RNA Polymerase III Transcription Factor IIIB Is a Target for Repression by Pocket Proteins p107 and p130 (original) (raw)
Abstract
RNA polymerase III (Pol III) transcription is subject to repression by the retinoblastoma protein RB, both in vitro and in vivo (R. J. White, D. Trouche, K. Martin, S. P. Jackson, and T. Kouzarides, Nature 382:88–90, 1996). This is achieved through a direct interaction between RB and TFIIIB, a multisubunit factor that is required for the expression of all Pol III templates (C. G. C. Larminie, C. A. Cairns, R. Mital, K. Martin, T. Kouzarides, S. P. Jackson, and R. J. White, EMBO J. 16:2061–2071, 1997; W.-M. Chu, Z. Wang, R. G. Roeder, and C. W. Schmid, J. Biol. Chem. 272:14755–14761, 1997). p107 and p130 are two closely related proteins that display 30 to 35% identity with the RB polypeptide and share some of its functions. We show that p107 and p130 can both repress Pol III transcription in transient transfection assays or when added to cell extracts. Pull-down assays and immunoprecipitations using recombinant components demonstrate that a subunit of TFIIIB interacts physically with p107 and p130. In addition, endogenous TFIIIB is shown by cofractionation and coimmunoprecipitation to associate stably with both p107 and p130. Disruption of this interaction in vivo by using the E7 oncoprotein of human papillomavirus results in a marked increase in Pol III transcription. Pol III activity is also deregulated in fibroblasts derived from p107 p130 double knockout mice. We conclude that TFIIIB is targeted for repression not only by RB but also by its relatives p107 and p130.
The retinoblastoma protein RB has two close relatives, called p107 and p130, to which it is 30 to 35% identical (reviewed in references 13 and 27). These three are often referred to as the pocket proteins, because most of their homology lies within a bipartite region called the pocket domain. They can each inhibit cell growth and proliferation when overexpressed in tumor cells, an effect that is associated with G1-specific cell cycle arrest (7, 30, 44, 45). A number of common target proteins have been found to interact with the pocket domains of RB, p107 and p130, including members of the E2F family of cellular transcription factors and the oncoprotein products of several DNA tumor viruses (reviewed in references 10, 15, 27, and 36). As a consequence, there is significant redundancy between the various pocket proteins. This is particularly marked for p107 and p130, which are much more closely related to each other (∼50% amino acid identity) than they are to RB (30 to 35% identity). A clear demonstration of the redundancy between p107 and p130 was provided by the phenotypes of knockout mice. Thus, animals lacking either p107 or p130 develop normally, whereas animals lacking both of these pocket proteins die within hours of being born (8). In contrast, _Rb_−/− mice die at midgestation, displaying defects in both proliferation and differentiation of certain cell lineages (6, 17, 23). This suggests that at least some functions of RB cannot be performed by p107 or p130. This contention is supported by the fact that many tumors contain mutations in Rb, whereas the genes encoding p107 and p130 are not targeted for inactivation in cancers.
One of the functions of RB that has been demonstrated relatively recently is the regulation of RNA polymerase (Pol) III transcription (reviewed in references 21 and 39). The synthesis of tRNA and 5S rRNA by Pol III in vivo was found to be fivefold more active in primary fibroblasts from _Rb_−/− mice than in the corresponding cells from Rb+/+ mice (43). Furthermore, recombinant RB will inhibit the transcription of a range of Pol III templates both in vitro and in transfected cells (5, 22, 43). These effects can be explained by the ability of RB to bind and inactivate a general Pol III factor called TFIIIB (5, 22). Immunoprecipitation and pull-down experiments demonstrated a physical interaction between recombinant RB and TFIIIB (5, 22). Furthermore, a stable association between endogenous RB and TFIIIB was shown by cofractionation and coimmunoprecipitation (5, 22). Substitutions in RB that prevent it from binding to TFIIIB also prevent it from repressing Pol III transcription (5). Functional assays with purified factors confirmed that TFIIIB is inactivated specifically by its interaction with RB (5, 22).
TFIIIB is a multisubunit complex that includes the TATA-binding protein (TBP) and an essential subunit called BRF (reviewed in references 14, 31, and 40). It is required for the expression of all class III genes, serving to recruit Pol III to a promoter and position it over the initiation site (18). This recruitment involves an interaction between BRF and Pol III (19, 38). The ability of RB to target TFIIIB provides it with the opportunity to regulate all Pol III-transcribed genes. This can be expected to have a major effect upon nuclear activity.
Since the three pocket proteins share a number of functions, it was important to determine whether p107 and p130 are also able to regulate Pol III transcription. A combination of in vitro and in vivo experiments indicate that this is the case. Recombinant p107 and p130 will both bind to TFIIIB and repress a variety of class III genes in cell extracts and in transfected cells. Furthermore, endogenous p107 and p130 can be shown by cofractionation and coimmunoprecipitation analyses to associate stably with endogenous TFIIIB. Specific inactivation of these pocket proteins by using the E7 oncoprotein of human papillomavirus will stimulate Pol III transcription in vivo. In addition, Northern blot analysis shows that Pol III activity is deregulated in fibroblasts derived from _p107_−/− _p130_−/− double knockout mice. We conclude that p107 and p130 share with RB the ability to bind and repress the general Pol III factor TFIIIB.
MATERIALS AND METHODS
Cell culture and transfection.
