Variations of Bacterial Populations in Human Feces Measured by Fluorescent In Situ Hybridization with Group-Specific 16S rRNA-Targeted Oligonucleotide Probes (original) (raw)
Abstract
Six 16S rRNA-targeted oligonucleotide probes were designed, validated, and used to quantify predominant groups of anaerobic bacteria in human fecal samples. A set of two probes was specific for species of the Bacteroides fragilis group and the species Bacteroides distasonis. Two others were designed to detect species of the Clostridium histolyticum and the Clostridium lituseburense groups. Another probe was designed for the genera Streptococcus and Lactococcus, and the final probe was designed for the species of the Clostridium coccoides-Eubacterium rectale group. The temperature of dissociation of each of the probes was determined. The specificities of the probes for a collection of target and reference organisms were tested by dot blot hybridization and fluorescent in situ hybridization (FISH). The new probes were used in initial FISH experiments to enumerate human fecal bacteria. The combination of the two _Bacteroides_-specific probes detected a mean of 5.4 × 1010 cells per g (dry weight) of feces; the Clostridium coccoides-Eubacterium rectale group-specific probe detected a mean of 7.2 × 1010 cells per g (dry weight) of feces. The Clostridium histolyticum, Clostridium lituseburense, and Streptococcus-Lactococcus group-specific probes detected only numbers of cells ranging from 1 × 107 to 7 × 108 per g (dry weight) of feces. Three of the newly designed probes and three additional probes were used in further FISH experiments to study the fecal flora composition of nine volunteers over a period of 8 months. The combination of probes was able to detect at least two-thirds of the fecal flora. The normal biological variations within the fecal populations of the volunteers were determined and indicated that these variations should be considered when evaluating the effects of agents modulating the flora.
The intestinal tract of humans contains a complex bacterial ecosystem, usually referred to as the normal flora, consisting mainly of obligatory anaerobic bacteria (6, 14). The composition of the normal flora plays an important role in human health and disease through its involvement in nutrition, pathogenesis, and immunology of the host. The normal flora provides colonization resistance and might stimulate immune responses to potentially pathogenic bacteria (7, 9, 16, 18, 31). To understand these phenomena and to be able to evaluate the effect of treatments aimed at modulating the normal flora, it is important to have accurate means to enumerate the various microbial populations. Conventional methods for determining the flora composition rely on the cultivation of bacteria on anaerobic selective media. However, many bacteria are difficult to culture or are unculturable (33, 34), and often media are not truly specific or are too selective for certain bacteria (28).
Molecular tools introduced in microbial ecology have made it possible to study the composition of intestinal flora in a culture-independent way based on the detection of rRNA (3). Detection of bacterial 16S rRNAs, or the corresponding genes, in the gastrointestinal tracts of humans and different mammals by using PCR amplification or dot blot hybridization with specific probes has been successful for tracking pathogenic or probiotic strains and studying the gut ecology (17, 19–21, 24, 32, 35). The biodiversity of the intestinal flora has been demonstrated by the analysis of a clone library with partial 16S rRNA genes amplified from DNA isolated from fecal samples (34). This allowed an oligonucleotide probe to be designed for the detection of an unknown group of gram-positive bacteria (Low G+C #2) related to members of the Clostridium leptum group that was difficult to culture but represented a significant proportion of the 16S rRNA extracted from fecal samples. Molecular analysis has also been used to show the diversity and dynamics of Bifidobacterium and Lactobacillus strains isolated from fecal samples (18, 27). A more direct method for the identification of single cells within complex ecosystems is fluorescent in situ hybridization (FISH) with specific 16S rRNA-based oligonucleotide probes (2, 3, 8, 10). This technique has been used to quantify bifidobacteria in the human gut (22) and for detection of salmonellae in the mouse gut (23). Depending on the probes chosen, FISH can be used to detect bacteria on different phylogenetic levels. Specific oligonucleotide probes and primers have been designed for many bacterial species which are known to be present in the intestinal tract (12, 13, 21, 29, 35). However, the complex microflora of the human gut is difficult to study with probes on a species level due to the diversity of this ecosystem. Therefore, it is more convenient to have probes specific for major genera and groups present in the gut, although the number of such probes is still limited (1). For the genus Bifidobacterium, four probes have been designed and extensively tested (17, 22). These bifidobacteria form a coherent monophyletic group for which specific probes are easily designed. Other probes detect more than one genus, such as the probe for Bacteroides which also detects Prevotella (26). For intertwined genera such as Eubacterium and Clostridium, genus-specific probes cannot be designed and probes for phylogenetic groups or clusters have to be considered.
Applying the FISH technique to human fecal samples to detect bifidobacteria has demonstrated that this group of bacteria could be enumerated at least as accurately by this technique as by conventional cultivation techniques (22). Here, an extension of this study is presented, and the design and validation of new probes for major groups of anaerobic bacteria are described. These probes were subsequently applied in conjunction with other group-specific probes to enumerate different bacterial populations in the feces of healthy volunteers. To gain insight into the normal biological fluctuations of these populations, their variations within and between the volunteers were determined.
MATERIALS AND METHODS
Organisms and culture conditions.
All reference strains used in this study are listed in Table 1. The strains were obtained from different sources, as indicated in the table (Deutsche Sammlung von Mikroorganismen und Zellkulturen [Braunschweig, Germany] [DSM], American Type Culture Collection [Rockville, Md.] [ATCC], Netherlands Institute for Dairy Research [Ede, The Netherlands] [NIZO], and Laboratory for Medical Microbiology [Groningen, The Netherlands] [MMB]). DSM or ATCC strains were cultured on the media described in the respective catalogs. All other strains were cultivated in anoxic peptone-yeast extract-glucose (PYG) medium (15) under anaerobic conditions at 37°C or in the case of facultative anaerobes on brain heart infusion agar (Oxoid, Basingstoke, United Kingdom). All MMB strains are clinical or human fecal isolates from local and regional public health laboratories that were identified by using normal routine procedures.
