Human Fas-Associated Factor 1, Interacting with Ubiquitinated Proteins and Valosin-Containing Protein, Is Involved in the Ubiquitin-Proteasome Pathway (original) (raw)
Abstract
Human Fas-associated factor 1 (hFAF1) is a novel protein having multiubiquitin-related domains. We investigated the cellular functions of hFAF1 and found that valosin-containing protein (VCP), the multiubiquitin chain-targeting factor in the degradation of the ubiquitin-proteasome pathway, is a binding partner of hFAF1. hFAF1 is associated with the ubiquitinated proteins via the newly identified N-terminal UBA domain and with VCP via the C-terminal UBX domain. The overexpression of hFAF1 and a truncated UBA domain inhibited the degradation of ubiquitinated proteins and increased cell death. These results suggest that hFAF1 binding to ubiquitinated protein and VCP is involved in the ubiquitin-proteasome pathway. We hypothesize that hFAF1 may serve as a scaffolding protein that regulates protein degradation in the ubiquitin-proteasome pathway.
Fas-associated factor 1 (FAF1) was originally identified by Chu et al. (5) as a Fas-associating molecule with the ability to enhance but not initiate apoptosis when it is overexpressed in murine L cells. Later, Ryu et al. (35) reported that human FAF1 (hFAF1) overexpression without any treatment can initiate apoptosis in BOSC23 cells. Recently, hFAF1 was also reported to be a member of the Fas death-inducing signaling complex (36) and as a suppressor of NF-κB activity (30). Unlike other Fas-associating proteins, hFAF1 does not contain a death domain but has several homologous domains based on amino acid sequence analysis, two ubiquitin homologous domains (Ub's), one UAS domain homologous with Caenorhabditis elegans open reading frame C281.1, and another domain homologous with proteins involved in the ubiquitin pathway (UBX) (3). Although hFAF1 seems to have multiple functions related to the apoptosis and ubiquitin pathway, its role in this regard is not clear.
In the ubiquitination pathway, free ubiquitin is activated by the Ub-activating enzyme (E1) through the formation of a thioester between a cysteine in E1 and the C terminus of Ub. The thioester is subsequently transferred to members of the Ub-conjugating enzyme (E2). Ub-protein ligase (E3) has been shown to be responsible for substrate recognition and for promoting Ub ligation to substrate. These multiubiquitin-tagged substrates are recognized and degraded by the 26S proteasome (10, 15, 38). The S5a subunit of the 19S proteasomal cap binds to a multiubiquitin chain, but there are no proteolytic defects in cells lacking the yeast S5a subunit, suggesting that other mechanisms may be involved in the transfer of substrates from the ubiquitination machinery to the proteasome. Recently, it was reported that valosin-containing protein (VCP), a member of the AAA family (ATPase associated with different cellular activities), and proteins containing UBA domain bind to multiubiquitinated substrates and regulate their proteolysis as a postubiquitination event (34). Although hFAF1 seems to have multiple functions related to the apoptosis and ubiquitin pathway, its role in this regard is not clear. Because of its multiple ubiquitin-related domains, we examined whether hFAF1 is involved in the ubiquitin-proteasome pathway. Employing immunochemical techniques combined with mass spectrometric analysis, we found that hFAF1 binds to VCP through its C-terminal UBX domain and recruits multiubiquitinated substrates through its N-terminal UBA domain. Transient overexpression of hFAF1 results in the accumulation of ubiquitinated substrates via the UBA domain and inhibits the degradation of these ubiquitinated proteins. We hypothesize that hFAF1 plays a role as a scaffolding protein in the ubiquitin-proteasome pathway and regulates the degradation of ubiquitinated proteins in proteasomes.
MATERIALS AND METHODS
Cell culture, cDNA expression, and antibodies.
The 293 human embryonic kidney epithelial (HEK293T) cells were grown and maintained in high-glucose Dulbecco's modified Eagle's medium, and Jurkat T lymphoma cells (E6-1 clone) were grown in RPMI 1640 supplemented with 10% fetal bovine serum at 37°C and 5% CO2.
For expression in mammalian cells, the constructs encoding pFlag-CMV-2-hFAF1 full, pFlag-CMV-2-FAF1(1-81), pFlag-CMV-2-FAF1(1-201), pFlag-CMV-2-FAF1(1-345), pFlag-CMV-2-FAF1(181-381), pFlag-CMV-2-FAF1(366-650), pFlag-CMV-2-FAF1(567-650), pFlag-CMV-2-FAF1(82-650), and glutathione _S_-transferase (GST)-hFAF1 were prepared as previously described (36). Flag-VCP and GST-VCP were cloned into the EcoRI and SalI sites of the pFlag-CMV-2 vector and pGEX-4T-1 vector by using a VCP cDNA clone obtained from a human cDNA library. UbG76V-green fluorescent protein (GFP) vector cloned in the EcoRI and BamHI sites of the EGFP-N1 vector was provided by N. P. Dantuma (Karolinska Institutet, Stockholm, Sweden) (8, 21). Monoclonal anti-Flag antibody (M2), monoclonal anti-Flag M2 agarose cross-linking affinity gel, and a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit were purchased from Sigma (St. Louis, Mo.), and polyubiquitin chains (Ub2-7) were purchased from Affiniti Research Products (BIOMOL International LP, Exeter, United Kingdom). Monoclonal antitubulin, anti-GFP, and anti-IκBα antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), monoclonal antiubiquitin antibody was purchased from Chemicon (Temecula, Calif.), monoclonal anti-VCP antibody was purchased from Research Diagnostics (Flanders, N.J.), and tumor necrosis factor alpha (TNF-α) was purchased from R&D Systems (Minneapolis, Minn.). Polyclonal antiserum of hFAF1 was raised in mouse against recombinant human FAF1 (36).
Transient transfection.
HEK293T cells were transfected with expression plasmid using the calcium phosphate precipitation method. Cells were seeded in 100-mm-diameter plates 1 day before transfection at a density of 1.2 × 106 cells and transiently transfected with 6 to 12 μg of expression plasmid by the calcium phosphate method. Jurkat cells were transfected by use of nucleofactor solution provided by an Amaxa apparatus (Cologne, Germany). For using the Amaxa system, 5 × 106 Jurkat cells were harvested and then resuspended in 100 μl of specified electroporation buffer with 5 μg of plasmid DNA. Resuspended cells were transferred to a cuvette and nucleofected with an Amaxa Nucleofector apparatus.
Immunoprecipitation.
Cells were disrupted with a lysis buffer containing protease inhibitors (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM dithiothreitol [DTT], 5 μg of aprotinin/ml, 10 μg of pepstatin A/ml, 10 μg of leupeptin/ml) for 30 min on ice. The lysates were centrifuged at 14,000 × g for 1 h, and the supernatant was incubated for 3 h at 4°C with monoclonal anti-Flag M2 agarose cross-linking affinity beads (Sigma) at 4°C. For immunoprecipitating endogenous FAF1, cell lysates were incubated with mouse immunoglobulin G or polyclonal anti-FAF1 antibody and then with protein G beads. The immunoprecipitated beads were washed at least three times with 1 ml of lysis buffer containing 0.5% NP-40. The pellet was used for immunoblotting. For ubiquitin binding, the cell pellet was lysed in 400 μl of hypotonic buffer containing protease inhibitors (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM PMSF, 0.2 mM DTT, 5 μg of aprotinin/ml, 10 μg of pepstatin A/ml, 10 μg of leupeptin/ml, pH 7.9) for 30 min on ice and centrifuged at 16,000 × g for 15 min. The supernatant was incubated for 3 h at 4°C with monoclonal anti-Flag M2 agarose cross-linking affinity beads. Beads were washed three times with 1 ml of lysis buffer containing 0.5% NP-40.
Two-dimensional gel electrophoresis.
The protein samples were mixed with a buffer containing 9.5 M urea, 2% Triton X-100, 5% β-mercaptoethanol, 1 mM PMSF, 5 μg of aprotinin/ml, 10 μg of pepstatin A/ml, 10 μg of leupeptin/ml, 1 mM EDTA, 10 mM Na3VO4, and 10 mM NaF for 30 min at room temperature and electrofocused in 7-cm-long Immobiline DryStrips (pH 4 to 7) with the Amersham IPGphor. The focusing protocol, with 50 μA per strip at 20°C, was used: (i) rehydration for 16 h, (ii) 500 V for 1 h (step and hold), (iii) 1,000 V for 1 h (step and hold), and (iv) 8,000 V for 3 h (step and hold). After electrofocusing, the strips were shaken for 15 min with equilibration buffer (1.5 M Tris-Cl, pH 8.8, 6 M urea, 30% glycerol, 2% sodium dodecyl sulfate [SDS], 10 mg of DTT/ml) and loaded onto Bio-Rad mini two-dimensional SDS-polyacrylamide gels.