Embryonic fibroblasts (MEFs) derived from p107+/− p130+/− and _p107_−/− _p130_−/− mice were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal calf serum, as described previously (16). MEFs were made quiescent by culture in DMEM containing 0.5% fetal calf serum. The human osteosarcoma cell line SAOS2 and the murine fibroblast line NIH 3T3 were cultured in DMEM with 10% fetal calf serum, as described previously (20, 45). Cell lines were transiently transfected by the calcium-phosphate precipitation method. DNA precipitates were left on the plates overnight, and then the cells were washed with phosphate-buffered saline and cultured for 24 h before being harvested. Total RNA was extracted with TRI reagent (Sigma), according to the manufacturer’s instructions. It was then analyzed by primer extension with both VAI-specific (5′-CACGCGGGCGGTAACCGCATG-3′) and chloramphenicol acetyltransferase (CAT)-specific (5′-CGATGCCATTGGGATATATCA-3′) labelled primers. Primer extension reactions were conducted as previously described (43).
Northern blotting.
Total cellular RNA was extracted by using TRI reagent (Sigma), according to the manufacturer’s instructions. Agarose gel electrophoresis, Northern transfer, and hybridization were carried out as described previously (3). The B2 gene probe was a 240-bp _Eco_RI-_Pst_I fragment from pTB14. The acidic ribosomal phosphoprotein (ARPP) P0 probe was a 1-kb _Eco_RI-_Hin_dIII fragment from the mouse cDNA (16).
Plasmids.
The pVAI, pHu5S3.1, and pGlu6 plasmids contain the adenovirus VAI gene, a human 5S rRNA gene, and a human tRNAGlu6 gene, respectively, and have been described by White et al. (41). Plasmid pTB14 contains a mouse B2 gene (42). pCMV-p107 and pCMV-p130 contain full-length p107 and p130, respectively, in the pCDNA3 vector (45). pCMV-RBΔ22 contains a null mutant version of full-length RB, in which exon 22 is deleted, in the pCDNA3 vector (30). pCAT (Promega) contains the CAT gene driven by the simian virus 40 promoter and enhancer. Expression constructs encoding the E7 oncoprotein of human papillomavirus type 16 and its mutant derivatives Δ21–35 and GLY26 have been described previously (20).
Preparation of extracts and protein fractions.
Glutathione _S_-transferase (GST) fusion proteins were expressed in bacteria and purified on glutathione-agarose as described previously (22). GST-p107 contains residues 249 to 936 of p107, and GST-p130 contains residues 372 to 1139 of p130 (9).
Whole-cell extracts were prepared from exponentially growing cells according to the method of Manley et al. (25). Nuclear extracts were purchased from the Computer Cell Culture Center (Mons, Belgium). PC-B is the 0.1 to 0.35 M KCl step fraction from a phosphocellulose column and contains both TFIIIB and Pol III (32). PC-C is the 0.35 to 0.6 M KCl step fraction from a phosphocellulose column and contains both TFIIIC and Pol III (32). QS-PC-B contains TFIIIB that has been fractionated by gradient chromatography on Q-Sepharose followed by phosphocellulose step chromatography, as described previously (3). DNA affinity purification of TFIIIC was carried out by applying a PC-C fraction to a B-block oligonucleotide resin, as described previously (41). A25(0.15) contains TFIIIB, and A25(1.0) contains Pol III; both were prepared by applying a PC-B fraction to DEAE-Sephadex, as described previously (41). The CHep-1.0 fraction contains TFIIIC that has been prepared by heparin-Sepharose chromatography of a PC-C fraction, as described previously (41). Gradient chromatography of a PC-B fraction on heparin-Sepharose was carried out as described previously (22).
Transcription assays.
Transcription reactions were carried out as described previously (42), except that pBR322 was not included and the incubations were for 60 min at 30°C. Radiolabelled transcripts were resolved on 7% polyacrylamide sequencing gels and detected by autoradiography.
Pull-down assays.
Reticulocyte lysate (Promega) was used to synthesize BRF in the presence of [35S]Met and [35S]Cys, according to the manufacturer’s specifications. Lysate (20 μl) was then incubated at 4°C on an orbital shaker with 20 μl of glutathione-agarose beads carrying equivalent amounts (as estimated by Coomassie blue staining) of immobilized GST, GST-RB, GST-p107, or GST-p130. After 3 h, the samples were pelleted, supernatants were removed, and the beads were washed five times with 700 μl of LDB buffer (20 mM HEPES-KOH [pH 7.9], 17% glycerol, 100 mM KCl, 12 mM MgCl2, 0.1 mM EDTA, 2 mM dithiothreitol). Material that remained bound to the beads was then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Immunoprecipitation.
Whole-cell extract (150 μg) was incubated at 4°C on an orbital shaker with 20 μl of protein A-Sepharose beads carrying equivalent amounts of prebound immunoglobulin G. Samples were then pelleted, supernatants were removed, and the beads were washed five times with 150 μl of LDB buffer. The bound material was analyzed by Western blotting. In the experiment shown in Fig. 4, reticulocyte lysate (15 μl) containing BRF translated in the presence of [35S]Met and [35S]Cys was mixed with nuclear extract (150 μg) during immunoprecipitation. In this case, the precipitated material was analyzed by autoradiography rather than Western blotting.
