi: 10.1128/jvi.18.1.151-155.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt’s lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 65.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamm I, Folkers K, Shunk C H, Horsfall F L. Inhibition of influenza virus multiplication by N-glycosides of benzimidazoles. J Exp Med. 1954;99:227–250. doi: 10.1084/jem.99.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamm I, Sehgal P B. Halobenzimidazole ribosides and RNA synthesis of cells and viruses. Adv Virus Res. 1978;22:187–258. doi: 10.1016/s0065-3527(08)60775-7. [DOI] [PubMed] [Google Scholar]
- 68.Townsend L B. Benzimidazole ribonucleosides: design, synthesis and evaluation of 2,5,6-trichloro-1-(β-d-ribofuranosyl) benzimidazole (TCRB), 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl) benzimidazole (BDCRB), and some structurally related analogs as agents for human cytomegalovirus infection. Presented at the Fifth International Conference on Antiviral Research, 8–13 March 1992, Vancouver, British Columbia, Canada. 1992. [Google Scholar]
- 69.Townsend L B, Devovar R V, Turk S R, Nassiri M R, Drach J C. Design, synthesis and antiviral activity of certain 2,5,6-trihalo-1-(β-d-ribofuranosyl) benzimidazoles. J Med Chem. 1995;38:4098–4105. doi: 10.1021/jm00020a025. [DOI] [PubMed] [Google Scholar]
- 70.Tsurumi T. Purification and characterization of the DNA-binding activity of the Epstein-Barr virus DNA polymerase accessory protein BMRF1 gene products, as expressed in insect cells by using the baculovirus system. J Virol. 1993;67:1681–1687. doi: 10.1128/jvi.67.3.1681-1687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J Virol. 1993;67:7648–7653. doi: 10.1128/jvi.67.12.7648-7653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsurumi T, Kobayashi A, Tamai K, Daikoku T, Kurachi R, Nishiyama Y. Functional expression and characterization of the Epstein-Barr virus DNA polymerase catalytic subunit. J Virol. 1993;67:4651–4658. doi: 10.1128/jvi.67.8.4651-4658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Underwood M R, Harvey R J, Stanat S C, Hemphill M L, Miller T, Drach J C, Townsend L B, Biron K K. Inhibition of HCMV DNA maturation by a benzimidazole ribonucleotide is mediated through the UL89 gene product. J Virol. 1998;72:717–725. doi: 10.1128/jvi.72.1.717-725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L H, Yogendran L, Weller S, Sallabank P, Wiggs R, Bye A. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. A phase I trial evaluating the tolerability and pharmacokinetics of 1263W94 following single oral administration of escalating doses in normal healthy volunteers, abstr. H28; p. 168. [Google Scholar]
- 75.Watanabe N, Sakakibara J, Hovanessian A, Taniguchi T, Fujita T. Activation of IFN-β promoter element by IRF-1 requires a post-translational event in addition to IRF-1 synthesis. Nucleic Acids Res. 1991;16:4421–4428. doi: 10.1093/nar/19.16.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong K-M, Levine A J. Identification and mapping of Epstein-Barr virus early antigens and demonstration of a viral gene activator that functions in trans. J Virol. 1986;60:149–156. doi: 10.1128/jvi.60.1.149-156.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yajima Y, Tanaka A, Nonoyama M. Inhibition of productive replication of Epstein-Barr virus DNA by phosphonoacetic acid. Virology. 1976;71:352–354. doi: 10.1016/0042-6822(76)90119-7. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Q, Holley-Guthrie E, Ge J-Q, Dorsky D, Kenney S. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV ori-Lyt. Virology. 1997;230:22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Q, Hong Y, Dorsky D, Holley-Guthrie E, Zalani S, Elshiekh N A, Kiehl A, Kenney S. Functional and physical interaction between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]