In-gel digestion and mass spectrometric analysis.
The gel spots were excised with a scalpel, crushed, and destained by washing with 25 mM ammonium bicarbonate-50% acetonitrile. The silver-stained gel was destained by washing with 15 mM potassium ferricyanide-50 mM sodium thiosulfate, crushed, dehydrated by addition of acetonitrile, rehydrated by adding 10 to 20 μl of 25 mM ammonium bicarbonate with 10 ng of sequencing-grade trypsin (Promega)/μl, and incubated at 37°C for 12 to 15 h. Peptides were extracted three times with 30 μl of a solution containing 60% acetonitrile-0.1% trifluoroacetic acid and finally with 20 μl of acetonitrile. The extracts were pooled and evaporated to dryness in a SpeedVac vacuum centrifuge. The dried peptides were mixed with saturated matrix solution α-cyano-4-hydroxycinnamic acid in 60% acetonitrile-0.1% trifluoroacetic acid and analyzed with matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Voyager-DE STR; Applied Biosystems, Inc.) or electrospray ionization quadrupole (ESI-q)-TOF tandem mass spectrometry (Micromass/Waters Corp.) in conjunction with the MS-Fit program available on the World Wide Web site of the University of California at San Francisco (http://prospector.ucsf.edu/).
In vitro binding assay.
GST-hFAF1 deletion mutant fusion proteins and GST-VCP were expressed in Escherichia coli BL21(DE3) cells and bound to glutathione-Sepharose 4B beads. The beads were washed three times with phosphate-buffered saline containing 0.1% Triton X-100. GST-hFAF1 bound to glutathione-Sepharose 4B beads was incubated with recombinant VCP. GST-VCP bound to glutathione-Sepharose 4B beads was incubated with recombinant hFAF1 in binding buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mg of bovine serum albumin/ml) with rocking for 3 h at 4°C. For ubiquitin binding, GST-hFAF1 bound to beads was incubated with cytoplasmic extract from HEK293T cells or polyubiquitin chains (Ub2-7) in binding buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mg of bovine serum albumin/ml) for 3 h at 4°C. The beads were washed five times with 1 ml of 0.1% Triton X-100 in binding buffer and then resuspended in 2× gel sample buffer.
Quantifying the degradation of ubiquitinated proteins.
Proteolysis of ubiquitinated proteins was quantified by using UbG76V-GFP as a substrate (8, 21). HEK293T cells were cotransfected with expression plasmid, 200 ng of UbG76V-GFP or EGFP-N1 vector, and 1 μg of various truncated Flag-hFAF1 constructs in 35-mm-diameter dishes by using the calcium phosphate precipitation method. After 24 h, the cells were washed with phosphate-buffered saline, and the fluorescence of UbG76V-GFP in 10,000 cells was measured with a fluorescence-activated cell sorter (FACS) flow cytometer (Becton Dickinson). Mean fluorescence intensity was calculated with Cellquest software.
Cell viability assay.
An MTT conversion assay was employed to assess cell viability. Jurkat cells transiently transfected with expression plasmid by use of an Amaxa electroporator were seeded in 96-well plates at a density of 105 cells/well. Ten microliters of MTT (Sigma) was added and then incubated for 2 h to allow metabolization of MTT to 3-[4,5-dimethyldiazol-2-yl]-2,5-diphenylformazan. The latter was solubilized with acidified isopropanol, and absorbance was measured at 570 nm in a microplate reader.
RESULTS
hFAF1 interacts with VCP in vitro and in vivo.
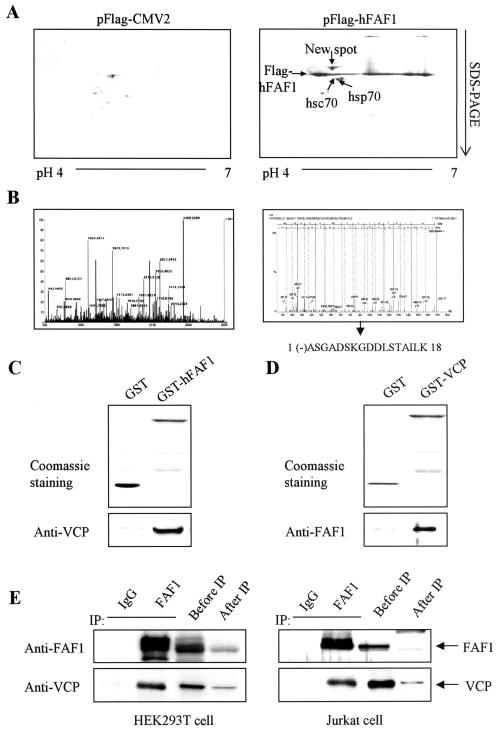
We attempted to identify the proteins interacting with hFAF1 as leads for understanding how hFAF1 functions. Flag-hFAF1 was transiently overexpressed in HEK293T cells. HEK293T cell lysates were immunoprecipitated with monoclonal anti-Flag M2 agarose cross-linking affinity beads, and the immunoprecipitates were analyzed by two-dimensional gel electrophoresis. One fraction was transferred to a polyvinylidene difluoride membrane for Western blot analysis, and the other was silver stained for visualization. Flag-hFAF1 was detected at 80 to 85 kDa with silver staining and confirmed by Western blotting (data not shown). In addition to hsp70, shown to interact with hFAF1 in an earlier study (18), we detected hsc70 and one new spot in the coprecipitates with Flag-tagged hFAF1 but not with control Flag. The new spot detected in the silver-stained gel was subjected to in-gel digestion with trypsin and analyzed by use of MALDI-TOF MS and ESI-q-TOF tandem MS. The protein interacting with hFAF1 was identified as VCP (NCBI accession number 6005942) by peptide mass fingerprinting using MALDI-TOF MS and confirmed by sequencing using ESI-q-TOF tandem MS (Fig. 1B).
FIG. 1.
hFAF1 interacts with VCP in vitro and in vivo. (A) HEK293T cells transfected with pFlag-CMV-2 vector and pFlag-hFAF1 were labeled with 1 μCi of [35S]methionine/ml. Immunoprecipitation in cell lysates was carried out with anti-Flag M2 agarose cross-linking affinity beads (Sigma). Precipitates were analyzed by two-dimensional gel electrophoresis and autoradiographed with BAS2500. (B) Protein spots detected in the autoradiograph were cut out from the corresponding silver-stained gel and subjected to in-gel digestion with trypsin. Mass peptide fingerprint analyses were conducted by MALDI-TOF MS (left spectrum), and peptides were sequenced by using ESI-q-TOF tandem MS (right spectrum). (C and D) GST-hFAF1 (C) and GST-VCP (D) were immobilized on glutathione beads and incubated with purified recombinant VCP (C) and purified recombinant hFAF1 (D). Binding proteins were detected on SDS-PAGE with Coomassie staining and immunoblotted by anti-VCP antibody (C) and anti-FAF1 antibody (D). (E) For the interaction between endogenous FAF1 and VCP, HEK293T cells and Jurkat cell lysates were immunoprecipitated with mouse immunoglobulin G control (lane 1) and anti-FAF1 polyclonal antibodies (lane 2). The amount of endogenous FAF1 and VCP in 10% of cell lysates before immunoprecipitation was compared with the amounts after immunoprecipitation. Immunoprecipitates were analyzed by Western blotting using anti-FAF1 and anti-VCP antibodies.
To examine whether hFAF1 interacts with VCP directly, we performed reciprocal in vitro binding assays. GST-hFAF1 was immobilized to glutathione-Sepharose beads and incubated with purified recombinant VCP. We observed direct interaction between GST-hFAF1 and VCP by using anti-VCP antibody (Fig. 1C). In the reciprocal experiment, GST-VCP that was immobilized on glutathione beads was incubated with purified recombinant hFAF1, and hFAF1 that was bound to GST-VCP was detected with anti-FAF1 antibody (Fig. 1D).