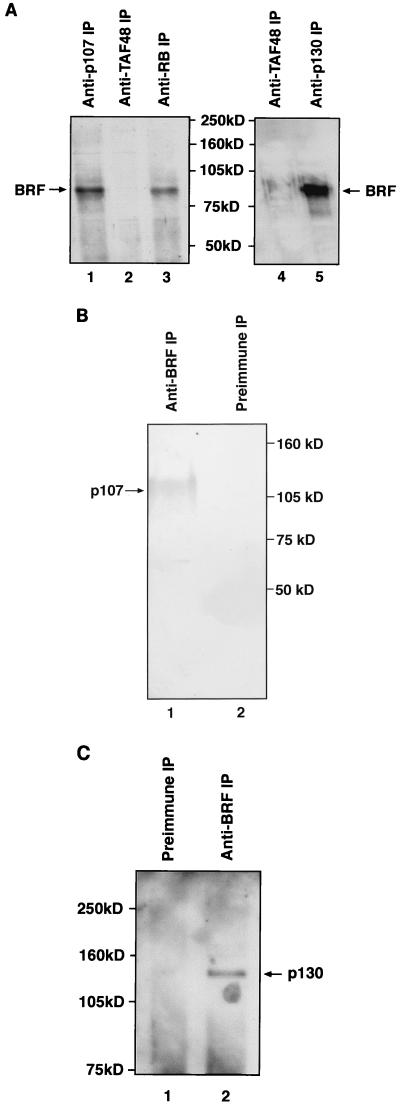
FIG. 4.
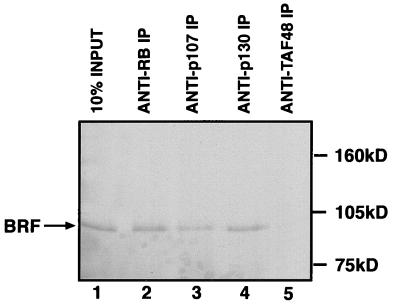
The BRF subunit of TFIIIB coimmunoprecipitates with endogenous RB, p130, and p107. Reticulocyte lysate (15 μl) containing in vitro-translated BRF was immunoprecipitated (IP) in the presence of 150 μg of HeLa whole-cell extract by using anti-RB antibody C-15 (lane 2), anti-p107 antibody C-18 (lane 3), anti-p130 antibody C-20 (lane 4), or anti-TAFI48 antibody M-19 (lane 5). Proteins retained after extensive washing were resolved on an SDS–7.8% polyacrylamide gel and then visualized by autoradiography. Lane 1 shows 10% of the input reticulocyte lysate containing in vitro-translated BRF.
Antibodies and Western blotting.
The antibodies used in this study were C-15 (Santa Cruz) and G99-549 (Pharminogen) against RB, SD9 (Santa Cruz) and C-18 (Santa Cruz) against p107, anti-Rb2 (Transduction Laboratories) and C-20 (Santa Cruz) against p130, M-19 (Santa Cruz) against TAFI48, 128 against BRF (2, 3), and Ab2 against TFIIICα (33). Western immunoblot analysis was performed as previously described (41).
RESULTS
The pocket proteins p107 and p130 inhibit Pol III transcription in transfected cells.
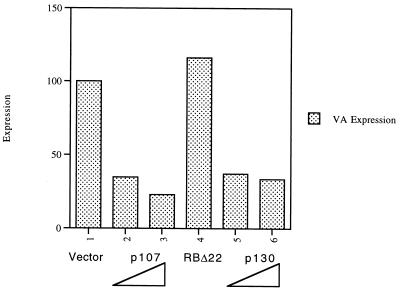
In order to examine whether p107 and p130 can inhibit Pol III transcription in vivo, the human osteosarcoma cell line SAOS2 was transfected with expression vectors encoding these pocket proteins. Primer extension analysis was used to determine the levels of transcription of a cotransfected VAI gene, which is used as a Pol III reporter. A control plasmid in which the CAT gene is driven by the simian virus 40 early promoter was also included in order to normalize for transfection efficiency. After correction for CAT RNA levels, p107 and p130 were found to repress VAI expression by three- to fourfold. This effect is specific, since no repression was obtained by using an RB null mutant (Δ22), from which the pocket region encoded by exon 22 had been deleted (Fig. 1). Similar results were obtained in transfections using the C33A human cervical carcinoma cell line, where VAI was again repressed by p107 and p130, but not by the RB Δ22 mutant (4). We conclude that the RB-related pocket proteins p107 and p130 can inhibit Pol III transcription in vivo.
FIG. 1.
Overexpression of p107 and p130 inhibits Pol III transcription in vivo. SAOS2 cells were transfected with pVAI (0.5 μg), pCAT (4 μg), and pCDNA3 vector (2 μg in lane 1, 1.5 μg in lanes 2 and 5), pCMV-p107 (0.5 μg in lane 2, 2 μg in lane 3), pCMV-RBΔ22 (2 μg in lane 4), or pCMV-p130 (0.5 μg in lane 5, 2 μg in lane 6). VAI and CAT RNA levels were assayed by primer extension and then quantitated by using a phosphorimager. The values shown for VAI have been normalized to the levels of CAT to correct for transfection efficiency and are expressed relative to the value obtained for the pCDNA3 control (arbitrarily designated 100%).
Recombinant p107 and p130 inhibit Pol III transcription in vitro.