TABLE 1.
Organisms used in dot blot specificity studies and results of FISH specificity tests
| Straina | Origin | Dot blotb and FISHc results with indicated probe | |||||
|---|---|---|---|---|---|---|---|
| Bfra602 | Bdis656 | Erec482 | Chis150 | Clit135 | Strc493 | ||
| Bacteroides fragilis | DSM 2151 | +, P | −, N | −, N | −, N | −, N | −, N |
| Bacteroides vulgatus | DSM 1447 | +, P | −, N | −, N | −, N | −, N | −, N |
| Bacteroides eggerthii | DSM 20697 | +, P | −, N | −, N | −, N | −, N | −, N |
| Bacteroides sp. (B. fragilis) | MMB | +, P | −, N | − | − | − | − |
| Bacteroides sp. (B. thetaiotaomicron) | MMB | +, P | −, N | − | − | − | − |
| Bacteroides sp. (B. vulgatus) | MMB | +, P | −, N | − | − | − | − |
| Bacteroides sp. (B. eggerthii) | MMB | + | − | − | − | − | − |
| Bacteroides sp. (B. uniformis) | MMB | + | − | − | − | − | − |
| Cytophaga xylanolytica | DSM 6779 | +/−, P | −, N | −, N | −, N | −, N | −, N |
| Bacteroides distasonis | DSM 20701 | −, N | +, P | −, N | −, N | −, N | −, N |
| Bacteroides sp. (B. distasonis) | MMB | −, N | +, P | − | − | − | − |
| Clostridium oroticum | DSM 1287 | −, N | −, N | +, P | −, N | −, N | −, N |
| Clostridium sp. (C. nexile) | MMB | − | − | +, P | − | − | − |
| Ruminococcus hansenii | DSM 20583 | − | − | +, P | − | − | − |
| Ruminococcus productus | DSM 2950 | − | − | +, P | − | − | − |
| Eubacterium ventriosum | DSM 3988 | − | − | + | − | − | − |
| Clostridium sp. (C. clostridiiforme) | MMB | − | − | + | − | − | − |
| Clostridium histolyticum | DSM 2158 | −, N | −, N | −, N | +, P | −, N | −, N |
| Clostridium sp. (C. beijerinckii) | MMB | − | − | − | + | − | − |
| Clostridium sp. (C. butyricum) | MMB | − | − | − | +, P | −, N | − |
| Clostridium sp. (C. perfringens) | MMB | − | − | − | +, P | −, N | − |
| Clostridium putrificum | DSM 1734 | − | − | − | +, P | − | − |
| Clostridium sp. (C. cadaveris) | MMB | − | − | − | +, P/N | − | − |
| Clostridium difficile | ATCC 9688 | −, N | −, N | −, N | −, N | +, P/N | −, N |
| Eubacterium tenue | DSM 20695 | − | − | − | −, N | +, P | −, N |
| Clostridium bifermentans | NIZO B529 | − | − | − | −, N | +, P | −, N |
| Clostridium sp. (C. sordellii) | MMB | − | − | − | − | + | − |
| Peptostreptococcus (P. anaerobius) | MMB | − | − | − | −, N | −, N | − |
| Fusobacterium nucleatumd | DSM 20482 | −, N | −, N | −, N | −, N | −, N | −, N |
| Eubacterium plautii | DSM 4000 | −, N | −, N | −, N | −, N | −, N | −, N |
| Eubacterium sp. (E. cylindroides) | MMB | − | − | − | − | − | − |
| Streptococcus sanguis | DSM 20567 | −, N | −, N | −, N | −, N | −, N | +, P |
| Streptococcus oralis | DSM 20627 | −, N | −, N | −, N | −, N | −, N | +, P |
| Streptococcus intermedius | DSM 20573 | − | − | − | − | − | +, P |
| Lactococcus lactis subsp. cremoris | DSM 20069 | −, N | −, N | −, N | −, N | −, N | +, P |
| Streptococcus sp. (S. mitis) | MMB | −, N | −, N | −, N | −, N | −, N | +, P |
| Leuconostoc lactis | NIZO B630 | − | − | − | − | − | +, P |
| Streptococcus sp. (S. bovis) | MMB | − | − | − | − | − | +, P |
| Streptococcus sp. (S. equi subsp. equi) | MMB | − | − | − | − | − | + |
| Streptococcus mutans | DSM 20523 | − | − | − | − | − | + |
| Streptococcus sp. (S. sanguis) | MMB | − | − | − | − | − | + |
| Streptococcus sp. (S. salivarius) | MMB | − | − | − | − | − | + |
| Streptococcus sp. (S. equinus) | MMB | − | − | − | − | − | + |
| Streptococcus sp. (S. pyogenes) | MMB | − | − | − | − | − | + |
| Enterococcus faecalis | DSM 20478 | −, N | −, N | −, N | −, N | −, N | −, N |
| Enterococcus gallinarum | DSM 20628 | − | − | − | − | − | − |
| Lactobacillus acidophilus | NIZO B228 | − | − | − | − | − | −, N |
| Weissella kandleri | DSM 20593 | N | |||||
| Lactobacillus fermentum | DSM 20052 | − | − | − | − | − | − |
| Vagococcus fluvialis | DSM 5731 | −, N | −, N | −, N | −, N | −, N | −, N |
| Bifidobacterium infantis | ATCC 15697 | −, N | −, N | −, N | −, N | −, N | −, N |
| Bifidobacterium dentium | ATCC 27678 | − | − | − | − | − | − |
| Bifidobacterium sp. (B. longum) | MMB | −, N | −, N | − | − | − | − |
| Bifidobacterium adolescentis | NIZO B659 | − | − | − | − | − | − |
| Bifidobacterium pseudolongum | NIZO B669 | − | − | − | − | − | − |
| Escherichia coli | DSM 25922 | −, N | −, N | −, N | −, N | −, N | −, N |
| Carnobacterium divergens | DSM 20623 | −, N | −, N | −, N | −, N | −, N | −, N |
| Lactobacillus maltaromicus | DSM 20342 | − | − | − | − | − | − |
| Salmonella sp. (S. typhi) | MMB | − | − | − | − | − | − |
| Enterobacter sp. (E. aerogenes) | MMB | − | − | − | − | − | − |
| Serratia sp. (S. marcescens) | MMB | − | − | − | − | − | − |
| Proteus sp. (P. vulgaris) | MMB | − | − | − | − | − | − |
| Klebsiella sp. (K. pneumoniae) | MMB | − | − | − | − | − | − |
Design of oligonucleotide probes.