In order to see whether endogenous hFAF1 interacts with endogenous VCP in vivo, we performed immunoprecipitation studies with polyclonal anti-hFAF1 antibody in HEK293T cells and Jurkat cells. In both HEK293T and Jurkat cells, endogenous FAF1 coimmunoprecipitated with VCP, as detected with anti-VCP antibody (Fig. 1E). This result indicates that endogenous FAF1 interacts with VCP in vivo as well.
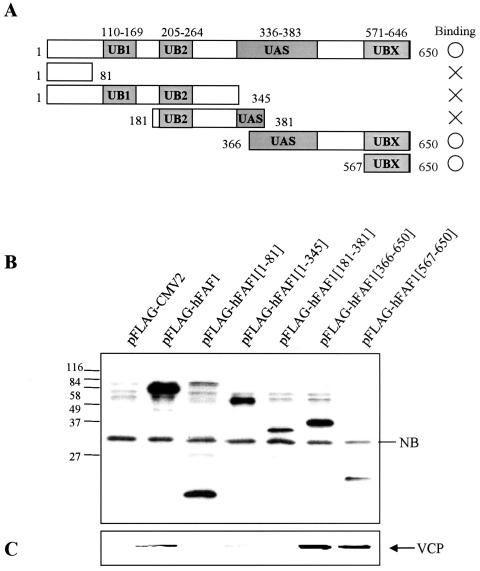
hFAF1 interacts with VCP via the UBX domain.
To characterize the domains of interaction between hFAF1 and VCP, various truncated Flag-hFAF1s were overexpressed in HEK293T cells, and their interactions with endogenous VCP were examined. As shown in Fig. 2A, overexpressed truncated Flag-hFAF1s were immunoprecipitated with anti-Flag M2 agarose cross-linking affinity beads and immunoblotted with anti-Flag antibody (Fig. 2B) and anti-VCP antibody (Fig. 2C). As shown in Fig. 2, endogenous VCP binds to Flag-hFAF1 full form and to hFAF1 fragments containing amino acids 366 to 650 and 567 to 650. This result suggests that VCP interacts with hFAF1 via the UBX domain located in the amino acid sequence 567 to 650. These results are consistent with previous reports that p47, identified as a major protein in complex with VCP and whose UBX domain is very similar in its structure to the UBX domain from hFAF1 (20, 43), interacts with VCP through the UBX domain. It appears that hFAF1 is another protein that interacts with VCP through its UBX domain.
FIG. 2.
hFAF1 interacts with VCP via the UBX domain. (A) Diagram of various truncated forms of Flag-hFAF1. UB1 and UB2, ubiquitin homologous domains; UAS domain, homologous with Caenorhabditis elegans open reading frame C281.1; UBX, domain present in ubiquitin regulatory proteins. (B and C) HEK293T cells transfected with pFlag-CMV-2 vector or truncated Flag-hFAF1 were lysed and immunoprecipitated with monoclonal anti-Flag M2 agarose cross-linking affinity gel (Sigma). Precipitates were analyzed by Western blotting using anti-Flag (B) and anti-VCP (C) antibodies. NB indicates a nonspecific protein band.
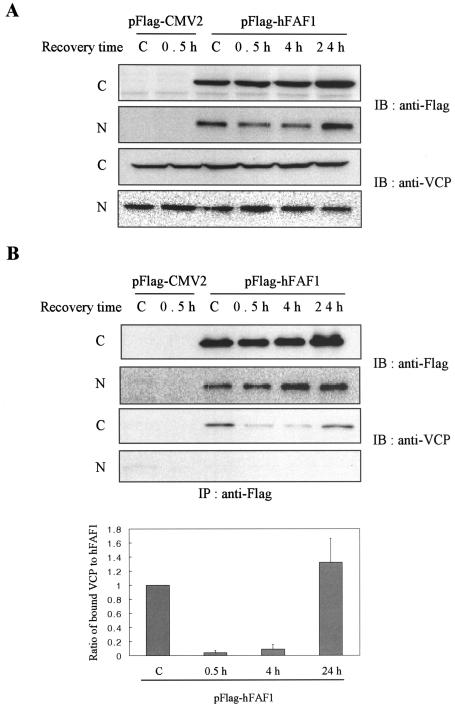
hFAF1 interacts with VCP in cytosol.
In previous reports, quail FAF1 was shown to have a novel nuclear localization signal and to localize in the nucleus (12). We examined whether hFAF1 interacts with VCP in the cytosolic or nuclear fractions and whether heat shock-inducing ubiquitination can influence this interaction. HEK293T cells transfected with the Flag-hFAF1 were exposed to heat shock stress at 45°C for 45 min and fractionated into cytosolic and nuclear fractions. As shown in Fig. 3A, both Flag-hFAF1 and VCP were present in cytosolic and nuclear fractions, but VCP was coimmunoprecipitated with Flag-hFAF1 only in the cytosolic fraction (Fig. 3B). The amount of VCP coimmunoprecipitating with Flag-hFAF1 significantly decreased from heat shock and increased during recovery, while the total amount of VCP in the cytosol remained unchanged. This result suggests that the interaction of hFAF1 with VCP occurred only in cytosol in a stress-dependent manner (Fig. 3B) and may be involved in the degradation of proteins in cytosol.
FIG. 3.
hFAF1 interacts with VCP in cytosol. HEK293T cells transiently transfected with pFlag-CMV-2 vector or Flag-hFAF1 were heat shocked at 45°C for 45 min and recovered for the indicated times. “C” lane headings indicate untreated cells. At each time point, cells were fractionated into cytosolic (C) and nuclear (N) fractions and Western blotted with anti-Flag and anti-VCP antibodies (A). For examining the binding, each fraction was immunoprecipitated with monoclonal anti-Flag M2 agarose cross-linking affinity gel, and precipitates were analyzed by Western blot analysis using anti-Flag and anti-VCP antibodies. Western analysis of panel B was quantified with ImageQuant (Amersham Biosciences). The relative ratio of bound VCP to Flag-hFAF1 at control is arbitrarily presented as 1. Bars show means ± standard deviations (SD) from three independent experiments (B).
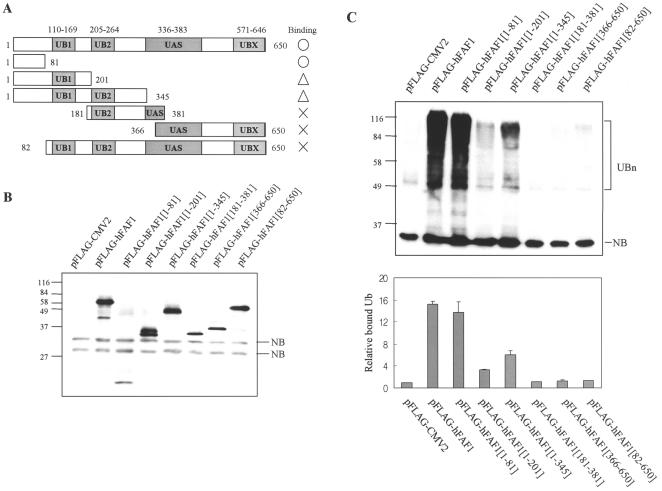
hFAF1 binds to ubiquitinated substrates in vitro and in vivo.
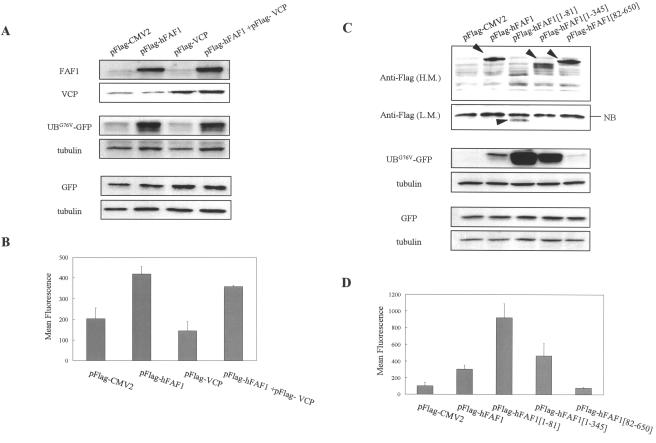
Because hFAF1 has multiple ubiquitin-related domains and also interacts with VCP, which is known to function in the ubiquitin-proteasome pathway (22, 42), we investigated whether hFAF1 is involved in the ubiquitin-proteasome pathway. Various truncated Flag-hFAF1s listed in Fig. 4A were overexpressed in HEK293T cells, immunoprecipitated with anti-Flag M2 agarose cross-linking affinity beads, and immunoblotted with antiubiquitin antibodies. As shown in Fig. 4C, full Flag-hFAF1 bound to multiubiquitinated proteins. N-terminal fragments containing amino acids 1 to 81 seem to be the region of hFAF1 involved in binding to a degree equal to or better than that of full hFAF1. Flag-hFAF1(1-201) and Flag-hFAF1(1-345) also bound to multiubiquitinated proteins, but the binding was weaker than with the wild type.