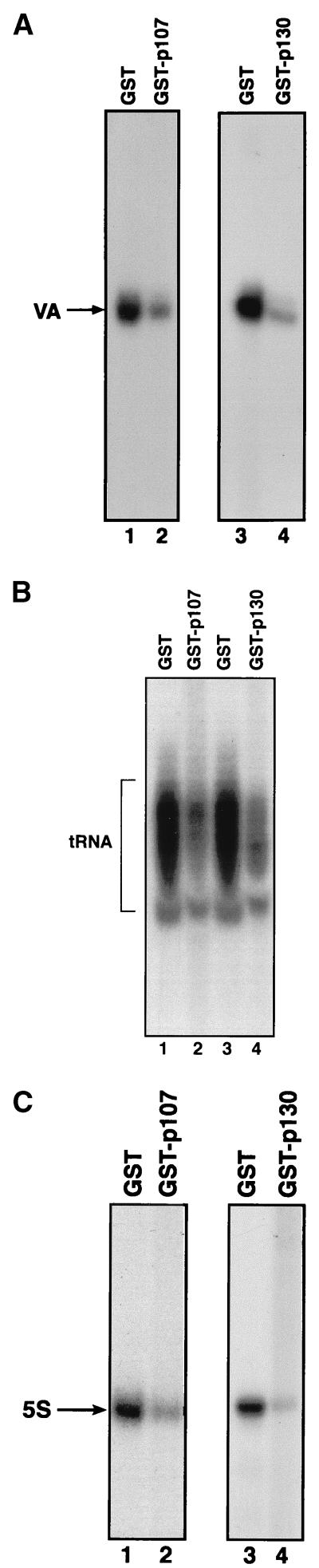
p107 and p130 were expressed in bacteria as fusions with GST and then purified by using glutathione-agarose beads. When added to cell extracts, these fusion proteins were found to inhibit the transcription of a range of Pol III templates (Fig. 2). For example, GST-p107 and GST-p130 both repressed VAI (Fig. 2A), tRNA (Fig. 2B), and 5S rRNA (Fig. 2C) genes by two- to fourfold relative to levels of expression obtained in the presence of an equal amount of GST alone. This response is comparable with that obtained in vivo (Fig. 1). In addition to these templates, p107 and p130 fusion proteins were also found to inhibit expression of Alu and U6 snRNA genes (4). We conclude that these pocket proteins can serve as general repressors of Pol III transcription.
FIG. 2.
Recombinant p107 and p130 inhibit transcription of a range of Pol III templates in vitro. (A) pVAI template (VA [250 ng]) was transcribed by using 10 μg of HeLa cell extract that had been preincubated for 15 min at 30°C with 250 ng of GST (lanes 1 and 3), GST-p107 (lane 2), or GST-p130 (lane 4). (B) pGlu6 template (250 ng) was transcribed by using 10 μg of HeLa cell extract that had been preincubated for 15 min at 30°C with 250 ng of GST (lanes 1 and 3), GST-p107 (lane 2), or GST-p130 (lane 4). The cluster of bands obtained reflects processing of the primary tRNA transcript. (C) pHu5S3.1 template (250 ng) was transcribed by using 10 μg of HeLa cell extract that had been preincubated for 15 min at 30°C with 250 ng of GST (lanes 1 and 3), GST-p107 (lane 2), or GST-p130 (lane 4).
Recombinant p107 and p130 interact with the BRF subunit of TFIIIB.
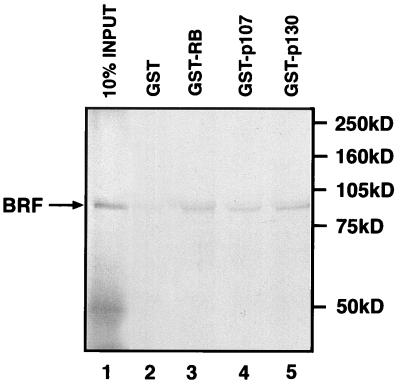
RB has been shown previously to associate with TFIIIB to form a stable complex that includes BRF (5, 22). To test whether the same is true of p107 and p130, these pocket proteins were expressed as GST fusions, purified and immobilized on glutathione-agarose beads. They were then incubated with radiolabelled BRF that had been translated in vitro by using a reticulocyte lysate. Proteins that remained bound to the beads after extensive washing were resolved by SDS-PAGE and then visualized by autoradiography (Fig. 3). Beads that carry GST-p107 or GST-p130 were found to retain BRF with an efficiency that is comparable to the binding obtained by using GST-RB. This effect is largely dependent on the presence of pocket protein sequences, since beads carrying an equal amount of GST alone retained only background levels of BRF. The data suggest that each of the known pocket proteins can associate with the BRF subunit of human TFIIIB. The interaction with BRF may be direct, but we cannot exclude an involvement of other proteins that are present in the reticulocyte lysate.
FIG. 3.
The BRF subunit of TFIIIB binds to recombinant RB, p107, and p130. Reticulocyte lysate containing in vitro-translated BRF was incubated in the presence of glutathione beads carrying equal amounts of GST (lane 2), GST-RB (lane 3), GST-p107 (lane 4), or GST-p130 (lane 5). Proteins retained after extensive washing were resolved on an SDS–7.8% polyacrylamide gel and then visualized by autoradiography. Lane 1 shows 10% of the input reticulocyte lysate containing in vitro-translated BRF. The truncated species of ∼50 kDa that appears in lane 1 is likely the result of degradation or internal initiation.