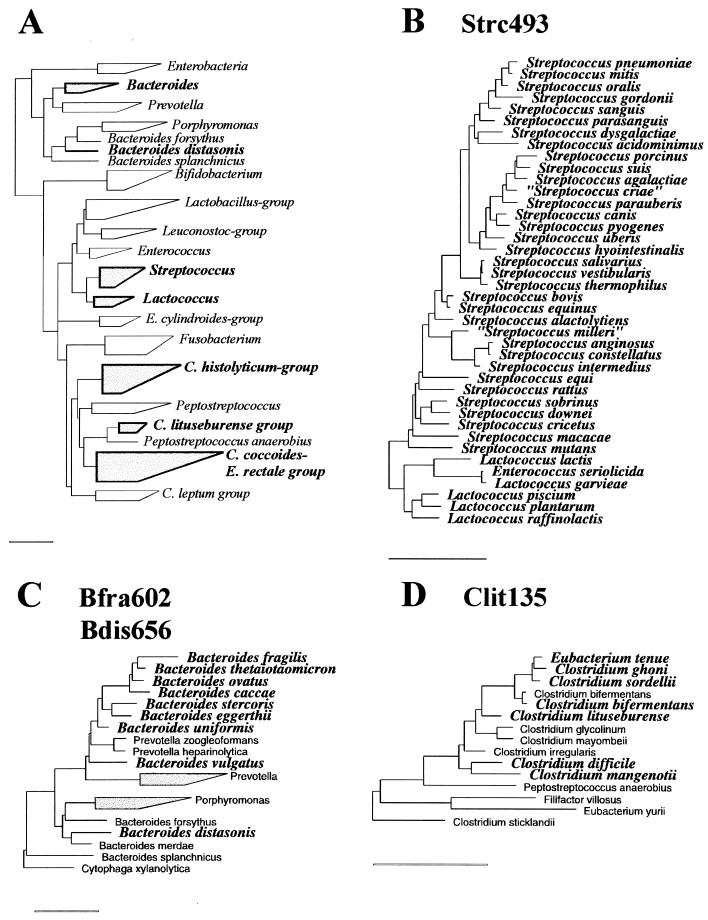
rRNA sequences of the target groups and reference organisms were obtained in an aligned form from the Ribosomal Database Project (RDP) (25). These aligned sequences were screened for group-specific target sequences which enabled discrimination between genera of anaerobic bacteria that are numerically important in the human intestinal flora. The selected oligonucleotide target sites (see Fig. 1) were tested for specificity against the 16S rRNA sequences available in the RDP by using the CHECK_PROBE analysis function of the RDP. Newly deposited sequences from GenBank were also screened for specificity. Phylogenetic trees, to illustrate the target groups and related species (see Fig. 2), were generated with the ARB program package (30) by extracting subtrees from a parsimony tree comprising around 5,000 sequences, which is provided by the authors.
FIG. 1.
Alignments of the probe sequences, their target sites, and sequences of corresponding sites in reference organisms. Probe names are in accordance with the Oligonucleotide Probe Database nomenclature (1) and are abbreviated in the text. Only those nucleotides that are different from the target sequences are shown. Y is a (C/T) wobbled nucleotide and R is an (A/G) wobbled nucleotide. Bold underlined nucleotides match the wobbled nucleotide of the probe. C., Clostridium; B., Butyrivibrio. The numerals following some species names refer to sequence accession numbers.
FIG. 2.
Phylogenetic trees illustrating the target groups of the new probes and related species. (A) Tree showing the relationship among the major genera, groups, and organisms known be present in the human gut. The heights of the boxes reflect the numbers of sequences used in the tree construction, and the horizontal lengths reflect the phylogenetic depths of the corresponding groups. Boxes and group names in boldface type indicate the groups that are represented in more detail in succeeding panels (B to F). The trees in panels B to F show the target organisms in bold italic typeface and related but nontarget organisms in normal typeface. (B) The complete genera Streptococcus and Lactococcus, all species of which are targets for the Strc493 probe. (C) The genus Bacteroides and related genera, with the target organisms for the Bfra602 and Bdis656 probes indicated. (D) Clostridium lituseburense group as a part of Clostridium cluster XI (classification according to reference 5), with the target organisms for the Clit135 probe indicated. (E) Clostridium histolyticum group (Clostridium clusters I and II), with the targets of the Chis150 probe indicated. (F) Clostridium coccoides-Eubacterium rectale group (Clostridium cluster CIV), with the targets for the Erec482 probe indicated. All bars represent 10% sequence divergence.
Probe specificity studies.
Unlabeled and fluorescein-labeled oligonucleotides were synthesized and purified by Eurogentec (Seraing, Belgium). Unlabeled oligonucleotides were labeled at their 5′ ends by using T4 polynucleotide kinase (Eurogentec) and [γ-32P]ATP (Amersham, Buckinghamshire, United Kingdom).