FIG. 4.
hFAF1 binds to multiubiquitinated proteins in vivo. (A) Diagram of various truncate forms of Flag-hFAF1. (B and C) HEK293T cells transfected with pFlag-CMV-2 vector or truncated forms of Flag-hFAF1 were lysed and immunoprecipitated with monoclonal anti-Flag M2 agarose cross-linking affinity gel (Sigma). Precipitates were analyzed by Western blotting using anti-Flag (B) and antiubiquitin (C) antibodies. NB indicates a nonspecific band, and UBn indicates multiubiquitinated proteins. Western analysis of panel C was quantified with ImageQuant (Amersham Biosciences). The relative ratio of bound multiubiquitinated proteins to pFlag-CMV2 is arbitrarily presented as 1. Bars show means ± SD from three independent experiments.
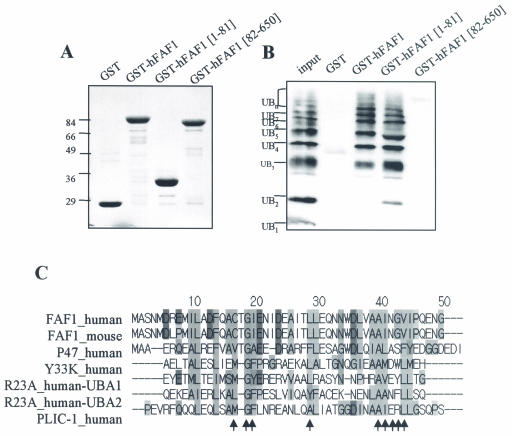
To examine whether hFAF1 directly interacts with multiubiquitinated proteins, we performed an in vitro binding assay. Various truncated GST-hFAF1s immobilized to glutathione-Sepharose beads were incubated with polyubiquitin chains (Ub2-7), and bindings were detected with immunoblotting using antiubiquitin antibody and synthetic Lys48-linked polyubiquitin chains (Ub2-7) (Affiniti Research), which mimic endogenous multiubiquitinated substrate. As shown in Fig. 5B, we found that full hFAF1 and its fragment containing amino acids 1 to 81 but not amino acids 82 to 650 bound to polyubiquitin chains (Ub2-7) in vitro. In addition, GST-hFAF1 full form and GST-hFAF1(1-81) interacted with multiubiquitin chains but not monoubiquitin. GST-hFAF1(1-81) interacted with di- and triubiquitin chains more strongly than GST-hFAF1 full form. However, both GST-hFAF1 full form and GST-hFAF1(1-81) have similar affinities to longer polyubiquitin chains. These results suggest that hFAF1 directly binds to multiubiquitinated proteins via amino acids 1 to 81. Structural differences probably account for the differences in the binding affinities of hFAF1(1-81) and full hFAF1.
FIG. 5.
hFAF1 binds to multiubiquitinated proteins in vitro. (A and B) Polyubiquitin chains (Ub2-7) were incubated with GST, GST-hFAF1, GST-hFAF1(1-81), and GST-hFAF1(82-650) proteins immobilized on glutathione-Sepharose in vitro. Various GST proteins immobilized to glutathione beads were detected on SDS-PAGE with Coomassie staining (A), and polyubiquitin chains associated with GST-hFAF1 were detected by immunoblotting using monoclonal antiubiquitin antibodies (B). (C) Analysis was performed with the programs ClustalW (http://www.ebi.ac.uk/clustalw/) and Jalview (http://www.ebi.ac.uk/jalview). Arrows indicate the amino acid residues occupying the conserved positions on the hydrophobic surface patch of the UBA domain (25). FAF1_human (FAS-associated factor 1 isoform a), NP_008982; FAF1_mouse (FAS-associated factor 1), P54731; P47_human (p47 protein isoform a), NP_057227; Y33K_human (UBA/UBX 33.3-kDa protein), Q04323; R23A_human (UV excision repair protein RAD23 homolog A), P54725; PLIC-1_human (PLIC-1), AAG02473.
When the amino acid sequence of hFAF1(1-81) was aligned with different proteins by using the Jalview program, we observed homology to the UBA domain, which is known to be involved in binding to multiubiquitinated proteins (Fig. 5C). The N-terminal amino acid sequence of hFAF1 seems to be well conserved among species. It is identical to that of mouse FAF1 and is similar to that of p47 and Y33K, all of which contain C-terminal UBX domains. Rad23 and PLIC are proteins having central and C-terminal UBA domains, which bind to multiubiquitinated proteins via the UBA domain (13, 39). The UBA domain has a conserved hydrophobic surface patch in solution structure, and the arrows in Fig. 5C indicate the amino acid residues that have been shown in the surface of the UBA domain (28). The hydrophobic surface patch of the UBA domain is important because it interacts with the hydrophobic surface on the five-stranded β-sheet of ubiquitin. Although the UBA domain of hFAF1 is not fully conserved with other UBA domains, the amino acid residues occupying the position on the hydrophobic surface patch in three-dimensional structure are well conserved (Fig. 5C). Based on a combination of experimental results and homology search, it appears that hFAF1 belongs to a new family of proteins binding to multiubiquitinated proteins via the N-terminal UBA domain.
hFAF1 binds to Lys48-linked and Lys63-linked ubiquitinated proteins via UBA domains.
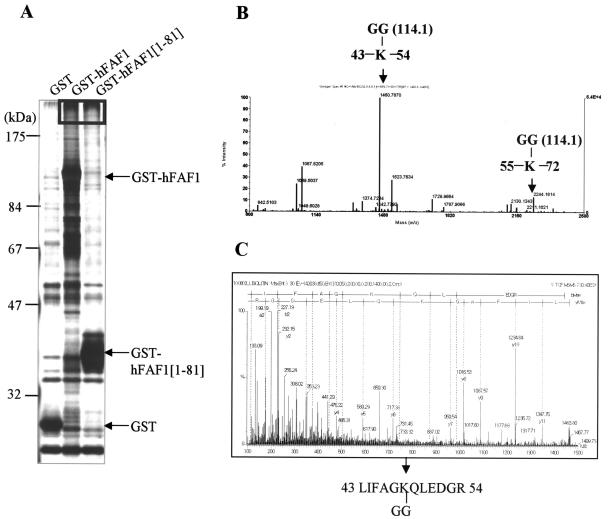
To determine which population of multiubiquitin chains can interact with hFAF1, we coupled GST-hFAF1 and GST-hFAF1(1-81) to glutathione-Sepharose beads and incubated them with HEK293T cell lysates. Interaction was detected by Western blot analysis using antiubiquitin antibodies (data not shown). High-molecular-weight multiubiquitinated proteins were analyzed by mass spectrometry (Fig. 6A). In the ubiquitin-proteasome pathway, multiubiquitin is chained through Lys48-Gly76 isopeptide bonds, with the proximal Ub linked to substrate lysine residue (31). Tryptic digestion of ubiquitin-conjugated protein produced a signature peptide at the ubiquitination site containing a two-residue remnant (Gly-Gly). This remnant was derived from the C terminus of ubiquitin and covalently attached to a lysine residue of the target protein via an isopeptide bond. This signature peptide has a mass shift of 114.1 Da at the lysine residue as well as a missed proteolytic cleavage, because trypsin cannot break at peptide bonds involving modified lysines (31). Using MALDI-TOF MS, we identified high-molecular-weight interacting proteins of hFAF1 as multiubiquitin chain (NCBI accession number 136670). In addition, we detected two increased 114.1-Da peaks (m/z, 1460.7870; m/z, 2244.1814) resulting from amino acid residues 43 to 54 and 55 to 72, respectively (Fig. 6B and Table 1). To verify that the 114.1-Da mass shifts occur because of isopeptides from a ubiquitin chain, we sequenced each peak by using ESI-q-TOF tandem mass spectrometry and identified them as Lys48-Gly-Gly (Fig. 6C) and Lys63-Gly-Gly isopeptides (data not shown). These results demonstrated that hFAF1 interacts with Lys48-linked and Lys63-linked multiubiquitin chains.