Endogenous p107 and p130 associate with the BRF subunit of TFIIIB.
Immunoprecipitation experiments were carried out to test whether BRF also interacts with endogenous cellular p107 and p130. Radiolabelled BRF was mixed with a HeLa cell extract that contains each of the pocket proteins in a wild-type state. The mixture was then immunoprecipitated by using a range of different antibodies immobilized on protein A beads, and the precipitates were examined for the presence of BRF by SDS-PAGE and autoradiography (Fig. 4). Antisera specific for RB, p107, and p130 were each found to coprecipitate readily detectable amounts of BRF. This effect is specific, since it was not observed by using a control antiserum against the TAFI48 subunit of the Pol I factor SL1. Similar results were obtained with alternative antisera specific for the pocket proteins (35).
Immunoprecipitations were also carried out to test whether endogenous TFIIIB can be found in association with endogenous p107 and p130. HeLa cell extracts containing wild-type pocket proteins were subjected to immunoprecipitation with a range of antibodies immobilized on protein A beads; the precipitated material was then resolved by SDS-PAGE and probed for the presence of BRF by Western blotting (Fig. 5A). Antibodies that specifically recognize either RB, p107, or p130 were each found to coprecipitate BRF. In contrast, no BRF was detected in immunoprecipitates obtained with a control antibody against the TAFI48 subunit of SL1. The presence of BRF in the material that coprecipitated with the pocket proteins was confirmed by using a second antiserum raised against a different region of the BRF polypeptide (35). As an additional test of specificity, we examined whether the Pol III factor TFIIIC2 could also be coprecipitated with the various pocket proteins. However, the α subunit of TFIIIC2 was not detected in immunoprecipitates obtained with antisera specific for RB, p107, or p130 (35). We conclude that the association observed between endogenous TFIIIB and the endogenous pocket proteins is a specific phenomenon.
FIG. 5.
Endogenous pocket proteins coimmunoprecipitate specifically with endogenous TFIIIB. (A) HeLa cell extract (150 μg) was immunoprecipitated (IP) by using anti-p107 antibody C-18 (lane 1), anti-TAFI48 antibody M-19 (lanes 2 and 4), anti-RB antibody C-15 (lane 3), and anti-p130 antibody C-20 (lane 5). Precipitated material was resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western blotting with anti-BRF antiserum 128. (B) HeLa cell extract (150 μg) was immunoprecipitated by using anti-BRF antiserum 128 (lane 1) or preimmune serum (lane 2). Precipitated material was resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western blotting with anti-p107 antibody SD9. (C) HeLa cell extract (150 μg) was immunoprecipitated by using anti-BRF antiserum 128 (lane 2) or the corresponding preimmune serum (lane 1). Precipitated material was resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western blotting with anti-Rb2 antibody against p130.
We also performed the converse experiment, in which the immunoprecipitations were carried out by using a BRF antiserum or the corresponding preimmune control, and the coprecipitated material was probed for the presence of pocket proteins by Western blotting with monoclonal antibodies. Both p107 (Fig. 5B) and p130 (Fig. 5C) could be readily detected in immunoprecipitates obtained with the BRF antiserum, but not with the preimmune control. The association of endogenous pocket proteins with endogenous TFIIIB has been confirmed by using two different antibodies against p107, two different antibodies against p130, and two different antibodies against BRF (35). In serial immunodepletion experiments, we have found up to ∼30% of the endogenous BRF in complexes with these pocket proteins (35). However, this figure may vary considerably according to cell type and growth conditions.
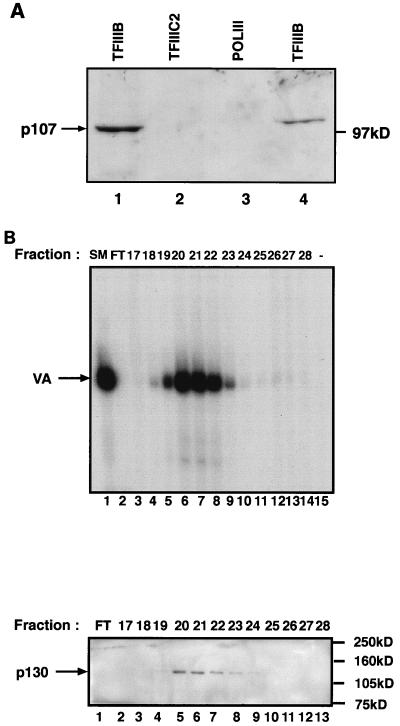
We have shown previously that a population of endogenous RB molecules copurifies consistently with TFIIIB during chromatography of cell extracts. This cofractionation is strongly suggestive of a stable interaction. We tested whether the same is true of p107 and p130. Fractions containing various components of the Pol III transcription apparatus were probed for p107 by Western blotting with a monoclonal antibody (Fig. 6A). p107 was readily detected in fractions containing TFIIIB that had been partially purified by a combination of a phosphocellulose step and a Q-Sepharose gradient (lane 1) or alternatively by a combination of phosphocellulose and DEAE-Sephadex steps (lane 4). In contrast, p107 was not found to copurify with either Pol III or TFIIIC2 (lanes 2 and 3). p130 was also detected in these TFIIIB fractions, but not in the fractions containing partially purified Pol III or TFIIIC2 (4). As a further test for a stable interaction, we examined whether endogenous p107 and p130 would cofractionate with TFIIIB during gradient chromatography on heparin-Sepharose. A phosphocellulose fraction containing TFIIIB was applied to a heparin-Sepharose column, and the bound proteins were eluted with a linear salt gradient. Individual fractions were then probed for the presence of TFIIIB (Fig. 6B, upper panel) and p130 (Fig. 6B, lower panel). Both TFIIIB and p130 were found to peak in fractions 20 to 22 and then tail off sharply. Thus, p130 cofractionates closely with TFIIIB on a heparin-Sepharose salt gradient. The same was found to be true of p107, and the results were confirmed by using alternative antisera against both pocket proteins (1). This consistent cofractionation, along with the coimmunoprecipitation data, suggests that endogenous TFIIIB associates stably and specifically with endogenous p107 and p130.
