The Replicative Capacities of Large E1B-Null Group A and Group C Adenoviruses Are Independent of Host Cell p53 Status (original) (raw)
Abstract
Recent reports suggest that an early region 1B (E1B) 55,000-molecular-weight polypeptide (55K)-null adenovirus type 5 (Ad5) mutant (_dl_1520) can replicate to the same extent as wild-type (wt) Ad5 in cells either deficient or mutated in p53, implicating p53 in limiting viral replication in vivo. In contrast, we show here that the replicative capacity of Ad5 _dl_1520 is wholly independent of host cell p53 status, as is the replicative capacity of comparable Ad12 E1B 54K-null adenoviruses (Ad12 _dl_620 and Ad12 _hr_703). Furthermore, we show that there is no requirement for complex formation between p53 and Ad5 E1B 55K or Ad12 E1B 54K for a productive infection, such that wt Ad5 and wt Ad12 will both replicate in cells which are null for p53. In addition, we find that these Ad5 and Ad12 mutant viruses induce S phase irrespective of the p53 status of the cell and that, therefore, S-phase induction does not correlate with the replicative capacity of the virus. Interestingly, the replicative capacities of the large E1B-null adenoviruses correlated positively with the ability to express E1B 19K and were related to the ability to repress premature adenovirus-induced apoptosis. Infection of primary human cells indicated that Ad5 _dl_1520, wt Ad5, and wt Ad12 replicated better in cycling normal human skin fibroblasts (HSFs) than in quiescent HSFs. Thus, the cell cycle status of the host cell, upon infection, also influences viral yield.
The ability of adenovirus early region 1 (E1) DNA to transform mammalian cells is well established (21, 22, 69), as is the essential requirement for E1 proteins in viral replication (5, 10, 38, 45, 53, 57). E1 comprises two transcription blocks: E1A encodes two homologous proteins, translated from 12S and 13S mRNAs (respectively, 243 and 289 amino acids [aa] in adenovirus type 5 [Ad5] and 235 and 266 aa in Ad12), which are identical except for the presence of a short region located towards the C terminus of the larger, 13S, molecule (CR3 [for conserved region 3]), while the E1B gene encodes two distinct polypeptides of molecular weight 19,000 (19K) and 55K/54K (in Ad5/Ad12). E1B 19K is dispensable for efficient viral replication (20), while E1B 55K/54K is required, in certain cell types, for a productive infection (5, 10, 38, 45, 53, 57).
E1A interacts with a number of cellular proteins important in regulating cell growth (8, 29, 49). They function predominantly as transcriptional adapters or are integral components of the transcriptional machinery, regulating transcription from both viral early and cellular promoters (11, 37). For example, E1A binds the retinoblastoma tumor suppressor protein Rb1 and the Rb-related proteins p107 and p130 through interactions with CR1 and CR2 (19, 42, 68). E1A also binds to the related proteins CBP and p300 through interaction with CR1 and the N terminus (17, 70) and binds mammalian SUG1 through an as-yet-unidentified region (30). The larger, 13S, protein also interacts with TATA box-binding protein (TBP) through CR3 (24). p53 functions as a negative regulator of cell growth. It regulates genes critical in controlling cycle progression, DNA repair, and apoptosis (41). E1A can, in the absence of cooperating oncogenes, up-regulate p53, inducing apoptosis (14, 43). Stabilization of the p53 molecule by E1A is achieved primarily by increasing the half-life of the protein (32, 43) and is dependent to some extent on E1A binding to either p300 (13) or p300 and/or Rb (48), requiring the up-regulation of p19ARF (15).
The E1B-encoded proteins play a major role in protecting infected and transformed cells from E1A-induced apoptosis (63, 64). The large E1B proteins perform this function during infection primarily by down-regulating the expression of cellular proteins and up-regulating the synthesis of viral proteins, thus inhibiting cellular mRNA transport from the nucleus and facilitating viral mRNA transport into the cytoplasm (2, 3). Ad5 E1B 55K is targeted to the nucleus during infection by E4 open reading frame 6 (ORF6), which possesses an arginine-rich nuclear localization signal; in the absence of E4 ORF6, E1B 55K is cytoplasmic (27). In contrast, Ad12 E1B 54K does not require the expression of other viral proteins for its localization to the nucleus (23, 31, 32); a putative nuclear localization signal for Ad12 E1B 54K which lies between aa 228 and 239 has recently been described (28). The ability of Ad5 E1B 55K to increase viral mRNA transport into the cytoplasm is dependent upon its association with E4 ORF6; the E1B 55K–E4 ORF6 complex functions as a nucleocytoplasmic transporter (16), with E4 ORF6 possessing a nuclear export signal similar to that found in Rev and Rex retroviral proteins.
As previously mentioned, expression of the early region genes E1A and E1B 54K/55K is essential for adenovirus replication in that mutant adenoviruses lacking one or both of these genes are replication defective. Infection with an Ad5 mutant that expresses the 13S E1A protein but not the 12S molecule suggests that 12S is dispensable for viral replication in HEK (human embryo kidney) cells and HeLa cells (45). Studies using an Ad12 E1B 54K-null adenovirus suggest that E1B 54K is required for viral DNA replication in both HeLa and KB cells (10, 57). Similarly, mutant adenoviruses that fail to express Ad5 E1B 55K (5, 38, 53) are replication defective in HeLa cells but not in Ad5 HEK 293 cells (where E1B 55K is endogenously expressed) or normal HEK cells (5, 35). It has therefore been suggested that HEK cells have an E1B 55K-like activity that facilitates viral replication (5). Ad12 E1B 54K-null viruses do not replicate in HEK cells (12). In transformed cells, Ad5 E1B 55K forms a tight association with p53 (72), whereas only a fraction of cellular p53 is found complexed with Ad12 E1B 54K (32); Ad5 E1B 55K and Ad12 E1B 54K, however, inhibit p53 transcriptional activity to the same extent (71). Specific complex formation between Ad5 E1B 55K and p53 has been observed in Ad5-infected rodent cells (9), and it has been postulated that formation of this complex is necessary for efficient viral replication (51), such that in cells expressing a mutant p53, or lacking p53, wild-type (wt) Ad5 is replication defective (51).
During infection, E1B 19K serves primarily to protect against the cytocidal phenotype and the degradation of both viral and host cell DNA (47, 58, 60, 67) induced by E1A (65, 66). Ad5/Ad12 E1B 19K, like Bcl-2, function as negative regulators of apoptosis by binding to and inhibiting the function of Bax, a p53-inducible and death-promoting protein (34). The ability of E1B 19K to serve as an inhibitor of apoptosis is dependent upon its localization to the nuclear lamins (50). Isolation of adenoviruses mutated in E1B 19K, however, has demonstrated that E1B 19K is not required for efficient viral replication in HEK or KB cells (20).
It has recently been proposed that p53 is one of the cellular factors crucial in limiting viral replication in vivo (6). Studies suggest that a mutant adenovirus (Ad5 _dl_1520) which does not express E1B 55K (4) and fails to replicate in cells expressing wt p53 can undergo viral replication in cells deficient, or mutated, in p53 (6, 36). This has been claimed to offer a potential therapeutic strategy for specifically destroying cancer cells that lack functional p53 (6). In this study, we examine this hypothesis further and determine the criteria that allow efficient Ad5 and Ad12 virus production. Specifically, we analyze the requirements for E1B-p53 complex formation, S-phase induction and cell cycle status, and repression of apoptosis in replication of both wt and E1B-null group A and group C adenoviruses.