FIG. 6.
hFAF1 binds to Lys48-linked and Lys63-linked ubiquitinated proteins via the UBA domain. (A) GST, GST-hFAF1, and GST-hFAF1(1-81) proteins were immobilized on glutathione beads, incubated with HEK293T cell lysates, and then detected on SDS-PAGE with silver staining. High-molecular-weight proteins indicated in the box in panel A were in-gel digested with trypsin and analyzed with MALDI-TOF MS (B). Arrows in panel B indicate ubiquitin peptides containing the isopeptide bond linkage through Lys48 (m/z, 1460.7870) and Lys63 (m/z, 2244.1814). (C) The peptide linked through Lys48 was fragmented by ESI-q-TOF MS, and the amino acid sequence, LIFAGK(-GG)QLEDGR, was determined.
TABLE 1.
Expected and observed peptide fragments obtained from trypsin cleavage of ubiquitin associated with GST-hFA1
| Tryptic peptide | Expected m/z | Observed m/z | Sequence | Modification |
|---|---|---|---|---|
| 12-27 | 1787.9278 | 1787.9066 | ||
| 30-42 | 1523.7818 | 1523.7634 | ||
| 34-42 | 1039.5172 | 1039.5037 | ||
| 43-54 | 1346.7432 | 1460.7870 | LIFAGK(-GG)QLEDGR | Lys-Gly-Gly |
| 55-72 | 2130.1559 | 2130.1343 | ||
| 55-72 | 2130.1559 | 2244.1814 | TLSDYNIQK(-GG)ESTLHLVLR | Lys-Gly-Gly |
| 64-72 | 1067.6213 | 1067.6204 |
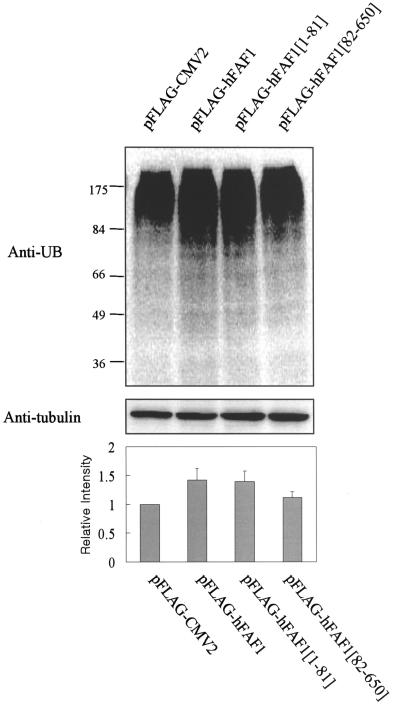
hFAF1 inhibits degradation of ubiquitin-dependent substrates.
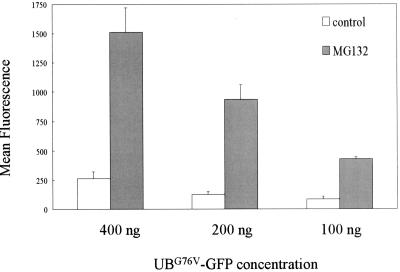
Figure 7 shows that the overexpression of Flag-hFAF1 and Flag-hFAF1(1-81) inhibited the degradation of ubiquitinated substrates, in contrast to the overexpression of pFlag-CMV vector and Flag-hFAF1(82-650). We hypothesize that the accumulation of the intracellular multiubiquitinated proteins following overexpression of hFAF1 occurred because hFAF1 complexed with them and prevented or inhibited their proteolytic degradation. To test this hypothesis, we decided to examine whether hFAF1 influences the proteolysis of ubiquitinated proteins. For this test, we employed the GFP-based proteasome substrate, UbG76V-GFP, used by Dantuma et al. for quantification of ubiquitin-proteasome-dependent proteolysis in living cells (8, 17, 21). We first confirmed that this system works under our experimental conditions (Fig. 8). We then transiently cotransfected HEK293T cells with 200 ng of UbG76V-GFP and 1 μg of Flag-hFAF1 or Flag-VCP and measured GFP fluorescence intensity by flow cytometry after proteasome degradation 24 h after transfection.
FIG. 7.
Overexpression of hFAF1 accumulates multiubiquitinated proteins. HEK293T cells transfected with pFlag-CMV-2 vector, Flag-hFAF1, Flag-hFAF1(1-81), and Flag-hFAF1(82-650) were separated on SDS-PAGE, and the multiubiquitinated proteins were detected with Western analysis using antiubiquitin antibody (upper panel). Protein concentrations were normalized with antitubulin antibody (middle panel). Western analysis was quantified with ImageQuant (Amersham Biosciences). The relative intensity of multiubiquitinated proteins when pFlag-CMV2 was overexpressed is arbitrarily presented as 1. Bars show means ± SD from three independent experiments (lower panel).
FIG. 8.
Effect of proteasome inhibitor on degradation of UbG76V-GFP. HEK293T cells transiently transfected with 100, 200, or 400 ng of UbG76V-GFP were incubated for 15 h with or without 10 μM MG132 (Calbiochem). At 24 h after transfection, the fluorescence of harvested cells was measured with a FACS flow cytometer (Becton Dickinson), and fluorescence data were analyzed with Cellquest software. The mean fluorescence intensities shown are the means ± SD from three independent experiments.
Overexpression of Flag-hFAF1 significantly reduced the UbG76V-GFP degradation detected with Western analysis (Fig. 9A) and raised the mean fluorescence intensity twofold compared to overexpression of the pFlag-CMV vector (Fig. 9B). To exclude the possibility that the difference in mean fluorescence intensity may be due to the transfection efficiency of UbG76V-GFP, we performed the same experiment using the EGFP-N1 vector. HEK293T cells were transiently cotransfected with the EGFP-N1 vector and Flag-hFAF1 or Flag-VCP. At 24 h after transfection, the GFP protein level was determined by Western analysis using GFP antibody and normalized with tubulin. As shown in Fig. 9A, GFP intensity based on the transfection efficiency remained unchanged, confirming that the increase in hFAF1 was specifically responsible for the decreased degradation of UbG76V-GFP reporter in proteasome.
FIG. 9.
hFAF1 inhibits ubiquitin-dependent protein degradation via the UBA domain. (A and B) Two hundred nanograms of UbG76V-GFP or EGFP-N1 vector was cotransfected with 1 μg of pFlag-CMV-2 vector, Flag-hFAF1, Flag-VCP, and a mixture of Flag-hFAF1 and Flag-VCP in HEK293T cells that had been seeded in 35-mm-diameter dishes at a density of 2 × 105 cells 1 day before. (C and D) Two hundred nanograms of UbG76V-GFP or EGFP-N1 vector was cotransfected with 1 μg of pFlag-CMV-2 vector, Flag-hFAF1, Flag-hFAF1(1-81), Flag-hFAF1(1-345), and Flag-hFAF1(82-650) in HEK293T cells that had been seeded in 35-mm-diameter dishes at a density of 2 × 105 cells 1 day before. At 24 h after transfection, cells were harvested and divided into two portions. One was used for detecting UBG76V-GFP by Western analysis (A and C), and the other was used for measuring the fluorescence of UBG76V-GFP by FACS analysis (B and D). (A and C) For confirming protein levels of hFAF1, VCP, and truncated hFAF1, cells were lysed, separated in SDS-PAGE, and immunoblotted by using anti-FAF1 (A), anti-VCP (A), and anti-Flag (C) antibodies. In the case of Flag-hFAF1(1-81), Western membrane was exposed for a long time [panel C, Anti-Flag (L.M.)] because transfection efficiency was lower than that for other truncated forms. The level of UbG76V-GFP or EGFP-N1 vector was detected with anti-GFP antibody, and protein concentration was normalized with antitubulin antibody. (B and D) The fluorescence of harvested cells was measured with a FACS flow cytometer (Becton Dickinson), and fluorescence data were analyzed with Cellquest software. The mean fluorescence intensities ± SD from three independent experiments are shown (P value, <0.05).