FIG. 6.
A population of endogenous p107 and p130 molecules cofractionate with endogenous TFIIIB. (A) Fractionated factors (20 μl), as indicated, were resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western immunoblotting with anti-p107 antibody SD9. The TFIIIB fractions in lanes 1 and 4 were QS-PC-B (10.5 μg) and A25(0.15) (1.6 μg), respectively. Lane 2 contained affinity-purified TFIIIC2 (3.8 μg). The Pol III fraction in lane 3 was A25(1.0) (2.2 μg). (B) p130 cofractionates with TFIIIB during gradient chromatography of a PC-B fraction on heparin-Sepharose. The upper panel shows TFIIIB activity of individual fractions, and the lower panel shows the p130 content of the same fractions. Fraction numbers are indicated. SM, starting material; FT, flowthrough. TFIIIB activity was assayed by using 4 μl of the indicated fraction, 2 μl of PC-C, and 250 ng of pVAI; after 15 min of incubation at 30°C, nucleotides were added to assay transcription. p130 content was assayed by using 15 μl of the indicated fractions, which were resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western immunoblotting with anti-Rb2 antibody specific for p130.
Pol III transcription is stimulated in vivo by the inactivation of endogenous p107 and p130.
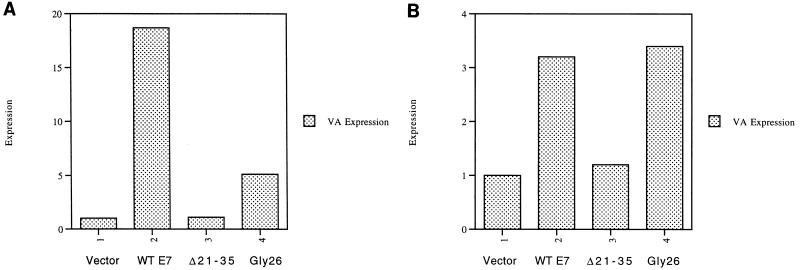
The E7 oncoprotein of human papillomavirus 16 has been shown to bind to the pocket proteins and prevent them from interacting with some of their cellular targets (11, 28, 37). Transfection of NIH 3T3 cells with an expression vector encoding wild-type E7 leads to a substantial increase in the levels of Pol III transcription of a cotransfected VAI gene (Fig. 7A). In contrast, no stimulation is observed with E7 mutant Δ21–35, which is unable to bind to any of the pocket proteins due to a deletion of residues 21 to 35 (20). This suggests that E7 is activating Pol III transcription by overcoming the repressive effects of the endogenous pocket proteins, all of which are functional in these fibroblasts. In order to determine whether p107 and p130 contribute to this repression, we made use of E7 mutant GLY26, which carries a single residue substitution at position 26; this mutant is unable to interact with RB, but retains its capacity to bind p107 and p130 (20). The GLY26 mutant was found to stimulate VAI transcription, which suggests that p107 and/or p130 is involved in repressing TFIIIB in these cells. However, endogenous RB appears to make the principal contribution to this repression, since the activation obtained with GLY26 is only 27% of that observed with wild-type E7. It is important to note that each of these E7 constructs is expressed at comparable levels (20). The results suggest that Pol III transcription in this fibroblast cell line is subject to repression not only by RB, but also by p107 and/or p130.
FIG. 7.
Transfected HPV E7 releases TFIIIB from repression by pocket proteins in vivo. (A) NIH 3T3 cells were transfected with pVAI (2 μg), pCAT (2 μg), and 6 μg of empty vector (lane 1), or vector encoding wild-type (WT) E7 (lane 2), Δ21–35 mutant E7 (lane 3), or GLY26 mutant E7 (lane 4). The values shown for VAI have been normalized to the levels of CAT expression to correct for transfection efficiency and are given relative to the value obtained for the vector control (arbitrarily designated 1). (B) SAOS2 cells were transfected with pVAI (2 μg), pCAT (2 μg), and 6 μg of empty vector (lane 1) or vector encoding wild-type E7 (lane 2), Δ21–35 mutant E7 (lane 3), or GLY26 mutant E7 (lane 4). The values shown for VAI have been normalized to the levels of CAT expression to correct for transfection efficiency and are given relative to the value obtained for the vector control (arbitrarily designated 1).