MATERIALS AND METHODS
Cells and viruses.
Human A549 cells were derived from a small cell lung carcinoma (25) and contain wt p53 (40). Human HCT116 cells were isolated from a colorectal carcinoma and also possess wt p53 (62). Human C33A cells derive from a cervical carcinoma and contain mutant p53 (aa 273, Arg to His [56]). HaCaT cells, which are a spontaneously transformed human epithelial cell line from adult skin (7), also express mutant p53 (aa 179, His to Arg; aa 282, Arg to Trp [39]). Human HT-29 cells also possess a mutant p53 (aa 273, Arg to His [52]). H1299 cells are derived from a lung carcinoma and are null for both alleles of p53 (59). JW2 cells, which are also null for both alleles of p53, were isolated from a colorectal carcinoma (46). Human skin fibroblasts (HSFs) were from an adult donor and express wt p53. All human cells were grown in HEPES-buffered Dulbecco’s modified Eagle’s medium containing 8% fetal calf serum (FCS) and supplemented with 2 mM glutamine, unless otherwise stated. wt Ad5 was from the American Type Culture Collection (ATCC). Ad5 _dl_1520, generated by deletional mutagenesis, is a large E1B-null adenovirus (4) that has introduced into its sequence a termination codon at position 3 and an additional deletion from nucleotides 2496 to 3323, with a linker insertion generating a second stop codon at nucleotide 3336 (4). The parental Ad12 used was Huie, obtained from the ATCC. Ad12 _dl_620, which contains a large in-frame deletion in the E1B 54K ORF from nucleotides 2129 to 2825, has been described previously (12, 29). Ad12 _hr_703, a host range mutant restricted for growth in A549 cells and HeLa cells, contains a point mutation at amino acid 134 of E1B 54K, converting a Gln to a termination codon (12), and Ad12 _pm_700, an E1B 19K-null adenovirus that expresses E1A and E1B 54K, has also been described previously (18). Ad5 viruses were grown on permissive Ad5 E1 HEK 293 cells, while Ad12 viruses were grown on permissive Ad12 E1-transformed HER3 cells, to produce high-titer stocks. The titers of the Ad5 and Ad12 stocks were determined by plaque assay on Ad5 293 cells and Ad12 HER3 cells, respectively.
Virus yield assay.
Cells were infected at a multiplicity of infection of 50 PFU/cell for 2 h, with agitation every 15 min. At appropriate times postinfection (p.i.), cells were harvested, resuspended in 1 ml of serum-free medium, and lysed by three freeze-thaw cycles, and the cell debris was sedimented by centrifugation. Viral replication was determined by plating serial diluents of cellular extracts with half-log dilutions on either Ad5 293 cells or Ad12 HER3 cells for Ad5 and Ad12, respectively. Titrations were carried out in 48-well cell culture clusters (Costar), and adenovirus cytopathic effects (CPE) were scored after 14 days (for Ad5) and 21 days (for Ad12).
Cell cycle analysis of Ad5- and Ad12-infected cells.
Cells were either mock infected or infected with the appropriate virus (50 PFU/cell) for 2 h, as described above. Cells were harvested at the appropriate times p.i. by trypsinization and fixed in 70% ethanol at −20°C for at least 30 min. Cells were then treated at room temperature with 2 M HCl (containing 2 mg of pepsin per ml) for 30 min and neutralized with 0.1 M sodium borate (pH 8.5) prior to treatment with RNase A and labelling with propidium iodide (final concentration, 5 μg/ml). Analysis was performed on a Coulter Epics XL flow cytometer. The percentages of cells in each phase of the cell cycle were determined by using Multicycle, version 2.53 (Phoenix).
Western blot analysis.
Cells were either mock infected or infected with the appropriate virus (50 PFU/cell) for 2 h. Cells were harvested at the appropriate times p.i., solubilized (in 9 M urea, 50 mM Tris [pH 7.4], 150 mM β-mercaptoethanol), and sonicated. Fifty-microgram aliquots of each sample were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and Western blotted with the appropriate primary antibodies, as follows. Ad12 E1A was detected with a sheep polyclonal antibody, and Ad12 E1B 54K was blotted with XPH9, a mouse monoclonal antibody. A rat polyclonal tumor bearer serum was used to recognize Ad12 E1B 19K. Ad5 E1A was detected with mouse monoclonal antibody M58 (Pharmingen). Ad5 E1B 55K was detected with 2A6, a mouse monoclonal antibody, and Ad5 E1B 19K was detected with a rat monoclonal antibody (Oncogene Science). The appropriate horseradish peroxidase-conjugated secondary antibody (Sigma) was used in each instance and developed by ECL (Amersham).
Acridine orange staining.
The fluorescent probe acridine orange (Molecular Probes) was prepared as a stock solution in phosphate-buffered saline (PBS) at 2 mg/ml and stored at 4°C prior to use. Infected cells were fixed at the appropriate time p.i. in 1% formaldehyde in PBS. Cells were stained with acridine orange at a final concentration of 5 μg/ml and placed on a coverslip, and the percentage of apoptotic cells was determined by using immunofluorescence microscopy. Apoptotic cells were identified by their condensed and fragmented nuclei (33).
RESULTS
Replication of wt Ad5 is independent of host cell p53 status.
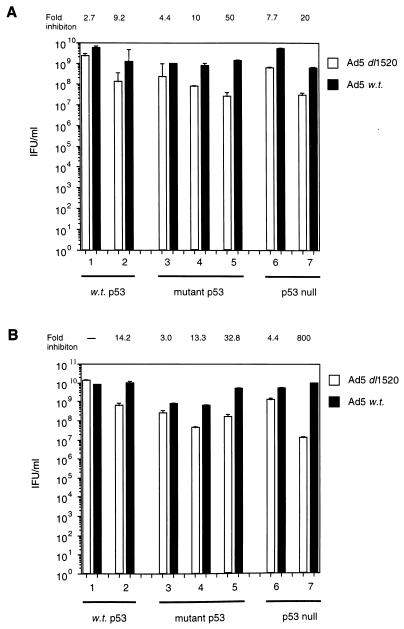
A recent report (51) has suggested that complex formation between Ad5 E1B 55K and p53 is essential for a productive infection by Ad5, such that wt Ad5 cannot replicate efficiently in cells lacking functional p53. We, however, have found that wt Ad5 replicated to similar extents in cells expressing either wt p53 (Fig. 1, filled bars, lanes 1 and 2) or mutant p53 (Fig. 1, filled bars, lanes 3, 4, and 5) or in cells lacking p53 (Fig. 1, shaded bars, lanes 6 and 7). Viral titers were typically between 109 and 1010 inclusion-forming units (IFU)/ml, with only slight variations between target cell lines. This variation did not correlate with host cell p53 status. These data indicate that complex formation between E1B 55K and p53 is not an essential prerequisite for replication of Ad5. This also suggests that large E1B-null adenoviruses might retain the ability to replicate in cells lacking functional p53.
FIG. 1.
Replicative capacities of Ad5 _dl_1520 and wt Ad5 in cells differing in p53 status. Virus yields were quantified as stated in Materials and Methods, by using permissive Ad5 HEK 293 cells. (A) Virus yield from Ad5 _dl_1520- and wt Ad5-infected cells at 48 h p.i. Lane 1, A549 cells (wt p53); lane 2, HCT116 cells (wt p53); lane 3, C33A cells (mutant p53); lane 4, HaCaT cells (mutant p53); lane 5, HT-29 cells (mutant p53); lane 6 H1299 cells (p53 null); lane 7, JW2 cells (p53 null). (B) Same as for panel A, except values were taken at 72 h p.i. The values above each panel reflect the fold decrease in replicative capacity of Ad5 _dl_1520 relative to wt Ad5. Results are means from two independent experiments.