Using flow cytometry and Western blotting, we found that overexpression of Flag-VCP decreased the level of UbG76V-GFP reporter (Fig. 9). This result was as expected because VCP is known to recruit ubiquitinated proteins and to promote their degradation in proteasomes (6, 7). When cotransfected with Flag-hFAF1 and Flag-VCP, the fluorescence level of the UbG76V-GFP reporter was in between the fluorescence levels of VCP and hFAF1. There are two possible explanations for this outcome. One is that this observed fluorescence is the sum of fluorescence levels of Flag-hFAF1 and Flag-VCP. The other explanation is that VCP might negatively regulate hFAF1 and cause the observed fluorescence. We attempted to determine which of these possibilities is in fact occurring and which domain of hFAF1 is necessary for stabilizing proteasomal substrates by inhibiting their degradation. For this determination, HEK293T cells were cotransfected with UbG76V-GFP reporter and various truncated forms of Flag-hFAF1. Using flow cytometry and Western blotting, we found that overexpression of full Flag-hFAF1 increased the level of UbG76V-GFP and that deletion of the UBA domain of hFAF1(82-650) had no effect (Fig. 9C and D). Flag-hFAF1(1-81) and Flag-hFAF1(1-345) containing the UBA domain but not the VCP binding domain increased fluorescence levels significantly over that of intact hFAF1 (Fig. 9C and D). These results indicate that the UBA domain of hFAF1 can recruit the ubiquitinated proteasomal substrates and that VCP, interacting with hFAF1 via the C terminus, can induce the degradation of ubiquitinated proteins recruited by hFAF1 in the proteasome degradation pathway.
hFAF1 interferes with the proteolytic degradation of endogenous ubiquitinated substrate.
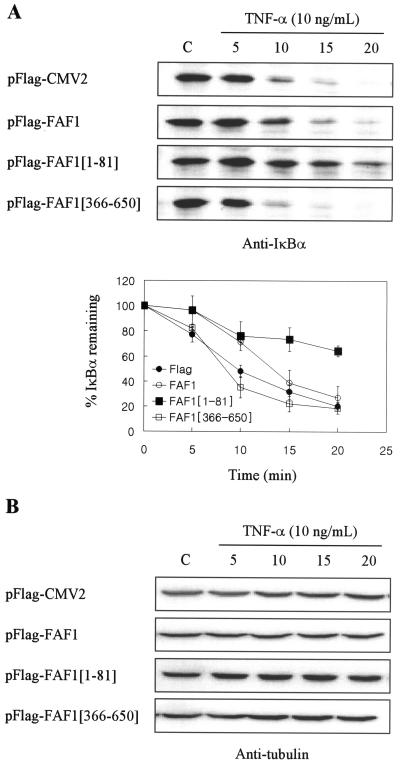
Since hFAF1 inhibited the degradation of UbG76V-GFP, we examined whether hFAF1 can also affect the degradation of endogenous ubiquitinated substrates. We selected IκBα as the endogenous substrate, because IκBα is known to be degraded by the ubiquitin-proteasome pathway (41) and also because VCP is associated with ubiquitinated IκBα when recruited to proteasomes (7). HEK293T cells transfected with Flag-hFAF1 or its truncated forms were exposed to TNF-α (10 ng/ml) for 5, 10, 15, and 20 min 24 h after transfection. As shown in Fig. 10, total cell lysates were immunoblotted using IκBα antibody and TNF-α, leading to the rapid degradation of IκBα, depending on the length of exposure. In untreated cells, IκBα content was almost the same irrespective of the overexpression of DNA. However, the overexpression of Flag-hFAF1(1-81) interfered with IκBα degradation, whereas Flag vector and Flag-hFAF1(366-650) did not (Fig. 10A). In the case of overexpression of Flag-hFAF1, IκBα degradation was attenuated slightly. IκBα content remained unchanged for up to 15 min during which Flag-hFAF1 was overexpressed and for up to 20 min during which Flag-hFAF1(1-81) was overexpressed. Total protein levels, normalized with tubulin, show that there was no degradation by TNF-α treatment itself (Fig. 10B). Consistent with the results obtained by using the UbG76V-GFP reporter, the effect of Flag-hFAF1(1-81) on IκBα degradation by TNF-α is stronger than that of Flag-hFAF1. It is possible that Flag-hFAF1(1-81) has an enhanced effect because VCP cannot bind to it. Besides, the absence of any difference in the total IκBα content suggests that hFAF1 affects only ubiquitinated protein. Taken together, these results indicate that the overexpression of hFAF1 interferes with the degradation of artificial as well as endogenous ubiquitinated proteins.
FIG. 10.
hFAF1 interferes with the degradation of endogenous ubiquitinated substrate. HEK293T cells transfected with pFlag-CMV-2 vector, Flag-hFAF1, Flag-hFAF1(1-81), and Flag-hFAF1(366-650) were exposed to 10 ng of TNF-α/ml for 5, 10, 15, and 20 min at 24 h after transfection. Cells were lysed, separated in SDS- PAGE, and immunoblotted with IκBα antibody. The amounts of IκBα remaining after TNF-α treatment were determined by ImageQuant (Amersham Biosciences). (A) Bars show means ± SD from three independent experiments. (B) Protein concentration was normalized with antitubulin antibody. C, untreated cells.
Cell death is induced by overexpression of hFAF1 and requires the UBA domain.
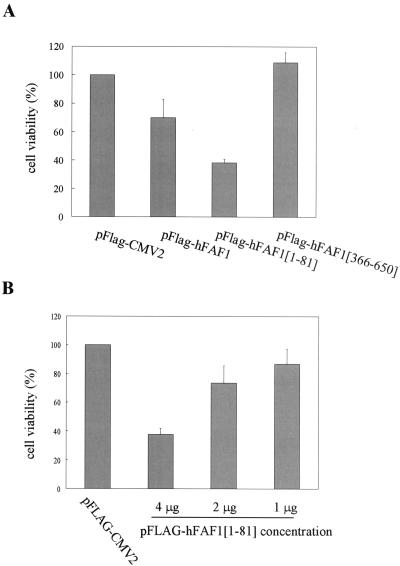
It has been reported that hFAF1 overexpression without any treatment can initiate apoptosis in BOSC23 cells and that hFAF1 interacts with the members of the Fas death-inducing signaling complex in Jurkat cells (35, 36). However, the relationship of ubiquitination and cell death by hFAF1 is not clear. We therefore examined whether the accumulation of ubiquitinated proteins caused by hFAF1 affects hFAF1-induced cell death. We used Jurkat cells in this study because earlier studies employed these cells, among others, to study cell death caused by FAF1 (35, 36). Jurkat cells were transiently transfected with Flag-hFAF1 or its truncated constructs by use of an Amaxa electroporator, and cell viability was assessed as described above. The overexpression of Flag-hFAF1 induced cell death, as previously reported (35, 36), and Flag-hFAF1(1-81) containing a UBA domain significantly increased cell death (Fig. 11A). The effect on cell death of hFAF1(1-81), which has only the UBA domain, is stronger than that of full hFAF1, consistent with GFP reporter experiments. In the case of Flag-hFAF1(366-650), cell viability rather increased compared to controls. To confirm the cell death effect of the UBA domain, we transiently transfected Jurkat cells with various amounts of Flag-hFAF1(1-81) by using an Amaxa electroporator and assessed cell viability by MTT assay. Cell death was induced by Flag-hFAF1(1-81) in a manner dependent on transfected DNA concentration (Fig. 11B), suggesting that the overexpression of the UBA domain of hFAF1 induces cell death by causing the accumulation of ubiquitinated proteins (Fig. 7 and 11).
FIG. 11.
Effect of hFAF1 overexpression on cell death. (A) Jurkat cells were transfected with pFlag-CMV-2 vector, Flag-hFAF1, Flag-hFAF1(1-81), and Flag-hFAF1(366-650) with an Amaxa electroporator. At 24 h after transfection, cell viability was assessed by using an MTT conversion assay. The relative viabilities shown are means ± SD from three independent experiments (P value, <0.05). (B) Jurkat cells were transfected with 1, 2, or 4 μg of Flag-hFAF1(1-81) with an Amaxa electroporator. At 24 h after transfection, cell viability was assessed by using an MTT conversion assay. The relative viabilities shown are means ± SD from three independent experiments.
DISCUSSION
These studies demonstrate the interaction of hFAF1 with VCP and multiubiquitinated proteins. We believe that these interactions may play a role in the regulation of proteolytic degradation in the proteasome.