Pure cultures (10 ml) of the reference strains were centrifuged (5,000 × g for 10 min at room temperature), and the pelleted cells were washed with phosphate-buffered saline (PBS; 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4, each per liter). Aliquots were fixed for FISH (see next section), and the rest of the cells were harvested by centrifugation (5,000 × g for 10 min). The cell pellets were stored at −20°C until nucleic acid extraction was performed. For nucleic acid extraction, the pellets were resuspended in 400 μl of extraction buffer (10 mM Tris-HCl [pH 7.5] and 1 mM EDTA [TE], supplemented with 1% sodium dodecyl sulfate [SDS]). Cells of gram-positive strains were boiled for 2 min in a microwave oven at 800 W. TE-buffered phenol (200 μl) was added to the extraction buffer, and the mixture was vortexed vigorously. The mixture was centrifuged at 13,000 × g, and the nucleic acids were purified from the aqueous phase by phenol-chloroform extraction and harvested by ethanol precipitation as described earlier (11).
Reference nucleic acids (approximately 100 ng of rRNA) were denatured and applied to nylon membranes (Hybond N+; Amersham) by using a dot blot format unit as previously described (10). The membranes were prehybridized with hybridization buffer (0.5 M phosphate buffer, 7% SDS, 1% bovine serum albumin, 1 mM EDTA [pH 7.2]) for 30 min in screw-cap hybridization tubes. The membranes were hybridized overnight at 40°C in hybridization buffer containing 5 ng of labeled probe · ml−1 and washed with 1× SSC (0.15 M NaCl plus 0.015 sodium citrate [pH 7])–1% SDS buffer initially at 40°C.
Dissociation temperature (Td) values of the oligonucleotides were determined as described previously (36), with some modifications. Filters with reference nucleic acids which were hybridized with radiolabeled probes were washed in 1× SSC–1% SDS at 3°C temperature increments (from 35 to 70°C) for 15 min at each increment. Probe released from the membrane at each wash temperature was quantified by using a Packard Tri-Carb scintillation counter (Canberra Packard, Tilburg, The Netherlands). The Td was determined as the temperature at which one-half of the hybridized probe remained on the filter under the conditions described above. The specificities of the oligonucleotide probes were tested by using “phylogrids,” membranes blotted with nucleic acids from the strains listed in Table 1. The blots were hybridized with either a radiolabeled or a fluorescein-labeled oligonucleotide, washed at a final temperature of 50°C, and visualized by 24 h of exposure to enhanced chemiluminescence (ECL) Hyperfilm (Amersham) or, in the case of the fluorescein-labeled probe, by using an anti-fluorescein antibody–horseradish peroxidase conjugate (Amersham), an ECL luminescence detection system (Amersham), and 0.5 h of exposure to the same film.
FISH specificity tests.
Cells of the reference strains were washed once in PBS, diluted 1:3 with 4% (wt/vol) paraformaldehyde in PBS (PFA), fixed at 4°C for 16 h, and stored in 50% (vol/vol) ethanol-PBS at −20°C until further use (2). Cells (1 μl) were applied to precleaned slides, dried, and hybridized overnight at 50°C in a humid chamber as described previously (2). The hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.1% SDS [wt/vol]) contained 5 ng of fluorescently labeled probe · μl−1. If more stringent conditions were needed, formamide was added to the hybridization buffer in concentrations ranging from 0 to 60% (vol/vol). Cells from Streptococcus and Lactococcus spp. were incubated prior to hybridization with 10 μl of a 1-mg · ml−1 concentration of lysozyme in 100 mM Tris-HCl (pH 7.2) for 10 min at room temperature. The cells were then fixed for the second time with PFA for 10 min at 4°C. For all hybridizations with the probes S.*.Chis-0150-a-A-23 (Chis150) and S-*-Clit-0135-a-A-19 (Clit135), 10% (wt/vol) dextran sulfate was added to the buffer to increase the diffusion rate of the probe to the target. Cells were washed in all cases at 50°C for 30 min in wash buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2]).
Collection and preparation of fecal samples.
Nine healthy volunteers ranging from 24 to 52 years of age provided fresh stools twice a month for an 8-month period. The four women and five men were not restricted in their normal western European diet nor did they take any antibiotics or other drugs known to influence the fecal flora composition for at least 4 weeks prior to sampling. Whole stools were collected in sterile bags and kept for no longer than 12 h, at 4°C, before processing. The stools were kneaded mechanically for 5 min at 4°C to distribute the sample evenly. Portions (0.5 g) of each stool were suspended in 4.5 ml of filtered (0.2-μm-pore-size filter) PBS and vortexed with a dozen glass beads for at least 3 min to homogenize the sample. The suspension was centrifuged at 700 × g for 1 min to remove debris. The supernatant was fixed with PFA as described above. Dry weights were determined by lyophilizing a precisely weighed portion of each kneaded fecal sample.
Enumeration of bacteria in the fecal samples by FISH.
The fixed samples were processed and FISH analysis was performed as described previously (22), with some modifications. In addition to the newly designed oligonucleotide probes, the bacterial probe S-D-Bact-0338-a-A-18 (Bact338) (2), the bifidobacterial probe Bif164 (22), and Lowgc2P, the probe for the Low G+C #2 group (34), were used. To the Lowgc2P probe, one extra nucleotide (G) was added at the beginning of the sequence to increase the Td, resulting in the sequence 5′-GGGACGTTGTTTCTGAGT. Cells from 1.5 ml of fixed suspension were washed twice in filtered PBS, resuspended in 300 μl of 50% (vol/vol) ethanol-PBS, and stored at −20°C until required but not longer than 2 months. The PBS-ethanol stocks were diluted 1:24 with 50°C hybridization buffer. For hybridizations with the S-*-Strc-0493-a-A-19 probe (Strc493), cells were centrifuged for 5 min at 12,000 × g before dilution, and the pellet was treated with 150 μl of a 1-mg · ml−1 concentration of lysozyme in 100 mM Tris-HCl (pH 7.2) for 10 min at room temperature. After a second centrifugation step, the cells were resuspended in 50°C hybridization buffer to the same dilution as those of the other suspensions. The diluted cell suspensions (45-μl aliquots) were added to 5 μl of a 50-ng · μl−1 concentration of oligonucleotide probes and hybridized overnight at 50°C, except for the Lowgc2P probe, which was hybridized at 45°C. An aliquot of 5 to 50 μl of this hybridized cell suspension was washed at 50°C (45°C for Lowgc2P) for 30 min in 5 ml of wash solution (hybridization buffer without SDS) before being filtered onto 0.2-μm-pore-size GTTP polycarbonate filters (Millipore, Etten-Leur, The Netherlands). For total cell counts, 4′,6-diamidino-2-phenylindole (DAPI) was added to the wash solution to a final concentration of 100 ng · ml−1. Filters were mounted on clean microscope slides with Vectashield (Vector Laboratories, Burlingame, Calif.). Digital images of the slides, viewed with a Leica (Wetzlar, Germany) DMRXA epifluorescence microscope, were taken with a Kodak Megaplus 1.4 charge-coupled device camera. These images were analyzed and fluorescent cells were counted by using Quantimet HR600 image analysis software (Leica). Depending on the amount of fluorescent cells, 10 to 30 microscopic fields were counted.