The replicative capacity of Ad5 _dl_1520 is independent of host cell p53 status.
Recent reports suggest that large E1B-null Ad5 mutants replicate selectively in cells expressing mutant p53 or in cells null for p53 but not in cells expressing wt p53 (6, 36). A detailed analysis of the replicative capacity of the E1B 55K-null Ad5 _dl_1520, relative to that of wt Ad5, was thus undertaken in tumor cells differing in p53 status. It can be seen from Fig. 1 that in all instances and irrespective of p53 status, the replicative capacity of Ad5 _dl_1520 was reduced relative to a wt Ad5 infection. This reduction in replicative capacity was typically between 10- and 100-fold. These data help to confirm that there is no essential requirement for E1B 55K-p53 complex formation during replication. However, the ability of Ad5 _dl_1520 to replicate did not correlate with host cell p53 status. Thus, Ad5 _dl_1520 can replicate as well in cells possessing wt p53 as it can in cells lacking p53 (Fig. 1). We next investigated the replicative capacities of the corresponding Ad12 E1B 54K-null viruses to see if there were differences between group A and C viruses.
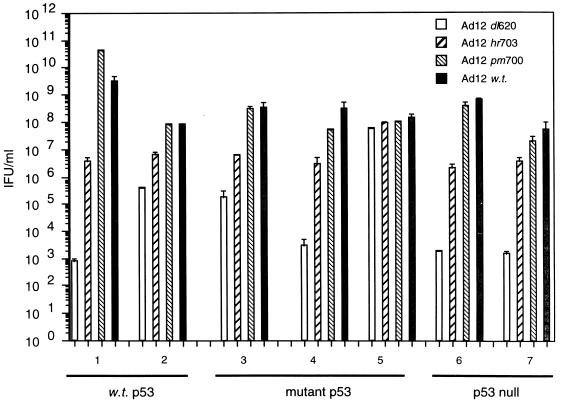
Host cell p53 status does not define the efficacy of replication of wt Ad12 or E1B-54K null Ad12 mutants.
wt Ad12 replicated to similar levels in all cell lines investigated, irrespective of the p53 status of the host cell (Fig. 2, dark-shaded bars, lanes 1 to 7). The viral titers were typically between 108 and 5 × 109 IFU/ml. As already shown for Ad5, there was no correlation between replicative capacity and the p53 status of the cell. Ad12 _pm_700 (E1B 19K-null) replicated to the same level as wt virus in all cell lines tested (Fig. 2, light-hatched bars, lanes 1 to 7), as previously demonstrated (18). Again, there were no constraints upon replicative capacity imposed by the host cell with respect to p53 status. In direct comparison with Ad5 _dl_1520, the replicative capacities of similar E1B 54K-null Ad12 mutants were investigated in cells differing in p53 status. In most circumstances, Ad12 E1B-54K null mutants were more restricted for growth, relative to a wt infection, than their Ad5 _dl_1520 counterpart (Fig. 2, hatched bars, lanes 1 to 7). Without exception, Ad12 _hr_703 replicated substantially better than Ad12 _dl_620. The replicative capacity of Ad12 _hr_703, however, was independent of p53 status (Fig. 2, dark-hatched bars, lanes 1 to 7). Ad12 _dl_620 was severely growth restricted in most cells studied (Fig. 2, open bars, lanes 1 to 7). There was, however, no correlation between the replicative capacity of Ad12 _dl_620 and the host cell p53 status.
FIG. 2.
Replicative capacities of Ad12 _dl_620, Ad12 _hr_703, Ad12 _pm_700 and wt Ad12 in cells differing in p53 status. Virus yields shown at 72 h p.i., were quantified as stated in Materials and Methods, by using permissive Ad12 HER3 cells. Lane 1, A549 cells (wt p53); lane 2, HCT116 cells (wt p53); lane 3, C33A cells (mutant p53); lane 4, HaCaT cells (mutant p53); lane 5, HT-29 cells (mutant p53); lane 6 H1299 cells (p53 null); lane 7, JW2 cells (p53 null). Results are means from two independent experiments.
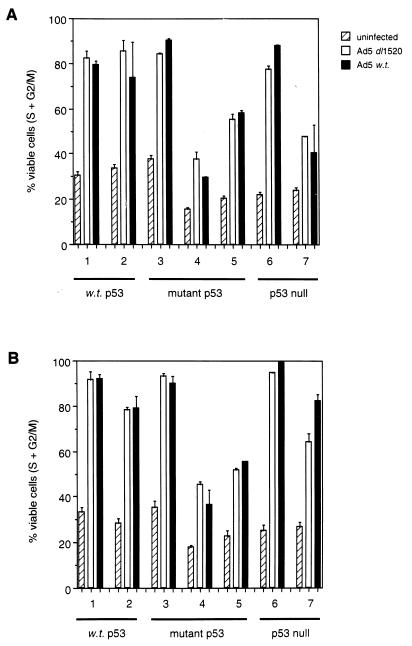
Ad5 _dl_1520 and wt Ad5 induce S phase, irrespective of p53 status.
Next, we investigated the ability of Ad5 viruses to induce S phase in cells differing in p53 status in an attempt to relate viral replication to the cell cycle status of the host cell. Since, these viruses are replication competent, however, the percentage of lysed (sub-G1) cells was omitted when the percentage of cells in S + G2/M was calculated. Moreover, the combined S and G2/M values, rather than S-phase values alone, were included in calculations since these viruses are capable of inducing polyploidy in certain cell lines (no correlation with p53 status [data not shown]), making S phase alone difficult to distinguish. It is possible that some of the observable polyploidy was due to propidium iodide-stained viral DNA from replicating viruses. Indeed, calculation suggests that at late times p.i. the DNA from replication-competent viruses may constitute half of the DNA present in an infected cell. However, fluorescence-activated cell sorting (FACS) analysis revealed a substantial depletion of host cells in G1 of the cell cycle at early time points (approximately 12 h p.i. depending upon cell type) during infection with either replication-competent or replication-defective viruses (data not shown). Moreover, there was no observable evidence of cells reentering G1 phase during the time course of the experiment (data not shown). Concomitant with the reduction in the number of cells in G1, there was a specific elevation at the protein level (and histone H1-directed kinase activities) of cyclins E and A, indicative of host cell entry into S phase (data not shown); this remained elevated during infection, indicative of failure of host cells to undergo mitosis. Both Ad5 _dl_1520 and wt Ad5 induced S phase to similar extents in all cell lines investigated, irrespective of p53 status (Fig. 3). There was, however, great variability between cell lines in the extent to which these viruses induced S phase. Moreover, since wt Ad5 can replicate better than Ad5 _dl_1520 in all but one of the cell lines investigated (Fig. 1), the ability of these viruses to replicate does not directly relate to the ability to induce S phase.
FIG. 3.
S-phase induction by Ad5 _dl_1520 and wt Ad5. (A) Percentages of cells in S + G2/M from Ad5 _dl_1520- and wt Ad5-infected cells at 48 h p.i. Lane 1, A549 cells (wt p53); lane 2, HCT116 cells (wt p53); lane 3, C33A cells (mutant p53); lane 4, HaCaT cells (mutant p53); lane 5, HT-29 cells (mutant p53); lane 6 H1299 cells (p53 null); lane 7, JW2 cells (p53 null). (B) Same as for panel A, except values were taken at 72 h p.i. Results are means from two independent experiments.