The effect of E7 on Pol III transcription was also examined in SAOS2 cells, which contain wild-type p107 and p130 but only a nonfunctional truncated mutant form of RB (34). Wild-type E7 was found to stimulate VAI transcription, and this was again due to pocket protein binding, since the Δ21–35 mutant had very little effect (Fig. 7B). The level of activation was much less than in NIH 3T3 cells, presumably due to the lack of functional RB; untransfected SAOS2 cells have been shown previously to have very high levels of Pol III activity (43). Unlike the situation in NIH 3T3 cells, the GLY26 mutant was no less efficient than wild-type E7 in stimulating VAI transcription when transfected into SAOS2 cells. This is consistent with the absence of functional RB in this osteosarcoma line and the ability of GLY26 to bind efficiently to p107 and p130 (20). Taken together with our evidence for an association with TFIIIB, these results suggest that endogenous p107 and/or p130 contributes to the repression of Pol III transcription in at least some mammalian cell types.
Pol III activity is deregulated in fibroblasts derived from p107 p130 double knockout mice.
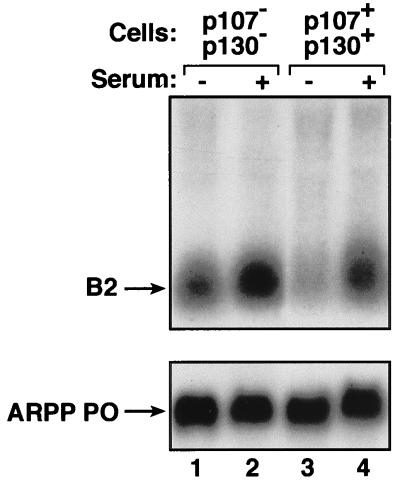
Specific gene disruption provides a more direct method of investigating the roles of endogenous pocket proteins. We compared the Pol III activities of MEFs from _p107_−/− _p130_−/− double knockout mice with those of matched heterozygotes expressing p107 and p130 (wild-type cells of the same genetic background were not available). Northern blot analysis was used to determine the levels of a Pol III transcript derived from the B2 middle repetitive gene family in RNA extracted from these MEFs (Fig. 8, upper panel). When growing rapidly in the presence of 10% serum, the p107 p130 null cells showed only slightly elevated B2 transcript levels compared with the corresponding MEFs expressing a full complement of pocket proteins (lanes 2 and 4, respectively). However, a more substantial difference was observed following serum withdrawal. When made quiescent through serum deprivation, the double knockout cells expressed B2 RNA at approximately twice the level of the matched heterozygotes (lanes 1 and 3, respectively). This effect was specific, since knockout of p107 and p130 did not alter the abundance of a Pol II transcript encoding acidic ribosomal phosphoprotein P0 (Fig. 8, lower panel). It has been demonstrated previously that p107 p130 null MEFs are not compromised in their ability to exit the cell cycle following serum withdrawal (16). This was confirmed in the present experiments by measuring [3H]thymidine incorporation as an indicator of DNA synthesis (26). Although the p107 p130 double knockouts are made quiescent in an apparently normal fashion, they are compromised in their ability to down-regulate Pol III. These observations provide additional evidence that endogenous p107 and/or p130 makes a significant contribution to the control of Pol III transcription in vivo.
FIG. 8.
p107 p130 double knockout fibroblasts display a specific increase in the expression of Pol III transcripts following serum withdrawal. Results represent Northern blot analysis of total RNA (10 μg) extracted from _p107_−/− _p130_−/− double knockout MEFs (lanes 1 and 2) or matched p107+/− p130+/− heterozygotes (lanes 3 and 4) that were actively growing in 10% serum (lanes 2 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 3). The upper panel shows the blot probed with a B2 gene, and the lower panel shows the same blot that has been stripped and reprobed with the ARPP P0 gene.
DISCUSSION
The data presented in this article suggest that the RB-related pocket proteins p107 and p130 can associate with TFIIIB and repress Pol III transcription both in vitro and in vivo. Expression of the VAI gene in transiently transfected cells can be inhibited by either p107 or p130. These pocket proteins will also repress a range of Pol III templates when added to cell extracts. These regulatory effects can be explained by a stable and specific interaction with TFIIIB. Thus, pull-down assays reveal that the BRF subunit of TFIIIB will associate with p107 and p130 that have been expressed in bacteria as GST fusion proteins. Furthermore, endogenous BRF can be coimmunoprecipitated with endogenous p107 and p130. A significant increase in Pol III transcription is observed in vivo when the E7 oncoprotein is used to neutralize the endogenous p107 and p130. Furthermore, targeted disruption of the genes encoding p107 and p130 results in a specific increase in Pol III transcript levels in quiescent MEFs. We conclude that these pocket proteins are likely to constitute physiologically significant regulators of class III gene expression.
Analyses of several deletion and substitution mutations have shown that the pocket domain is required for RB to regulate Pol III activity both in vitro and in vivo (5, 43). This conclusion is reinforced by the effect observed here with E7, a protein which has been shown to bind to the pocket by crystallographic analysis (24). Since the pocket domain is the principal region of homology between RB and its relatives p107 and p130, it is not unexpected that all three of these proteins can interact with TFIIIB. Nevertheless, this was by no means a foregone conclusion, since there is substantial sequence divergence, even in the pocket region (12). Furthermore, many of the genes regulated in murine fibroblasts by p107 and p130 are different from the set of targets that are controlled by RB (16).