RESULTS
Probe design.
To discriminate major groups of anaerobic gut bacteria, six different oligonucleotides were designed based on comparative analysis of all small-subunit rRNA sequences from the RDP and newly deposited sequences from GenBank. Alignments of all relevant 16S rRNA sequences were screened for signature sequences that enabled discrimination of the genera Bacteroides, Clostridium, Eubacterium, Streptococcus, and Lactococcus. Figure 1 shows the selected oligonucleotide sequences, their target sites, and alignments of the corresponding sites of target organisms and related reference organisms which have one or more mismatches with the target sites. Figure 2 shows the target groups of probes comprising all organisms possessing the target sequences and related species which have one or more mismatches. The first probe, S-*-Bfra-0602-a-A-19 (Bfra602), is specific for the Bacteroides fragilis group, and the second probe, S-*-Bdis-0656-a-A-18 (Bdis656), is specific for the species Bacteroides distasonis. These two groups comprise the major Bacteroides species known to be present in the human gut. The third probe, Clit135, is specific for part of the Clostridium lituseburense group (Clostridium cluster XI; according to the classification described before [5]), and the fourth, Chis150, is specific for most species of the Clostridium histolyticum group (Clostridium clusters I and II). The fifth probe, S-*-Erec-0482-a-A-19 (Erec482), is specific for most of the clostridia and eubacteria belonging to the Clostridium coccoides-Eubacterium rectale group (Clostridium clusters XIVa and XIVb). The last probe, Strc493, is specific for almost all streptococci and lactococci.
Specificity of the probes.
The selected oligonucleotide sequences were chosen in such a way that they possess a theoretical Td just over 50°C. The _Td_s were experimentally determined by using one target 16S rRNA and one rRNA with one or two mismatches for each probe. The determined values are listed in Fig. 1. Although the _Td_s were a few degrees higher than 50°C, for practical reasons, 50°C was required for optimal washing conditions during dot blot hybridizations. Therefore, the probes were checked for specificity by using a phylogrid dot blot containing the nucleic acids of the 64 target and nontarget organisms mentioned in Table 1. The identical phylogrids were hybridized with the six new probes and the bacterial probe Bact338 as a control. This control probe hybridized to the nucleic acids of all bacteria as expected (data not shown). All of the newly designed probes hybridized to their corresponding target organisms and did not show cross-hybridization with nontarget organisms, with a few exceptions (Table 1). Cytophaga xylanolytica hybridized under standard conditions with probe Bfra602, since its 16S rRNA has only one mismatch with this probe. Probe Chis150, which has two nucleotides at position 6 of the oligonucleotide (C/T wobbled sequence), is specific for both variants of the target sequences although it could not discriminate under our conditions between target sequences and the one mismatch of Clostridium cadaveris rRNA. Probe Strc493 did not detect two-mismatch target organisms, such as, for example, Enterococcus faecalis, but it did hybridize under our conditions with the one-mismatch organism Leuconostoc lactis.
FISH of reference strains.
FISH was performed on a selection of relevant strains from those listed in Table 1. All hybridizations were performed under standard conditions, that is, at 50°C with no formamide. Under these conditions, all probes hybridized with only their target organisms, with two exceptions. The Clostridium cadaveris strain gave hybridization signals with the Chis150 probe, despite the single mismatch at the end of the probe, and probe Bfra602 hybridized with Cytophaga xylanolytica, with which it has one mismatch in the middle of the sequence. This is all in agreement with the dot blot experiments. Discrimination between Cytophaga xylanolytica and Bacteroides species was attempted by adding increasing amounts of formamide to the hybridization buffer and thus increasing the stringency. With a concentration of 30% formamide, the hybridization signals at 50°C with Cytophaga xylanolytica became fainter, while Bacteroides fragilis signals remained the same. With 60% formamide, neither strain gave a signal. However, the differences in signals were never so large that they would enable easy discrimination in environmental samples. All streptococci, enterococci, and lactobacilli needed permeabilization treatment with lysozyme. Weissella kandleri, which has two mismatches with the Strc493 probe, was not included in the dot blot experiments, but in situ hybridizations showed that the probe did not hybridize with this strain under standard conditions. Hybridization of Clostridium histolyticum and Clostridium putrificum cells with the specific probes was established under standard conditions, but the signals increased after the addition of dextran sulfate to the hybridization buffer. Clostridium difficile hybridized only weakly to the Clit135 probe, probably due to poor growth of the culture and thus low rRNA content of the cells. Lysozyme treatment could not increase these poor hybridization signals.
Enumeration of fecal bacteria with the new probes.