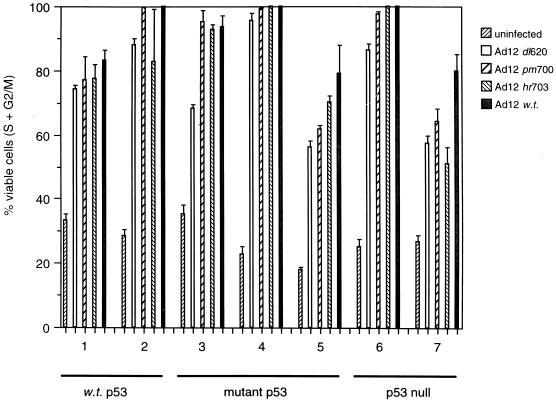
The ability of Ad12 mutants to induce S phase does not correlate with the ability to replicate.
The E1B 54K-null virus Ad12 _dl_620 was largely replication defective in all cell lines investigated, whereas Ad12 _hr_703 was partially defective and Ad12 _pm_700 replicated to the same extent as wt Ad12 (Fig. 2). Ad12 _dl_620, however, was still capable of inducing S phase, to levels similar to the replication-competent Ad12 viruses, irrespective of the p53 status of the target cell (Fig. 4). Similarly, Ad12 _hr_703 induced S phase to an extent similar to Ad12 _pm_700 and wt Ad12, suggesting that S-phase induction per se does not create a permissive environment and that other factors determine whether the virus can replicate or not.
FIG. 4.
S-phase induction by Ad12 _dl_620, Ad12 _hr_703, Ad12 _pm_700, and wt Ad12. Percentages of cells in S + G2/M at 72 h p.i. are shown. Lane 1, A549 cells (wt p53); lane 2, HCT116 cells (wt p53); lane 3, C33A cells (mutant p53); lane 4, HaCaT cells (mutant p53); lane 5, HT-29 cells (mutant p53); lane 6 H1299 cells (p53 null); lane 7, JW2 cells (p53 null). Results are means from two independent experiments.
Ad5 E1B 19K expression is reduced in most instances of Ad5 _dl_1520 infection, relative to a wt Ad5 infection.
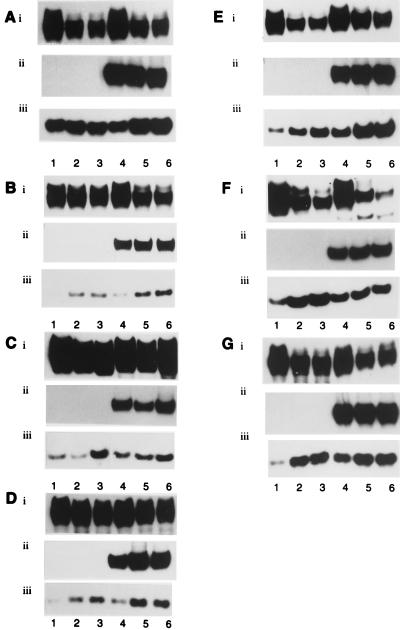
The pattern of adenovirus early gene expression was monitored to determine whether the phenotype of Ad5 _dl_1520 differed from wt Ad5 (Fig. 5). As expected, Ad5 _dl_1520-infected cells did not express Ad5 E1B 55K (Fig. 5A to G, panels ii, lanes 1 to 3). The levels of Ad5 E1B 55K in wt Ad5-infected cells were comparable between lines (Fig. 5A to G, panels ii, lanes 4 to 6). The levels of Ad5 E1A were comparable in Ad5 _dl_1520- and wt Ad5-infected cells (Fig. 5A to G, panels i). There were noticeable differences in the levels of E1B 19K expression, however, between Ad5 _dl_1520- and wt Ad5-infected cells (Fig. 5A to G, panels iii). In all but one of the lines, E1B 19K expression was temporally delayed in Ad5 _dl_1520-infected cells relative to wt Ad5-infected cells; this is most obvious after 24 h of infection (Fig. 5B to G, panels iii). In two instances (C33A and H1299), however, Ad5 _dl_1520 was able to express E1B 19K at levels comparable to wt Ad5 after prolonged infection (Fig. 5C and F, panels iii). In all other instances, however, Ad5 _dl_1520 infection resulted in reduced expression of Ad5 E1B 19K relative to wt Ad5 infection. There was no obvious relationship between expression of Ad5 E1B 19K and the p53 status of the cell.
FIG. 5.
Expression of adenovirus early gene products at 24, 48, and 72 h p.i. in Ad5 _dl_1520- and wt Ad5-infected cells. (A) A549 cells (wt p53). (B) HCT116 cells (wt p53). (C) C33A cells (mutant p53). (D) HaCaT cells (mutant p53). (E) HT-29 cells (mutant p53). (F) H1299 cells (p53 null). (G) JW2 cells (p53 null). Lanes 1 to 3, Ad5 _dl_1520-infected cells; lanes 4 to 6, wt Ad5-infected cells. Panels i, Ad5 E1A; panels ii, Ad5 E1B 55K; panels iii, Ad5 E1B 19K.
Ad12 E1B 19K expression is dramatically reduced upon infection with Ad12 E1B 54K-null adenoviruses.
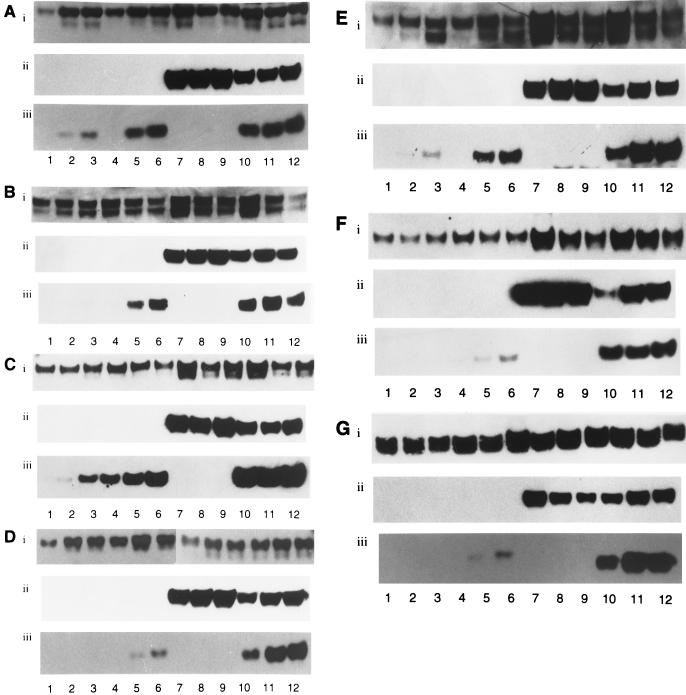
Similarly, Ad12 early gene expression was examined (Fig. 6) to determine phenotypic differences between Ad12 mutants and the parental virus and for comparison with Ad5. As expected, the Ad12 E1B 54K-null viruses Ad12 _dl_620 and Ad12 _hr_703 did not express Ad12 E1B 54K upon infection, whereas Ad12 _pm_700, in most instances, expressed Ad12 E1B 54K to higher levels than wt Ad12-infected cells (Fig. 6A to G, panels ii, lanes 1 to 12). Interestingly, Ad12 E1B 19K expression was substantially reduced both early in infection and after prolonged periods of infection in Ad12 _dl_620- and Ad12 _hr_703-infected cells, relative to wt Ad12-infected cells (Fig. 6A to G, panels iii). Ad12 _dl_620 was more restricted for Ad12 E1B 19K expression than Ad12 _hr_703 in all instances. As expected, Ad12 _pm_700 did not express Ad12 E1B 19K (Fig. 6A to G, panels iii). Interestingly, the ability of Ad5 and Ad12 large E1B-null mutants to replicate correlated positively with the ability to express E1B 19K; i.e., Ad5 _dl_1520 replicated better than Ad12 _hr_703, which in turn replicated better than Ad12 _dl_620. We therefore addressed whether E1B 19K was able to protect cells from premature adenovirus-induced apoptosis during infection with large E1B-null viruses.