VCP is a member of the AAA family (ATPase-associated family with different cellular activities). VCP's yeast homolog, cdc48p, was identified as a cell division cycle protein (25, 27). VCP participates in various cellular processes, including membrane fusion, vesicle-mediated transport, and peroxisome assembly (1, 22) through binding to specific cofactors. p47 is a cofactor of VCP in membrane fusion during postmitotic reassembly of the Golgi stack and mediates the binding of VCP to syntaxin 5, analogous to α-SNAP (34). Also, Ufd1 and Npl4 have been implicated as cofactors of VCP in the proteasome-mediated processing of endoplasmic reticulum-bound proteins as well as cytosolic proteins (24, 42). Furthermore, cdc48p has been shown to bind to several proteins, such as Ufd3p and Ufd2p, implicated in the ubiquitin fusion degradation pathway (14, 19). VCP's role in targeting IκBα to the proteasome for degradation (6, 7, 9) has already been noted. These findings suggest that VCP may act as a chaperone that targets many substrates to the proteasome for degradation.
UBA domains have about 45 amino acid residues and are small (16). UBA domains are present in many proteins of the ubiquitination pathway, including UBP, E2, and E3; UV excision repair proteins, such as Rad23 and Dsk2; and protein kinases, such as p62, involved in cell signaling pathways and cell cycle control (28, 37). Mueller and Feigon presented the three-dimensional solution structure of UBA domain by nuclear magnetic resonance spectroscopy (28). The hydrophobic surface patch is a common protein-interacting surface present in diverse UBA domains, and it was suggested that the hydrophobic surface patch of the UBA domain interacts with the hydrophobic surface on the five-stranded β-sheet of ubiquitin. In fact, it has been reported that Rad23- and Dsk2-containing UBA domains bind to mono-, di-, tetra-, and multiubiquitin chains. In addition, UBA domain proteins can interact with ubiquitinated target proteins and regulate their proteolysis (4, 13, 39).
The newly identified N-terminal UBA domain of hFAF1 in this study seems to be important for recruiting and stabilizing the ubiquitinated proteasomal substrates. We found that VCP bound to the C-terminal UBX domain of hFAF1 enhances the degradation of ubiquitinated proteins in proteasomes. Since hFAF1 has many ubiquitin pathway-related domains, a relationship between hFAF1 and the ubiquitin pathway was proposed (2). Our studies described here are the first to confirm experimentally that hFAF1 is involved in the ubiquitin-proteasome pathway.
hFAF1 interacts with VCP via its C-terminal UBX domain, composed of 80 amino acid residues. The UBX domain reveals a structural relationship with ubiquitin and has been shown to be present in p47, Y33K, FAF1, UBXD1, and Rep-8 proteins (3). Of these, the UBX domain in p47 is the only one whose function has been described so far. Apparently, p47 interacts with VCP through its UBX domain (43) and plays a role in membrane fusion processes as an adapter for VCP (20). It is possible that VCP exerts its various cellular functions, including participation in the ubiquitin pathway, by interacting with different proteins in a similar fashion.
Our studies also demonstrate that hFAF1 associates with multiubiquitinated proteins of HEK293T cells in vivo via its N-terminal UBA domain and that hFAF1 interacts in vitro with polyubiquitin chains (Ub2-7) linked by Lys48. As shown in Fig. 5, hFAF1 does not interact with monoubiquitin but interacts strongly with multiubiquitin chains longer than the tetraubiquitin chain. This finding is in agreement with previous reports that the UBA domain has a higher affinity for tetraubiquitin (Kd, 30 nM) than for monoubiquitin (Kd, >1 mM) (39). Our finding that the UBA domain of hFAF1 is aligned with the UBA peptide sequence of different proteins, depicted in Fig. 5C, suggests that the UBA domain of hFAF1 is not fully conserved with other UBA domains, while the amino acid residues occupying the position on the hydrophobic surface patch in three-dimensional structure, which interacts with ubiquitin, are well conserved (Fig. 5C). We conclude that hFAF1 might represent a new family of proteins with a distinct UBA domain.
hFAF1 was originally identified as a Fas-associated protein, and its amino acid sequence from 1 to 201 was characterized as the region binding to Fas death domain (35). In addition, hFAF1 was shown to interact with Hsp70 via amino acid residues 82 to 180 (18). We found that Flag-hFAF1(1-201) and Flag-hFAF1(1-345) bound to multiubiquitinated proteins, but these interactions were weaker than those of full Flag-hFAF1 and Flag-hFAF1(1-81) (Fig. 4). Also, amino acid residues 1 to 201 and 1 to 345 contain Fas- and Hsp70-interacting domains in addition to the UBA domain. It is possible that structural changes may explain why the truncated constructs show weaker interactions, but our results do suggest the possibility that hFAF1 interaction with multiubiquitinated proteins may be regulated by other binding proteins. Further work on the interactions among hFAF1, Fas, Hsp70, and ubiquitinated proteins is clearly required to rule in or out these and other alternate possibilities.
As part of our study of the involvement of hFAF1, interacting with multiubiquitinated proteins, in the degradation of proteasomal substrate, we found that overexpression of full hFAF1 and hFAF1(1-81) interfered with the degradation of artificial substrate, UbG76V-GFP, and endogenous substrate, IκBα. Constructs not containing the UBA domain did not inhibit the degradation of UbG76V-GFP and IκBα. Truncated hFAF1(1-81) and hFAF1(1-345) inhibited UbG76V-GFP degradation to a greater degree than full hFAF1 did (Fig. 9), although the binding of full hFAF1 to ubiquitinated proteins was similar to that of hFAF1(1-81) (Fig. 4). These results suggest that the C-terminal UBX domain is required to degrade the ubiquitinated proteins binding to the UBA domain.
Our studies using the method developed by Dantuma et al. and Lindsten et al. (8, 21) for the quantification of ubiquitin-proteasome-dependent proteolysis in living cells confirmed that the overexpression of hFAF1 causes inhibition of ubiquitin-proteasome-dependent proteolysis of endogenous as well as artificial substrate (Fig. 10).
Recent studies implicate ubiquitin and ubiquitination in many biological processes, including protein degradation, endocytosis, protein sorting, subnuclear trafficking, and gene expression, among others (32). Lys48-linked multiubiquitin chains are the principal proteasomal targeting signal, while Lys63-linked chains appear to be nonproteolytic signals (11, 32). Raasi and Pickart, using synthetic peptides, found that the C-terminal UBA domain of Rad23 binds to Lys48-linked polyubiquitin chains in preference to Lys63- or Lys29-linked chains (33). Peng et al. reported that Lys6, Lys11, Lys27, Lys33, Lys48, and Lys63 residues are involved in the ubiquitin chain formation, even though the relative abundance of other Lys-linked chains is less than that of Lys48- and Lys63-linked chains (31). Our studies showing that the N-terminal UBA domain of hFAF1 interacts with both Lys48- and Lys63-linked ubiquitin chains suggest that hFAF1 may have functions other than proteasomal degradation in the ubiquitination pathway. Studies designed to identify the ubiquitinated proteins interacting with hFAF1 are in progress in our laboratory. It is hoped that they will help to explain the cellular function of this interaction.
We found that the overexpression of hFAF1 itself causes cell death and that the UBA domain is responsible for it. Other studies also reveal a link between ubiquitination and apoptosis. Ubiquitinated proteins dramatically increase in several models of apoptosis, triggered variously by DNA-damaging agents, serum deprivation, and γ-irradiation (23, 28, 29). Blockers of protein degradation induce apoptosis in several types of cancer cells. In addition, the proteasome inhibitor PS-341 decreased the levels of several antiapoptotic proteins, such as Bcl-2 and FLIP, and up-regulated proteins implicated in proapoptotic cascades, such as Fas, Fas ligand, and caspase (26, 40).
In summary, our studies demonstrate that hFAF1, interacting with VCP, is involved in the ubiquitin pathway by recruiting multiubiquitinated proteins in the UBA domain. We hypothesize that the role of hFAF1 in this regard may be one of a scaffolding protein, regulating the stability of ubiquitinated proteins by interacting with ubiquitinated proteins and VCP via different domains.
Acknowledgments
We thank J. W. Seo and Y. H. Seo for the generation of ESI-q-TOF tandem MS results and N. P. Dantuma (Karolinska Institutet) for providing the UbG76V-GFP clone.
This work was supported by KOSEF through the Center for Cell Signaling Research at Ewha Women's University, by the 21C Frontier Functional Proteomics Center Project (MOST FPR03B3-04-110 and FPR02A7-32-110), and by the National R&D Program for Cancer Control (0420190-1). E.J. Song and S.-H. Yim were supported by the Brain Korea 21 Project.