To determine the usefulness of the validated probes, each probe was used to enumerate target bacteria in individual fecal samples provided by nine volunteers. The total hybridizable cell numbers obtained with the bacterial probe Bact338 were, on average for all volunteers, 2.6 × 1011 cells · g (dry weight) of feces−1. Counts of Bacteroides with a combination of the Bfra602 and Bdis656 probes were 5.4 × 1010 cells · g−1. Counts of the Clostridium coccoides-Eubacterium rectale group enumerated with the Erec482 probe were on average 7.2 × 1010 cells · g−1. Cells of the Clostridium histolyticum group counted with the Chis150 probe only occasionally reached the level at which quantification was reliable. The quantification level is defined here as one cell per microscopic field with a maximum of cells on the filter being about 108 cells · g−1. Clostridium histolyticum group numbers were estimated to be in the range of 0.1 × 108 to 7 × 108 cells · g−1, but reliable counts could not be made. Similar results were obtained for the counts of streptococci and lactococci with the Strc493 probe. Counts of the Clostridium lituseburense group were never high enough for a reliable quantification, although fluorescent cells were detected in the samples. Because of the low number of positive cells detected with the Chis150, Clit135, and Strc493 probes, these probes are not useful to enumerate these groups in fecal samples from healthy volunteers and thus were not included in the population dynamics study which is described below.
Methodological variations.
To study variations in the microbial populations between volunteers, and in individual volunteers over time, the variation due to the error in the FISH method needed to be determined. This variation (assay error) was quantified by processing a 0.5-g fecal sample from one volunteer as described above and preparing 10 separate PBS-ethanol stocks. Cells from these 10 stocks were analyzed by hybridization with the Bact338 probe and by DAPI staining. From this data set, coefficients of variation (i.e., the standard deviation divided by the mean, termed the CVassay values) were calculated. The magnitude of the CVassay values is determined only by fluctuations in the method itself and can therefore be used to describe the magnitude of the assay error. The CVassay for the DAPI staining technique was 0.20; that of the FISH with the Bact338 probe was 0.18.
Population dynamics of fecal bacteria in healthy volunteers.
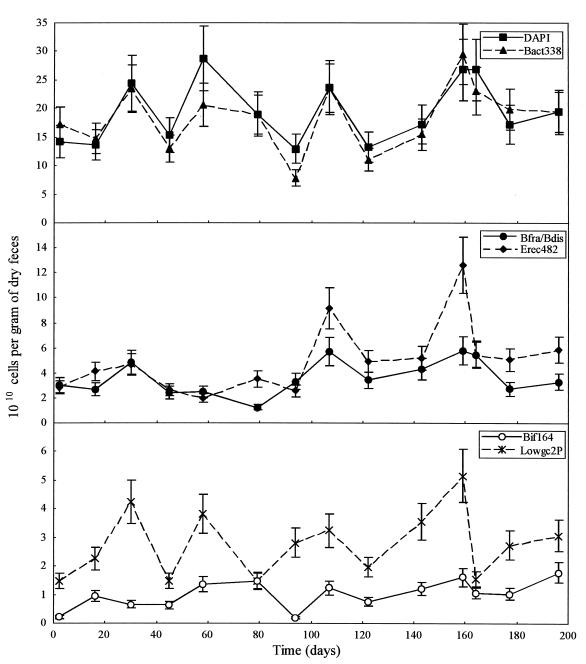
The total bacterial populations were enumerated for each of the nine volunteers by using FISH analysis with three of the newly designed probes (Bfra602, Bdis656, and Erec482). In addition, the bacterial probe Bact338 and two other specific probes, the Bifidobacterium probe Bif164 and the Lowgc2P probe for the Low G+C #2 group described elsewhere (22, 34), were included in the analysis to obtain better coverage of the composition. Total cell numbers in the fecal samples were determined by DAPI staining. Eight fecal samples from all but one of the volunteers were analyzed over the 8-month study period; 14 samples from the remaining volunteer (volunteer B) were measured. Table 2 shows the average number of bacteria in the fecal samples provided by the volunteers. The majority of bacteria in human feces were detectable by FISH analysis since 90 to 100% of the DAPI-stained cells hybridized to the Bact338 probe. The variation in the average cell numbers shown in Table 2, expressed as a standard deviation, is composed of the assay error and the variation due to normal biological fluctuations in the composition of the fecal microflora. This latter variation is referred to as CVlong and is discussed below. The changes in the fecal flora of the one individual studied in detail, volunteer B, during the 8-month study period are shown in Fig. 3. The longitudinal shifts in the bacterial groups can be measured significantly due to the previous calculation of the CVassay value (see previous section), which is represented in Fig. 3 by the error bars.
TABLE 2.
Mean counts of total bacteria and four major bacterial groups in fecal samples collected from nine healthy human volunteers over 8 months as determined by FISH
| Volunteer | Mean counts (SD)a | |||||
|---|---|---|---|---|---|---|
| Total bacteria (1011) | Bacteroides (1010) with Bfra-Bdis | C. coccoides-E. rectale (1010) with Erec482 | Low G+C #2 gram-positive bacteria (1010) with Lowgc2P | Bifidobacteria (109) with Bif164 | ||
| DAPI | Bact338 | |||||
| A | 2.9 (1.1) | 2.8 (0.9) | 4.8 (2.0) | 7.3 (2.6) | 2.5 (1.1) | 8.0 (7.5) |
| B | 2.0 (0.6) | 1.8 (0.6) | 3.6 (1.4) | 5.1 (2.8) | 2.8 (1.2) | 10.1 (4.8) |
| C | 2.5 (0.6) | 2.4 (0.8) | 3.2 (1.8) | 5.9 (3.4) | 2.1 (1.5) | 4.0 (2.1) |
| D | 3.5 (1.3) | 3.3 (1.1) | 3.1 (1.0) | 12.3 (3.4) | 4.2 (1.1) | 5.2 (3.5) |
| E | 2.7 (0.7) | 2.8 (0.9) | 6.6 (3.4) | 6.3 (1.8) | 3.5 (1.4) | 14.9 (8.1) |
| F | 2.3 (0.6) | 2.4 (0.9) | 8.2 (4.2) | 8.1 (3.3) | 3.9 (1.4) | 7.9 (3.7) |
| G | 3.8 (0.7) | 3.8 (0.7) | 11.6 (6.0) | 11.4 (5.0) | 3.1 (1.9) | 6.9 (6.0) |
| H | 2.6 (1.9) | 2.7 (2.2) | 6.5 (6.0) | 7.3 (9.9) | 1.7 (2.1) | 13.7 (7.2) |
| I | 3.4 (1.1) | 3.5 (1.2) | 5.6 (2.3) | 11.1 (3.5) | 6.2 (2.3) | 13.8 (10.3) |
FIG. 3.