FIG. 6.
Expression of adenovirus early gene products at 24, 48, and 72 h p.i. in Ad12 _dl_620-, Ad12 _hr_703-, Ad12 _pm_700-, and wt Ad12-infected cells. (A) A549 cells (wt p53). (B) HCT116 cells (wt p53). (C) C33A cells (mutant p53). (D) HaCaT cells (mutant p53). (E) HT-29 cells (mutant p53). (F) H1299 cells (p53 null). (G) JW2 cells (p53 null). Lanes 1 to 3, Ad12 _dl_620-infected cells; lanes 4 to 6, Ad12 _hr_703-infected cells; lanes 7 to 9, Ad12 _pm_700-infected cells; lanes 10 to 12, wt Ad12-infected cells. Panels i, Ad12 E1A; panels ii, Ad12 E1B 54K; panels iii, Ad12 E1B 19K.
Replication-defective adenovirus mutants induce apoptosis to a greater extent than do replication-competent adenovirus mutants.
The ability of Ad mutants to induce apoptosis was assayed by using the fluorescent probe acridine orange. Condensed and fragmented nuclei were visualized by immunofluorescence microscopy at 48 h p.i. (Table 1). It can be seen that in all instances of infection, Ad12 _dl_620, which expressed the lowest levels of E1B 19K, induced apoptosis to the greatest extent (Table 1). The levels of apoptosis attained during infection with Ad12 _hr_703 were reduced relative to Ad12 _dl_620, while Ad5 _dl_1520, which expresses E1B 19K to levels approaching those seen with wt Ad5, had the lowest observable levels of apoptosis (Table 1). At this time of infection, wt viruses showed no morphological evidence of undergoing apoptosis (data not shown). Interestingly, the levels of adenovirus-induced apoptosis were relatively low, particularly considering that the ability of Ad12 _dl_620 to replicate differs from wt Ad12 in some instances by 6 orders of magnitude (Fig. 2). This might suggest either that low levels of E1B 19K afford some protective effect from premature adenovirus-induced apoptosis or, alternatively, that the morphological changes associated with apoptosis are not yet apparent.
TABLE 1.
Induction of apoptosis by large E1B-null Ad5 and Ad12
| Cell line (phenotype) | % Apoptosisa | ||
|---|---|---|---|
| Ad12 _dl_620 | Ad12 _hr_703 | Ad5 _dl_1520 | |
| A549 (wt p53) | 13.9 | 3.6 | 1.6 |
| HCT116 (wt p53) | 13.2 | 6.5 | 2.6 |
| C33A (mutant p53) | 12.5 | 5.3 | 1.7 |
| HaCaT (mutant p53) | 9.9 | 3.4 | 1.2 |
| HT-29 (mutant p53) | 6.7 | 3.2 | 2.1 |
| H1299 (p53 null) | 17.3 | 10.5 | 0.5 |
| JW2 (p53 null) | 13.5 | 6.8 | 1.4 |
Ad5 and Ad12 replicate better in growing HSFs than in quiescent HSFs.
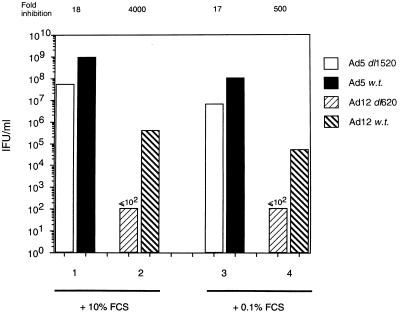
The ability of large E1B-null adenoviruses to replicate has so far been considered in highly proliferative, tumor-derived cell lines. We therefore considered it of value to study the replicative capacity of these viruses in primary, nontumorigenic cells with intact cell cycle checkpoints. Interestingly, Goodrum and Ornelles have recently proposed that Ad5 E1B 55K-null adenoviruses replicate with greater efficacy in host cells that are in S phase at the time of infection than in G1 phase (26). Furthermore, they demonstrated that wt Ad5 could replicate in any phase of the cell cycle without loss of efficacy, suggesting that S-phase factors compensated the virus for the lack of Ad5 E1B 55K (26). Because we previously showed that S-phase induction per se does not facilitate viral replication, we investigated the replicative capacities of Ad5 and Ad12 in exponentially growing primary HSFs and in HSFs that had been grown to confluence, maintained in low serum (0.1%), and allowed to quiesce over a 7-day period. At the time of infection, approximately 95% of quiescent cells were in G0/G1, whereas approximately 30% of growing cells were in S phase at this time (data not shown). It can be seen from Fig. 7 that both Ad5 _dl_1520 and wt Ad5 replicated better, by 10-fold in each instance, in growing cells than in cells that were quiescent. Similarly, wt Ad12 replicated better in exponentially growing cells than in cells that were quiescent (Fig. 7). These data suggest that actively cycling cells facilitate viral replication of both wt Ad5 and Ad5 _dl_1520 irrespective of Ad5 E1B 55K expression. Ad12 _dl_620, however, was wholly replication defective in both growing cells and quiescent cells (Fig. 7).
FIG. 7.
Replicative capacities of Ad5 and Ad12 in cycling and quiescent HSFs. Virus yields were determined at 72 h p.i., as described in Materials and Methods, by using permissive Ad5 HEK 293 cells and Ad12 HER3 cells for Ad5 and Ad12, respectively. Lanes 1 and 2, growing HSFs; lanes 3 and 4, quiescent HSFs; lanes 1 and 3, Ad5 _dl_1520 and wt Ad5; lanes 2 and 4, Ad12 _dl_620 and wt Ad12. The values above the figure reflect the fold decrease in replicative capacity of Ad5 _dl_1520 relative to wt Ad5 and of Ad12 _dl_620 relative to wt Ad12. Results are means from two independent experiments.
DISCUSSION
The tumor suppressor gene p53 has recently been implicated in limiting viral replication in vivo due to the ability of an Ad5 E1B 55K-null adenovirus (Ad5 _dl_1520) to replicate in cells which lack a functional p53 molecule (6). This mutant adenovirus can replicate in human C33A cervical carcinoma cells, which possess a mutant p53, to the same extent as a wt Ad5 virus but has 100-fold-reduced replicative capacity in human U2OS osteosarcoma cells where p53 is wt (6). Introduction of the Ad5 E1B 55K gene into U2OS cells subsequently rendered them sensitive to infection by Ad5 _dl_1520, whereas introduction of the E1B 55K gene lacking the p53-binding site into U2OS cells did not. Similarly, Ad5 _dl_1520-induced CPE was observed in RKO cells transfected with a dominant-negative p53 but not in parental RKO cells. These observations have subsequently been expanded to suggest that Ad5 _dl_1520 has a better replicative capacity in tumor cells per se than in normal primary cells (36), suggesting that p53 might be transcriptionally inactive in tumor cells (36).