Of the three known pocket proteins, RB appears to play the dominant role in controlling Pol III transcription in cycling murine fibroblasts. Thus, VAI expression in growing NIH 3T3 cells was stimulated much more strongly by wild-type E7 than by the GLY26 mutant that only targets p107 and p130. Furthermore, knockout of RB causes a substantial increase in Pol III transcription in growing MEFs (43), whereas knockout of p107 and p130 has little effect under the same circumstances. Hurford et al. (16) have shown previously that growing _p107_−/− _p130_−/− MEFs upregulate RB levels; this may explain why there is little effect on B2 gene expression, since overproduction of RB may compensate for the loss of p107 and p130. However, the upregulation of RB in _p107_−/− _p130_−/− MEFs is largely restricted to cycling cells and is not observed following serum starvation (16); accordingly, the effect of these pocket proteins on Pol III activity is revealed when the double knockouts become quiescent.
RB, p107, and p130 share the ability to inhibit cell growth (7, 30, 44, 45). Although unique regulatory functions may contribute to this capability (44), it seems likely that growth arrest by these homologous proteins involves the control of a number of common cellular targets. Since rapid growth requires high rates of synthesis of tRNA and rRNA, it has been suggested that the repression of Pol III transcription may contribute to the growth control function of RB (15, 21, 29, 39). A similar argument can now be applied to p107 and p130.
ACKNOWLEDGMENTS
We thank Peter Whyte and Xavier Mayol for pocket protein expression constructs, Roger Watson and Karen Vousden for E7 expression vectors, Fred Dick for ARPP P0, and Rene Bernards for MEFs.
This work was funded by project grant CO5766 to R.J.W. from the Biotechnology and Biological Sciences Research Council and by project grant 98-46 to R.J.W. from the Association for International Cancer Research. R.J.W. is a Jenner Research Fellow of the Lister Institute of Preventive Medicine.
J.E.S. and S.J.A. were supported by studentships from the Biotechnology and Biological Sciences Research Council and the Medical Research Council, respectively.
REFERENCES
- 1.Allison, S. J., and R. J. White. Unpublished observations.
- 2.Alzuherri H M, White R J. Regulation of a TATA-binding protein-associated factor during cellular differentiation. J Biol Chem. 1998;273:17166–17171. doi: 10.1074/jbc.273.27.17166. [DOI] [PubMed] [Google Scholar]
- 3.Cairns C A, White R J. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns, C. A., and R. J. White. Unpublished observations.
- 5.Chu W-M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 6.Clarke A R, Maandag E R, van Roon M, van der Lugt N M T, van der Valk M, Hooper M L, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 7.Claudio P P, Howard C M, Baldi A, De Luca A, Fu Y, Condorelli G, Sun Y, Colburn N, Calabretta B, Giordano A. p130/Rb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res. 1994;54:5556–5560. [PubMed] [Google Scholar]
- 8.Cobrinik D, Lee M H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 9.Corbeil H B, Whyte P, Branton P E. Characterization of transcription factor E2F complexes during muscle and neuronal differentiation. Oncogene. 1995;11:909–920. [PubMed] [Google Scholar]
- 10.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 11.Dyson N, Howley P M, Munger K, Harlow E. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 12.Ewen M E, Xing Y, Lawrence J B, Livingston D M. Molecular cloning, chromosomal mapping and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 13.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 15.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 16.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 17.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 18.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 19.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 20.Lam E W-F, Morris J D H, Davies R, Crook T, Watson R J, Vousden K H. HPV16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J. 1994;13:871–878. doi: 10.1002/j.1460-2075.1994.tb06330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larminie C G C, Alzuherri H M, Cairns C A, McLees A, White R J. Transcription by RNA polymerases I and III: a potential link between cell growth, protein synthesis and the retinoblastoma protein. J Mol Med. 1998;76:94–103. doi: 10.1007/s001090050196. [DOI] [PubMed] [Google Scholar]
- 22.Larminie C G C, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C J, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 24.Lee J-O, Russo A A, Pavletich N P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 25.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLees, A., and R. J. White. Unpublished observations.
- 27.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 28.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumour suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasmyth K. Another role rolls in. Nature. 1996;382:28–29. doi: 10.1038/382028a0. [DOI] [PubMed] [Google Scholar]
- 30.Qin X, Chittenden T, Livingston D M, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 31.Rigby P W J. Three in one and one in three: it all depends on TBP. Cell. 1993;72:7–10. doi: 10.1016/0092-8674(93)90042-o. [DOI] [PubMed] [Google Scholar]
- 32.Segall J, Matsui T, Roeder R G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980;255:11986–11991. [PubMed] [Google Scholar]
- 33.Shen Y, Igo M, Yalamanchili P, Berk A J, Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol Cell Biol. 1996;16:4163–4171. doi: 10.1128/mcb.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shew J-Y, Lin B T-Y, Chen P-L, Tseng B Y, Yang-Feng T L, Lee W-H. C-terminal truncation of the retinoblastoma gene product leads to functional inactivation. Proc Natl Acad Sci USA. 1990;87:6–10. doi: 10.1073/pnas.87.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliffe, J. E., and R. J. White. Unpublished observations.
- 36.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 37.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 38.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 39.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 40.White R J. RNA polymerase III transcription. Berlin, Germany: Springer-Verlag; 1998. [Google Scholar]
- 41.White R J, Gottlieb T M, Downes C S, Jackson S P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White R J, Stott D, Rigby P W J. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- 43.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 44.Woo M S-A, Sánchez I, Dynlacht B D. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol Cell Biol. 1997;17:3566–3579. doi: 10.1128/mcb.17.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]