REFERENCES
- 1.Bays, N. W., and R. Y. Hampton. 2002. Cdc48-ufd1-npl4: stuck in the middle with Ub. Curr. Biol. 12**:**R366-R371. [DOI] [PubMed] [Google Scholar]
- 2.Becker, K., P. Schneider, K. Hofmann, C. Mattmann, and J. Tschopp. 1997. Interaction of Fas(Apo-1/CD95) with proteins implicated in the ubiquitination pathway. FEBS Lett. 412**:**102-106. [DOI] [PubMed] [Google Scholar]
- 3.Buchberger, A., M. J. Howard, M. Proctor, and M. Bycroft. 2001. The UBX domain: a widespread ubiquitin-like module. J. Mol. Biol. 307**:**17-24. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L., and K. Madura. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22**:**4902-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, K., X. Niu, and L. T. A. Williams. 1995. Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc. Natl. Acad. Sci. USA 92**:**11894-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai, R. M., and C. C. H. Li. 2001. Valosin-containing protein is a multiubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3**:**740-744. [DOI] [PubMed] [Google Scholar]
- 7.Dai, R. M., E. Chen, D. L. Longo, C. M. Gorbe, and C. C. H. Li. 1998. Involvement of valosin-containing protein, an ATPase copurified with IκBα and 26S proteasome, in ubiquitin-proteasome mediated degradation of IκBα. J. Biol. Chem. 273**:**3562-3573. [DOI] [PubMed] [Google Scholar]
- 8.Dantuma, N. P., K. Lindsten, R. Glas, M. Jellen, and M. G. Masucci. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome dependent proteolysis in living cells. Nat. Biotechnol. 18**:**538-542. [DOI] [PubMed] [Google Scholar]
- 9.Doss-Pepe, E. W., E. W. Stenroos, W. G. Johnson, and K. Madura. 2003. Ataxin-3 interactions with Rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 23**:**6469-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsasser, S., D. Chandler-Militello, B. Muller, J. Hanna, and D. Finley. 2004. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279**:**26817-26822. [DOI] [PubMed] [Google Scholar]
- 11.Finley, D. 2001. Signal transduction. An alternative to destruction. Nature 412**:**285-286. [DOI] [PubMed] [Google Scholar]
- 12.Frohlich, T., W. Risau, and I. Flamme. 1998. Characterization of novel nuclear targeting and apoptosis-inducing domains in FAS associated factor 1. J. Cell Sci. 111**:**2353-2363. [DOI] [PubMed] [Google Scholar]
- 13.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99**:**745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghislain, M., R. Dohmen, F. Levy, and A. Varshavsky. 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15**:**4884-4899. [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67**:**425-430. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., and P. Bucher. 1996. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21**:**172-173. [PubMed] [Google Scholar]
- 17.Johnson, E. S., P. C. M. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270**:**17442-17456. [DOI] [PubMed] [Google Scholar]
- 18.Kim, H. J., E. J. Song, Y. S. Lee, E. Kim, and K. J. Lee. Human Fas-associated factor 1 interacts with heat shock protein 70 and negatively regulates chaperone activity. J. Biol. Chem., in press. [DOI] [PubMed]
- 19.Koegl, M., T. Hoppe, S. Schlenker, H. Ulrich, T. Mayer, and S. A. Jentsch. 1999. Novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96**:**635-644. [DOI] [PubMed] [Google Scholar]
- 20.Kondo, H., C. Rabouille, R. Newman, T. P. Levine, D. Pappin, P. Freemont, and G. Warren. 1997. p47 is a cofactor for p97-mediated membrane fusion. Nature 388**:**75-78. [DOI] [PubMed] [Google Scholar]
- 21.Lindsten, K., F. M. de Vrij, L. G. Verhoef, D. F. Fischer, F. W. van Leeuwen, E. M. Hol, M. G. Masucci, and N. P. Dantuma. 2002. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J. Cell Biol. 157**:**417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord, J. M., A. Ceriotti, and L. M. Roberts. 2002. ER dislocation: Cdc48p/p97 gets into the AAAct. Curr. Biol. 12**:**R182-R184. [DOI] [PubMed] [Google Scholar]
- 23.McBride, W. H., K. S. Iwamoto, R. Syljuasen, M. Pervan, and F. Pajonk. 2003. The role of the ubiquitin/proteasome system in cellular responses to radiation. Oncogene 22**:**5755-5773. [DOI] [PubMed] [Google Scholar]
- 24.Meyer, H. H., J. G. Shorter, J. Seemann, D. Pappin, and G. Warren. 2000. Complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19**:**2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, H. H., Y. Wang, and G. Warren. 2002. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21**:**5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsiades, N., C. S. Mitsiades, V. Poulaki, D. Chauhan, G. Fanourakis, X. Gu, C. Bailey, M. Joseph, T. A. Libermann, S. P. Treon, N. C. Munshi, P. G. Richardson, T. Hideshima, and K. C. Anderson. 2002. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA 99**:**14374-14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moir, D., S. Stewart, B. Osmond, and D. Bostein. 1982. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties and pseudoreversion studies. Genetics 100**:**547-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller, T. D., and J. Feigon. 2002. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J. Mol. Biol. 319**:**1243-1255. [DOI] [PubMed] [Google Scholar]
- 29.Orlowski, R. Z. 1999. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 6**:**303-313. [DOI] [PubMed] [Google Scholar]
- 30.Park, M. Y., H. D. Jang, S. Y. Lee, K. J. Lee, and E. Kim. 2004. Fas-associated factor-1 inhibits nuclear factor-κB (NF-κB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-κB. J. Biol. Chem. 279**:**2544-2549. [DOI] [PubMed] [Google Scholar]
- 31.Peng, J., D. Schwartz, J. E. Elias, C. C. Thoreen, D. Cheng, G. Marsischky, J. Roelofs, D. Finley, and S. P. Gygi. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 8**:**921-926. [DOI] [PubMed] [Google Scholar]
- 32.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8**:**499-504. [DOI] [PubMed] [Google Scholar]
- 33.Raasi, S., and C. M. Pickart. 2003. Rad23 UBA domains inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine48-linked polyubiquitin chains. J. Biol. Chem. 278**:**8951-8959. [DOI] [PubMed] [Google Scholar]
- 34.Rabouille, C., H. Kondo, R. Newman, N. Hui, P. Freemont, and G. Warren. 1998. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92**:**603-610. [DOI] [PubMed] [Google Scholar]
- 35.Ryu, S. W., S. K. Chae, K. J. Lee, and E. Kim. 1999. Identification and characterization of human Fas associated factor 1, hFAF1. Biochem. Biophys. Res. Commun. 262**:**388-394. [DOI] [PubMed] [Google Scholar]
- 36.Ryu, S. W., S. J. Lee, M. Y. Park, J. I. Jun, Y. K. Jung, and E. Kim. 2003. Fas-associated factor 1, FAF1, is a member of Fas death-inducing signaling complex. J. Biol. Chem. 278**:**24003-24010. [DOI] [PubMed] [Google Scholar]
- 37.Vadlamudi, R. K., I. Joung, J. L. Strominger, and J. Shin. 1996. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56 lck, belongs to a new class of ubiquitin-binding proteins. J. Biol. Chem. 271**:**20235-20237. [DOI] [PubMed] [Google Scholar]
- 38.Verma, R., R. Oania, J. Graumann, and R. J. Deshaies. 2004. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118**:**99-110. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson, C. R., M. Seeger, R. Hartmann-Petersen, M. Stone, M. Wallace, C. Semple, and C. Gordon. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3**:**939-943. [DOI] [PubMed] [Google Scholar]
- 40.Yang, Y., and X. Yu. 2003. Regulation of apoptosis: the ubiquitous way. FASEB J. 17**:**790-799. [DOI] [PubMed] [Google Scholar]
- 41.Yaron, A., A. Hatzubai, M. Davis, I. Lavon, S. Amit, A. M. Manning, J. S. Adhersen, M. Mann, F. Mercurio, and Y. Ben-Neriah. 1998. Identification of the receptor component if the IκBα-ubiquitin ligase. Nature 396**:**590-594. [DOI] [PubMed] [Google Scholar]
- 42.Ye, Y., H. H. Meyer, and T. A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414**:**652-656. [DOI] [PubMed] [Google Scholar]
- 43.Yuan, X., A. Shaw, X. Zhang, H. Kondo, J. Lally, P. S. Freemont, and S. Matthews. 2001. Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J. Mol. Biol. 311**:**255-263. [DOI] [PubMed] [Google Scholar]