Microbial composition of fecal samples of volunteer B. (Top panel) Total cells enumerated by DAPI staining and total bacteria counted by FISH with the Bact338 probe; (middle panel) Bacteroides cells enumerated with the Bfra602-Bdis656 probe combination and the Clostridium coccoides-Eubacterium rectale group cells enumerated with the Erec482 probe; (bottom panel) bifidobacteria and the Low G+C #2 group enumerated with the Bif164 probe and the Lowgc2P probe, respectively.
Table 3 lists the mean number of bacteria enumerated per gram (dry weight) of feces and the variations in bacterial populations from all the fecal samples analyzed. On average, the Bfra602-Bdis656 probe combination detected 5.7 × 1010 cells · g−1, or 20% of the total population, and Erec482 detected 29% of the total population (8.1 × 1010 cells · g−1); together, they accounted for almost half of the flora. The Lowgc2P probe detected 12% (3.3 × 1010 cells · g−1), whereas Bif164 detected only 3% (9.4 × 109 cells · g−1) of the population. The variation in the 78 samples from the nine different volunteers is represented by CVtotal. This variation includes the normal biological differences in bacterial numbers between the volunteers (CVinter), the changes in bacterial numbers within individual volunteers over time (CVlong), and the error in the assay (CVassay). The CVtotal determined with DAPI staining is similar to that determined by FISH with the Bact338 probe (0.40 compared to 0.43). The variations in the populations of the Clostridium coccoides-Eubacterium rectale and the Low G+C #2 gram-positive groups (0.60 and 0.61, respectively) are less changeable than those of the Bacteroides fragilis subgroup (0.73) and the bifidobacteria (0.74). The longitudinal variations (CVlong) and the interindividual variations (CVinter) were determined for each bacterial group (Table 3). These values have been corrected for the assay error and thus show to what extent the bacterial groups fluctuate over time and between the individual volunteers. The bifidobacteria show the greatest variation of all the groups examined, particularly in the case of longitudinal variation (CVlong).
TABLE 3.
Variations in fecal flora populations within each volunteer (CVlong) and between volunteers (CVinter)
| Population and stain or probe | Counta | CVtotala | CVlongb | CVinterc | |
|---|---|---|---|---|---|
| Mean | SD | ||||
| Total bacteria | |||||
| DAPI | 2.8 × 1011 | 1.1 × 1011 | 0.40 | 0.16 | 0.30 |
| Bact338 | 2.8 × 1011 | 1.2 × 1011 | 0.43 | 0.27 | 0.33 |
| Bacteroides/Bfra602-Bdis656 | 5.7 × 1010 | 4.2 × 1010 | 0.73 | 0.45 | 0.62 |
| C. coccoides-E. rectale/Erec482 | 8.1 × 1010 | 4.8 × 1010 | 0.60 | 0.39 | 0.50 |
| Low G+C #2 gram-positive bacteria/Lowgc2P | 3.3 × 1010 | 2.0 × 1010 | 0.61 | 0.44 | 0.56 |
| Bifidobacteria/Bif164 | 9.4 × 109 | 6.9 × 109 | 0.74 | 0.59 | 0.63 |
DISCUSSION
Six different probes were designed and used to quantify bacteria in human fecal samples. Used in combination, the Bfra602 and Bdis656 probes detected 20% of the fecal flora, identifying them as Bacteroides. The Clostridium coccoides-Eubacterium rectale group counted with the Erec482 probe constituted 29% of the flora. This makes these two groups of bacteria major constituents of the fecal flora, which is consistent with current knowledge (14). The other three probes for streptococci and lactococci, the Clostridium lituseburense group, and Clostridium histolyticum were responsible for detecting less than 1% of the normal flora and therefore were not useful in enumerating these groups in the feces of healthy people by FISH. However, these probes are useful to describe the full biodiversity of the gut flora by demonstrating the low numbers of these groups. Moreover, notorious pathogens known to become more dominant during gut disorders or disturbance by antibiotic treatment (4) are recognized by these probes. Thus, the probes will be important in investigations involving floras different from those of healthy adults, such as people suffering from gut disorders and newborn infants. The specificity of the new probes was demonstrated with only a few exceptions. Probe permeation of the gram-positive cell wall will remain problematic in FISH experiments, although permeabilization protocols were optimized for the gram-positive target groups. The probes are designed on the basis of currently available 16S rRNA sequences (25), and the specificity is not guaranteed for unknown gut bacteria. It is an advantage, then, that these probes are designed on a phylogenetic basis and that related species are more likely to have the same target sequences.
In the population dynamics study, it has been possible to detect about two-thirds of the fecal microflora of healthy volunteers. The Bacteroides and Clostridium coccoides-Eubacterium rectale bacteria constituted half of the fecal flora. The proportion of bifidobacteria enumerated (3%) was comparable to the counts previously obtained (22). The proportion of bacteria detected by the Lowgc2P probe (12%) was in accordance with the 1 to 10% proportion of total human fecal 16S rRNA detected by this probe in a previous study (34). Our future aim is to have a set of about 10 probes that can detect more than 90% of the flora, dividing it into large groups. After analyzing these large groups, refinements on a species level can be made to study the groups of interest. For detecting less abundant organisms and for discrimination on a species level, alternative molecular methods such as PCR will be required. Although the set of probes described here detects two-thirds of the normal flora, a proportion remains undetected. Probes need to be designed for groups like peptostreptococci, fusobacteria, enterobacteria, and other groups of microorganisms, known or unknown.