In contrast to these studies, however, we demonstrate that Ad5 _dl_1520 can replicate in cells irrespective of their p53 status (Fig. 1). In most instances, the replicative capacity of Ad5 _dl_1520 was reduced by only 10% of the wt yield. This suggests that wt p53 does not play a major role in limiting viral replication in vivo, since Ad5 _dl_1520 can replicate relatively efficiently in wt p53-expressing cells (Fig. 1) with yields as high as that seen for wt Ad12 (Fig. 2). In contrast to Ad5, Ad12 E1B 54K-null viruses are significantly growth restricted in cells, irrespective of p53 status. In all circumstances Ad12 _dl_620 was more restricted in growth than Ad12 _hr_703 (Fig. 2). No Ad12 E1B 54K immunogenic polypeptides were detected in either Ad12 _dl_620- or Ad12 _hr_703-infected cells by Western blotting with anti-54K antibodies. Similarly, no Ad5 E1B 55K immunogenic polypeptides were detected in Ad5 _dl_1520-infected cells with anti-55K antibodies, although expression of a truncated adenovirus E1B polypeptide should be theoretically possible. Indeed, E1B 54K/55K truncated proteins have previously been shown to be highly unstable. Interestingly, any truncated large E1B-products synthesized would not retain any p53-neutralizing activity, since the p53 binding site on Ad5 E1B 55K and Ad12 E1B 54K, which lies between aa 216 and 235 (28), lies C-terminal to the termination codons.
Goodrum and Ornelles (26) have also investigated the replicative capacity of an Ad5 E1B 55K-null virus, Ad5 _dl_338, in asynchronously growing cells possessing wt p53 (H460 and A549) or lacking p53 (H358 and Saos-2). They found that wt virus replicated better than the Ad5 _dl_338 virus in all instances, in good agreement with the data presented here (Fig. 1). Moreover, they also found that Ad5 _dl_338 replicated better in cells containing wt p53 than in cells null for p53 per se. However, a comparison of virus yields suggested that no direct correlation between p53 status and Ad5 _dl_338 replicative capacity could be drawn. These data are in accordance with those presented here but similarly contrast with those obtained previously with Ad5 _dl_1520 (6, 36).
It is evident from the data presented here that Ad12 E1B 54K-null viruses are more restrictive for growth than their Ad5 counterparts (cf. Fig. 1 and 2), as has been noted previously (10, 12). A major phenotypic difference between the Ad5 and Ad12 mutant viruses is the level of expression of E1B 19K. Although Ad5 E1B 19K levels are reduced early during Ad5 _dl_1520 infection relative to wt Ad5, the levels are not dramatically reduced after prolonged infection (Fig. 5). In contrast, Ad12 E1B 19K levels are dramatically reduced in Ad12 _dl_620-infected cells even after prolonged infection, and 19K expression can be detected only by overexposing the blots. Interestingly, levels of Ad12 E1B 19K attained during infection with Ad12 _hr_703 are greater than for Ad12 _dl_620 but are still lower than that for wt Ad12 (Fig. 6). The levels of E1B 19K attained during infection with large E1B-null adenoviruses directly correlates with the ability to replicate, such that Ad5 _dl_1520 replicates better than Ad12 _hr_703, which in turn replicates better than Ad12 _dl_620.
The ability of E1B 19K to protect cells from adenovirus E1A-induced apoptosis is well documented (64). Indeed, cells infected with a mutant adenovirus lacking a functional E1B 19K molecule have previously been shown to be susceptible to premature adenovirus-induced apoptosis (64–66). We therefore investigated the ability of these mutant adenoviruses to induce apoptosis (Table 1). Interestingly, we showed that the replicative capacities of the large Ad E1B-null viruses correlated well with the ability to repress premature adenovirus-induced apoptosis. Thus, infection with Ad12 _dl_620 resulted in significant amounts of apoptosis, whereas infection with Ad5 _dl_1520 resulted in substantially reduced levels (Table 1). Ad12 _hr_703 showed an intermediate phenotype (Table 1). There was no obvious relationship between the p53 status of the cell and the ability of large E1B-null viruses to induce apoptosis. Indeed, Ad E1A-induced apoptosis can occur through both p53-dependent and p53-independent pathways (13, 61). The p53-dependent pathway requires p53-mediated transcription (1, 54), whereas the p53-independent pathway requires E1A induction of the E4 region (44).
A role for p53-Ad5 E1B 55K complex formation in regulating adenovirus replication has recently been proposed (51). It was shown that virus mutants with defects in the Ad5 E1B 55K gene (i.e., Ad5 _dl_338) failed to replicate either in cells expressing wt p53 (A549 cells) or in cells expressing a mutant p53 (143B cells). In contrast, wt Ad5 was able to replicate in cells expressing wt p53 (A549 cells), while the replicative capacity of wt Ad5 was severely restricted in cells containing a mutant p53 (143B, J82, and T98G cells). The authors proposed a model whereby Ad5 E1B 55K binding to p53, in concert with Ad5 E4 ORF6 expression, is required for efficient shutoff of p53 synthesis. This expands on an earlier report from our laboratory that proposed that both E1B 55K and E4 expression are required for shutoff of p53 synthesis (32). Ridgway et al. (51), however, also proposed that complex formation and subsequent down-regulation of p53 allowed for efficient viral replication. In circumstances where p53 cannot be down-regulated, they suggested, viral replication does not occur.
In direct contrast to these findings, we demonstrate that both wt Ad5 and wt Ad12 can replicate efficiently in cells lacking functional p53 (Fig. 1 and 2). Similarly, we demonstrate that Ad5 _dl_1520 can replicate in cells irrespective of whether they express p53 (Fig. 1). These data suggest that complex formation between p53 and Ad5 E1B 55K is not essential for viral replication. The reasons for these anomalies are not clear at present, though they may reflect methodological differences in determining viral replication. Ridgway and colleagues analyzed expression of a viral E1B DNA sequence (51), while our study quantified virus yield and observed adenovirus CPE. Irrespective of virus titers calculated after infection, both infection of p53-null cells with wt virus and infection of wt p53-containing cells with Ad5 _dl_1520 gave clear and unambiguous Ad CPE (data not shown).
The dependence on the cell cycle of replication of wt Ad5 and E1B 55K-null adenoviruses (Ad5 _dl_338 and Ad5 _dl_1520) in HeLa cells has recently been explored (26). Goodrum and Ornelles (26) show that Ad5 _dl_338 replicates significantly better when infection takes place in cells synchronized in the S phase than in the G1 phase of the cell cycle. wt Ad5, in contrast, replicates slightly better in HeLa cells infected during G1 than during S phase. These data suggest that an asynchronously cycling population of cells may facilitate viral replication to a greater extent than quiescent cells. Data presented here suggest that wt Ad5, Ad5 _dl_1520, and wt Ad12 can replicate more efficiently per se in HSF cultures with a high level of cycling cells than in cultures of predominantly quiescent HSFs (Fig. 7). This implies that cells actively progressing through the cell cycle provide cellular factors that allow efficient virus replication irrespective of whether they express Ad5 E1B 55K. Cells that are initially in G0 upon infection do not, however, provide these factors; hence, the virus is less efficient at replicating. Interestingly, wt Ad5, Ad5 _dl_1520, and wt Ad12 all retain the ability to replicate in quiescent cells (Fig. 7), whereas Ad12 _dl_620 is severely restricted in replication (Fig. 7). These viruses do, however, possess the ability to induce S phase in cells irrespective of the p53 status of the cell (Fig. 3 and 4). Surprisingly, there is no direct relationship between the ability of the virus to induce S phase and its ability to replicate (cf. Fig. 1 and 2 with Fig. 3 and 4).
The relationship we observe between the replicative capacity of the virus and the cell cycle status upon infection may have implications for Ad5 _dl_1520 infection in vivo. Since replicative capacity is enhanced in growing cells relative to quiescent cells, it might not be a coincidence that Ad5 _dl_1520 replicates better in tumor cells per se. This might not relate to p53 inactivation, as suggested by Heise and coworkers (36), but may merely reflect the intrinsic cell cycle status of tumor cells. In situations where Ad5 _dl_1520 infects noncycling tumor cells in vivo, the virus’s replicative capacity might be substantially reduced but apoptotic pathways might still be activated. It is our opinion that the reasons for the tumoricidal effects of Ad5 _dl_1520 in vivo are more complex than previously thought in that a number of cell death mechanisms may be activated by the virus. A better understanding of Ad5 _dl_1520 interaction with tumor cells in vivo may permit the development of a virus with a stronger cytocidal phenotype and therefore a greater therapeutic index.
ACKNOWLEDGMENTS
We are grateful to the CRC for financial support. P.H.G. is a CRC Gibb Fellow.
We thank Jim Williams (Carnegie Mellon University, Pittsburgh, Pa.) for provision of Ad12 _hr_703, Keith Leppard (Department of Biological Sciences, University of Warwick, Coventry, United Kingdom) for provision of Ad5 _dl_1520, Christos Paraskeva and Ann Williams (Department of Pathology, University of Bristol, Bristol, United Kingdom) for HT-29 and JW2 cells, Norbert Fusenig (German Cancer Research Center, Heidelberg, Germany) for HaCaT cells, and Todd Waldmann and Bert Vogelstein (Howard Hughes Medical Institute, Baltimore, Md.) for HCT116 cells. We also thank colleagues from the CRC Institute for Cancer Studies, University of Birmingham, for considerable help. In particular, we thank Sally Roberts for help with confocal microscopy, Susan Williams for photography, Kim Bird for FACS analysis, and Sue Rookes for technical assistance.
REFERENCES
- 1.Attardi L D, Lowe S W, Brugarolas J, Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J Virol. 1984;50:202–212. doi: 10.1128/jvi.50.1.202-212.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babbiss L E, Ginsberg H S, Darnell J E. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 5.Bernards R, De Leeuw M G W, Houweling A, van der Eb A J. Role of the adenovirus early region 1B tumor antigens in transformation and lytic infection. Virology. 1986;150:126–139. doi: 10.1016/0042-6822(86)90272-2. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Nh L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 7.Boukamp P, Petrussevska R T, Breikreutz D, Hornung J, Markham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braithwaite A W, Cheetham B F, Li P, Parish C R, Waldron-Stevens L K, Bellet A J D. Adenovirus-induced alterations of the cell growth cycle: a requirement for E1A but not E1B. J Virol. 1985;45:192–199. doi: 10.1128/jvi.45.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braithwaite A W, Blair G E, Nelson C C, McGovern J, Bellett A J D. Adenovirus E1b-58kD antigen binds to p53 during infection of rodent cells: evidence for an N-terminal binding site on p53. Oncogene. 1991;6:781–788. [PubMed] [Google Scholar]
- 10.Breiding D E, Edbauer C A, Yong J T, Byrd P, Grand R J A, Gallimore P H, Williams J. Isolation and characterization of adenovirus type 12 E1 host-range mutants defective for growth in nontransformed human cells. Virology. 1988;164:390–402. doi: 10.1016/0042-6822(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 11.Brockmann D, Esche H. Regulation of viral and cellular gene expression by E1A proteins. Curr Top Microbiol Immunol. 1995;199:81–112. doi: 10.1007/978-3-642-79586-2_5. [DOI] [PubMed] [Google Scholar]
- 12.Byrd P J, Grand R J A, Breiding D, Williams J F, Gallimore P H. Host range mutants of adenovirus 12 E1 defective for lytic infection, transformation and oncogenicity. Virology. 1988;163:155–165. doi: 10.1016/0042-6822(88)90242-5. [DOI] [PubMed] [Google Scholar]
- 13.Chiou S K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 15.de Stanchina E, McCurrach M E, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signalling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobblestein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55kDa and E4 34kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kDa protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 18.Edbauer C, Lambert C, Tong J, Williams J. Adenovirus type 12 E1B 19-kilodalton protein is not required for oncogenic transformation in rats. J Virol. 1988;62:3265–3273. doi: 10.1128/jvi.62.9.3265-3273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faha B, Ewen M E, Tsai L-H, Livingston D M, Harlow E. Interaction between human cyclin A and adenovirus E1A-associated p107 protein. Science. 1992;255:87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- 20.Fukui Y, Saito I, Shiroki K, Shimojo H. Isolation of transformation-defective, replication-nondefective early region 1B mutants of adenovirus 12. J Virol. 1984;49:154–161. doi: 10.1128/jvi.49.1.154-161.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallimore P H, Byrd P J, Grand R J A. Adenovirus genes involved in transformation. What determines the oncogenic phenotype? Symp Soc Gen Microbiol. 1985;37:126–172. [Google Scholar]
- 22.Gallimore P H, Byrd P J, Whittaker J L, Grand R J A. Properties of rat cells transformed by DNA plasmids containing adenovirus 12 E1 DNA or specific fragments of the E1 region. Comparison of transforming frequencies. Cancer Res. 1985;45:2670–2680. [PubMed] [Google Scholar]
- 23.Gallimore P H, Lecane P S, Roberts S, Rookes S M, Grand R J A, Parkhill J. Adenovirus type 12 early region 1B 54K protein significantly extends the life span of normal mammalian cells in culture. J Virol. 1997;71:6629–6640. doi: 10.1128/jvi.71.9.6629-6640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisberg J V, Lee W S, Berk A J, Ricciardi R P. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TaTa box binding protein. Proc Natl Acad Sci USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giard D J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 26.Goodrum F D, Ornelles D A. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grand, R. J. A., J. Parkhill, T. Szestak, S. M. Rookes, S. Roberts, and P. H. Gallimore. Definition of a major p53 binding site on the Ad2 E1B 58K protein and a possible nuclear localization signal on the Ad E1B 54K protein. Oncogene, in press. [DOI] [PubMed]
- 29.Grand R J A, Ibrahim A P, Taylor A M R, Milner A E, Gregory C D, Gallimore P H, Turnell A S. Human cells arrest in S phase in response to adenovirus 12 E1A. Virology. 1998;244:330–342. doi: 10.1006/viro.1998.9102. [DOI] [PubMed] [Google Scholar]
- 30.Grand, R. J. A., A. S. Turnell, G. G. F. Mason, W. Wang, A. E. Milner, J. S. Mymryk, S. M. Rookes, A. J. Rivett, and P. H. Gallimore. Adenovirus early region 1A protein binds to mammalian SUG—a regulatory component of the proteasome. Oncogene, in press. [DOI] [PubMed]
- 31.Grand R J A, Owen D, Rookes S M, Gallimore P H. Control of p53 expression by adenovirus 12 early region 1A and early region 1B 54K proteins. Virology. 1996;218:23–34. doi: 10.1006/viro.1996.0162. [DOI] [PubMed] [Google Scholar]
- 32.Grand R J A, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 33.Gregory C D, Dive C, Henderson S, Smith C A, Williams G T, Gordon J, Rickinson A B. Activation of Epstein-Barr virus latent genes protects human B cells from death by apoptosis. Nature. 1991;349:612–614. doi: 10.1038/349612a0. [DOI] [PubMed] [Google Scholar]