The measurements made in this study reveal that the intestinal ecosystem is undergoing fluctuations in compositions detectable on a linear scale, unlike culturing data, which are logarithmically distributed. The CVinter and CVlong values (Table 3) show that relative differences in flora composition, between volunteers and over time, can be assessed objectively. This illustrates the usefulness of FISH with 16S rRNA-targeted probes to monitor bacterial populations, particularly when those groups are difficult to culture (e.g., Bacteroides and the Low G+C #2 group). Within each volunteer, bifidobacteria were the most variable population. This is interesting in that this is the group of bacteria that is being investigated due to its possible health-promoting activities. The dynamics of the bifidobacteria highlight the importance of understanding the composition of, and the changes in, the normal microflora before conclusions can be drawn about factors affecting intestinal microbial populations.
Applying FISH with the described probes has provided new information on the composition of normal human fecal flora and more accurately describes natural variations in bacterial populations. This research, and future molecular analysis, will help to refine current knowledge about the composition and dynamics of the flora and its changes in response to modulation.
ACKNOWLEDGMENTS
We thank R. H. J. Tonk for his assistance with computer systems and image analysis and A. C. M. Wildeboer-Veloo for assistance with the cultivation of the strains.
This work was supported by grant 901-14-167 to G.W.W. from the Netherlands Organization for Scientific Research (NWO) and the European Research Project Fair-CT-97-3035.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:299–306. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chrisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borriello, S. P. 1995. Clostridial disease of the gut. Clin. Infect. Dis. 20(Suppl. 2):S242–S250. [DOI] [PubMed]
- 5.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 6.Finegold S M, Sutter V L, Mathisen G E. Normal indigenous intestinal flora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press; 1983. pp. 3–31. [Google Scholar]
- 7.Gibson G R, Roberfroid M B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 8.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldin B R, Gorbach S L. Probiotics for humans. In: Fuller R, editor. Probiotics: the scientific basis. London, United Kingdom: Chapman & Hall; 1992. pp. 355–376. [Google Scholar]
- 10.Harmsen H J M, Prieur D, Jeanthon C. Group-specific 16S rRNA-targeted oligonucleotide probes to identify thermophilic bacteria in marine hydrothermal vents. Appl Environ Microbiol. 1997;63:4061–4068. doi: 10.1128/aem.63.10.4061-4068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen H J M, Kengen H M P, Akkermans A D L, Stams A J M. Phylogenetic analysis of two syntrophic propionate-oxidizing bacteria in enrichment cultures. Syst Appl Microbiol. 1995;18:67–73. [Google Scholar]
- 12.Hensiek R, Krupp G, Stackebrandt E. Development of diagnostic oligonucleotide probes for four Lactobacillus species occurring in the intestinal tract. Syst Appl Microbiol. 1992;15:123–128. [Google Scholar]
- 13.Hertel C, Ludwig W, Pot B, Kersters K, Schleifer K H. Differentiation of lactobacilli occurring in fermented milk products by using oligonucleotide probes and electrophoretic protein profiles. Syst Appl Microbiol. 1993;16:463–467. [Google Scholar]
- 14.Hill M J. The normal gut bacterial flora. In: Hill M J, editor. Role of gut bacteria in human toxicology and pharmacology. London, United Kingdom: Taylor and Francis Ltd.; 1995. pp. 3–17. [Google Scholar]
- 15.Holdeman L V, Cato E P, Moore W E C. Anaerobe laboratory manual. 4th ed. Blacksburg: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 16.Jansen G, Deddens B, Wilkinson M, van der Waaij D. The influence of Enterococcus faecalis on the morphology and the antibody-binding capacity of the intestinal bacteria of then healthy human volunteers. Infection. 1995;23:46–50. doi: 10.1007/BF01710058. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura K, McCartney A L, McConnell M A, Tannock G W. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl Environ Microbiol. 1997;63:3394–3398. doi: 10.1128/aem.63.9.3394-3398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klijn N, Weerkamp A H, de Vos W M. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl Environ Microbiol. 1995;61:2771–2774. doi: 10.1128/aem.61.7.2771-2774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kok R G, de Waal A, Schut F, Welling G W, Weenk G, Hellingwerf K J. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol. 1996;62:3668–3672. doi: 10.1128/aem.62.10.3668-3672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreader C A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol. 1995;61:1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licht T R, Krogfelt K A, Cohen P S, Poulsen L K, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, Raskin L, Stahl D A. Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol Ecol. 1997;22:281–294. [Google Scholar]
- 25.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 27.McCartney A L, Wenzhi W, Tannock G W. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl Environ Microbiol. 1996;62:4608–4613. doi: 10.1128/aem.62.12.4608-4613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson G M, George S E. Comparison of media for selection and enumeration of mouse fecal flora populations. J Microbiol Methods. 1995;22:293–300. [Google Scholar]
- 29.Schleifer K-H, Ehrmann M, Beimfohr C, Brockmann E, Ludwig W, Amann R. Application of molecular methods for the classification and identification of lactic acid bacteria. Int Dairy J. 1995;5:1081–1094. [Google Scholar]
- 30.Strunk O, Ludwig W. ARB, a software environment for sequence data. Munich, Germany: Technische Universität München; 1996. [Google Scholar]
- 31.Vollaard E J, Clasener H A L. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Cao W, Cerniglia C E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 34.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto T, Morotomi M, Tanaka R. Species-specific oligonucleotide probe for five Bifidobacterium species detected in human intestinal microflora. Appl Environ Microbiol. 1992;58:4076–4079. doi: 10.1128/aem.58.12.4076-4079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]