- 34.Han J H, Sabbatini P, Perez D, Rao L, Modha D, White E. The E1B 19K protein blocks apoptosis by interacting with and inhibiting the p53-inducible and death-promoting Bax protein. Genes Dev. 1996;10:461–471. doi: 10.1101/gad.10.4.461. [DOI] [PubMed] [Google Scholar]
- 35.Harrison T, Graham F L, Williams J. Host range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977;122:109–124. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- 36.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff D D, Kirn D H. Onyx-015, an E1B-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 37.Jones N. Transcriptional modulation by the adenovirus E1A gene. Curr Top Microbiol Immunol. 1995;199:59–80. doi: 10.1007/978-3-642-79586-2_4. [DOI] [PubMed] [Google Scholar]
- 38.Lassam N J, Bayley S T, Graham F L. Tumor antigens of human Ad 5 in transformed cells and in cells infected with transformation defective host range mutants. Cell. 1979;18:781–791. doi: 10.1016/0092-8674(79)90131-4. [DOI] [PubMed] [Google Scholar]
- 39.Lehman T A, Modali R, Boukamp P, Stanek J, Bennett W P, Welsh J A, Metcalf R A, Stampfer M R, Fusenig N, Rogan E M, Reddel R, Harris C C. p53 mutations in human immortalized epithelial-cell lines. Carcinogenesis. 1993;14:833–839. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]
- 40.Lehman T A, Bennett W P, Metcalf R A, Reddel R, Welsh J A, Ecker J, Modali R V, Ullrich S, Romano J W, Appella E, Testa J R, Gerwin B I, Harris C C. p53 mutations, ras mutations and p53-heat shock protein complexes in human lung cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- 41.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Graham C, Lacy S, Duncan A M V, Whyte P. The adenovirus E1A-associated 130kDa-protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 43.Lowe S W, Ruley H E. Stabilization of the p53 turnover suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 44.Marcellus R C, Teodoro J G, Wu T, Brough D E, Ketner G, Shore G C, Branton P E. Adenovirus type 5 early region 4 is responsible for E1A-induced p53-independent apoptosis. J Virol. 1996;70:6207–6215. doi: 10.1128/jvi.70.9.6207-6215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montell C, Fisher E F, Caruthers M H, Berk A. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982;295:380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- 46.Paraskeva C, Buckle B G, Sheer D, Wigley C B. The isolation and characterization of colo-rectal epithelial cell lines at different stages in malignant transformation from familial polyposis patients. Int J Cancer. 1984;34:49–59. doi: 10.1002/ijc.2910340109. [DOI] [PubMed] [Google Scholar]
- 47.Pilder S, Logan J, Shenk T. Deletion of the gene encoding the adenovirus 5 early region 1B 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J Virol. 1984;52:664–671. doi: 10.1128/jvi.52.2.664-671.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Querido E, Teodoro J G, Branton P E. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan M P, Grodzicker T. Adenovirus E1A 12S protein induces DNA synthesis and proliferation in primary epithelial cells in both the presence and absence of serum. J Virol. 1987;61:673–682. doi: 10.1128/jvi.61.3.673-682.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao L, Modha D, White E. The E1B 19K protein associates with lamins in vivo and its proper localization is required for inhibition of apoptosis. Oncogene. 1997;15:1587–1597. doi: 10.1038/sj.onc.1201323. [DOI] [PubMed] [Google Scholar]
- 51.Ridgway P J, Hall A R, Myers C J, Braithwaite A W. p53/E1b 58kDa complex regulates adenovirus replication. Virology. 1997;237:404–413. doi: 10.1006/viro.1997.8782. [DOI] [PubMed] [Google Scholar]
- 52.Rodrigues N R, Rowan A, Smith M E F, Kerr I B, Bodmer M F, Gannon J V, Lane D P. p53 mutations in colorectal cancer. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross S R, Levine A J, Galos R S, Williams J, Shenk T. Early viral proteins in HeLa cells infected with adenovirus type 5 host range mutants. Virology. 1980;103:475–492. doi: 10.1016/0042-6822(80)90205-6. [DOI] [PubMed] [Google Scholar]
- 54.Sabbatini P, Lin J Y, Levine A J, White E. Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev. 1995;9:2184–2192. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- 55.Sarnow P, Shih Ho Y, Williams J, Levine A J. Adenovirus E1B-58kD tumor antigen and SV40 large tumor antigen are physically associated with the same 54kD cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 56.Scheffner M, Munger K, Byrne J C, Howley P M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiroki K, Ohshima K, Fukui Y, Ariga H. The adenovirus type 12 early-region 1B 58,000-Mr gene product is required for viral DNA synthesis and for initiation of cell transformation. J Virol. 1986;57:792–801. doi: 10.1128/jvi.57.3.792-801.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian T, Kuppuswamy M, Mak S, Chinnadurai G. Adenovirus cyt+ locus, which controls cell transformation and tumorigenicity, is an allele of lp+ locus, which codes for a 19-kilodalton tumor antigen. J Virol. 1984;52:336–343. doi: 10.1128/jvi.52.2.336-343.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi, T., M. M. Nau, I. Chiba, M. J. Birrer, R. K. Rosenberg, M. Uncar, M. Levitt, H. Pass, R. F. Gazdar, and J. D. Minna. p53—a frequent target for genetic abnormalities in lung cancer. Science **246:**491–494. [DOI] [PubMed]
- 60.Takemori N, Cladaras C, Bhat B, Conley A J, Wold W S M. cyt gene of adenoviruses 2 and 5 is an oncogene for transforming function in early region E1B and encodes the E1B 19,000-molecular-weight polypeptide. J Virol. 1984;52:793–805. doi: 10.1128/jvi.52.3.793-805.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teodoro J G, Shore G C, Branton P E. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- 62.Waldmann T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5181–5190. [PubMed] [Google Scholar]
- 63.White E. Regulation of p53-dependent apoptosis by E1A and E1B. Curr Top Microbiol Immunol. 1995;199:33–58. [PubMed] [Google Scholar]
- 64.White E. Regulation of apoptosis by adenovirus E1A and E1B oncogenes. Semin Virol. 1998;8:505–513. [Google Scholar]
- 65.White E, Stillman B. Expression of the adenovirus E1B mutant phenotypes is dependent on the host cell and on synthesis of E1A proteins. J Virol. 1987;61:426–435. doi: 10.1128/jvi.61.2.426-435.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White E, Cipriani R, Sabbitini P, Denton A. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White E, Grodzicker T, Stillman B W. Mutations in the gene encoding the adenovirus early region 1B 19,000-molecular-weight tumor antigen cause degradation of chromosomal DNA. J Virol. 1984;52:410–419. doi: 10.1128/jvi.52.2.410-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between oncogene and anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 69.Williams J, Williams M, Lu C, Telling G. Assessing the role of E1A in differential oncogenicity of group A and group C of human adenoviruses. Curr Top Microbiol Immunol. 1995;199:149–175. doi: 10.1007/978-3-642-79586-2_8. [DOI] [PubMed] [Google Scholar]
- 70.Yaciuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300-kilodalton product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–5397. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 72.Zantema A, Schrier P I, Davis-Olivier A, van Laar T, Vaessen R T M, van der Eb A J. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]