Posttranscriptional Downregulation of c-IAP2 by the Ubiquitin Protein Ligase c-IAP1 In Vivo (original) (raw)
Abstract
Inhibitor of apoptosis proteins (IAPs) c-IAP1 and c-IAP2 were identified as part of the tumor necrosis factor receptor 2 (TNFR2) signaling complex and have been implicated as intermediaries in tumor necrosis factor alpha signaling. Like all RING domain-containing IAPs, c-IAP1 and c-IAP2 have ubiquitin protein ligase (E3) activity. To explore the function of c-IAP1 in a physiologic setting, c-IAP1-deficient mice were generated by homologous gene recombination. These animals are viable and have no obvious sensitization to proapoptotic stimuli. Cells from c-IAP1−/− mice do, however, express markedly elevated levels of c-IAP2 protein in the absence of increased c-IAP2 mRNA. In contrast to reports implicating c-IAPs in the activation of NF-κB, resting and cytokine-induced NF-κB activation was not impaired in c-IAP1-deficient cells. Transient transfection studies with wild-type and E3-defective c-IAP1 revealed that c-IAP2 is a direct target for c-IAP1-mediated ubiquitination and subsequent degradation, which are potentiated by the adaptor function of TRAF2. Thus, the c-IAPs represent a pair of TNFR-associated ubiquitin protein ligases in which one regulates the expression of the other by a posttranscriptional and E3-dependent mechanism.
Inhibitor of apoptosis proteins (IAPs) were identified in baculovirus because of their ability to prevent the death of infected cells (9). IAPs are defined by the presence of one or more (baculovirus IAP repeat) BIR domains, which in the context of flanking sequences directly bind to and inhibit caspases (reviewed in references 18 and 34). Eight mammalian IAPs have been identified (34), five of which contain a carboxyl-terminal RING domain that confers ubiquitin protein ligase (E3) activity (3, 22, 46). The highly homologous c-IAP1 and c-IAP2 are two such RING-containing proteins that were identified biochemically as part of the tumor necrosis factor receptor 2 (TNFR2) signaling complex (21, 30, 43). c-IAP1 and c-IAP2 do not bind directly to TNFR2 but rather bind the signaling molecules tumor necrosis factor receptor-associated factor 1 (TRAF1) and TRAF2, the latter of which binds the cytoplasmic tail of TNFR2. The association of these molecules with TRAFs depends on the IAP N-terminal BIR-containing domain and the TRAF C-terminal region (30).
The antiapoptotic activity of c-IAP1 and c-IAP2 has been attributed to their abilities to bind and inhibit caspase 3 and 7 (15, 32), to ubiquitinate the death-inducing protein Smac/Diablo (14), and to activate NF-κB (5, 41). Overexpression of c-IAP2 has been shown to inhibit apoptosis in a variety of cell types, including rat hepatocytes (5, 36). c-IAP1 and c-IAP2 have also been identified as possible oncogenes (10, 16), suggesting that their antiapoptotic activity may be involved in some cases of tumor progression. In contrast, cleaved products of c-IAP1 can induce apoptosis, suggesting that c-IAP1 can be proapoptotic at times (6). Consistent with this, we have found that c-IAP1 ubiquitinates TRAF2 in a TNFR2-dependent manner and sensitizes cells to tumor necrosis factor alpha (TNF-α)-induced cell death (19).
All of the work to date on the role of c-IAPs in various aspects of cell survival and signal transduction has been performed by overexpression in cell lines or analysis in cell-free systems. To determine how c-IAP1 functions in a physiologic setting, we generated and analyzed mice that lack c-IAP1. These animals are viable and fertile, allowing us to characterize the consequences of c-IAP1 loss at the molecular and cellular level. Notably, c-IAP1-deficient mice had markedly elevated levels of c-IAP2 protein despite normal levels of c-IAP2 mRNA. The results show that c-IAP1 ubiquitinates c-IAP2 in a multimeric complex with adaptor TRAF proteins, ultimately leading to c-IAP2 degradation.
MATERIALS AND METHODS
Reagents.
The primers used to generate a cDNA probe of the c-IAP1 genomic sequence outside of the homologous recombination arm were as follows: 5′-GTTTTAAAACCAGCTTGGGTTATATTG-3′ and 5′-GTTCCTCACCCAACCAGTCTACTTAG-3′. Mouse c-IAP1 and c-IAP2 expression vectors and anti-c-IAP antiserum (rabbit anti-rat c-IAP2, which cross-reacts on mouse c-IAP1 and c-IAP2) (13) were provided by Peter Liston (University of Ottawa, Ottawa, Ontario). c-IAP1mut and c-IAP2mut were made by substituting an alanine for the Zn2+-coordinating histidine residues in the RING domains of c-IAP1 (amino acid residue 582) and c-IAP2 (amino acid residue 570) by the use of a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The c-IAP1 and c-IAP2 mutagenic primers were as follows: c-IAP1mut sense (5′-CGTGTTCATTCCCTGTGGCGCTCTGGTCGTGTGCAAAG-3′); c-IAP1mut antisense (5′-CTTTGCACACGACCAGAGCGCCACAGGGAATGAACACG-3′); c-IAP2mut sense (5′-GTGTTCATTCCCTGTGGCGCTCTGGTCGTGTGCAAAG-3′); and c-IAP2mut antisense (5′-CTTTGCACACGACCAGAGCGCCACAGGGAATGAACAC-3′). Site-directed mutagenesis was confirmed by direct sequencing. Flag-c-IAP2 and Flag-c-IAP2mut were generated by subcloning into the pCMV-Tag2B vector (Stratagene). Glutathione _S_-transferase (GST)-c-IAP1 was generated by subcloning into the pGEX-6P vector (Amersham BiSciences, Piscataway, N.J.). The reading frames were confirmed by direct sequencing. The expression vector for the hemagglutinin (HA)-tagged TRAF2 mutant lacking the RING domain, amino acid residues 87 to 501 (HA-ΔTRAF2), was provided by Chuanjin Wu (National Cancer Institute, Bethesda, Md.). His-tagged TRAF1 (amino acids 189 to 416) and TRAF2 (272-502) were generated by PCR. The primers used for creating these truncation mutants were as follows: for TRAF1 (189-416), 5′-GCGCGGGATCCGAGCTGGAGGGGAAGCTG-3′ and 5′-GCGCGGTCGACCTAAGTGCTGGTCTCCAC-3′; and for TRAF2 (272-502), 5′-GCGCGGGATCCTGCGAGAGCCTGGAGAAG-3′ and 5′-GCGCGAAGCTTTTAGAGCCCTGTCAGGTC-3′. The PCR products were subcloned into pET28a (Novagen, EMD Biosciences, Madison, Wis.). The reading frames and sequences were confirmed by direct sequencing. c-IAP1 protein expression in eukaryotic cells was detected using an anti-c-IAP1 polyclonal antibody (R & D Systems), whereas c-IAP1 and c-IAP2 were detected simultaneously using the anti-c-IAP antiserum. The following antibodies were used: anti-XIAP polyclonal antibody (R & D Systems), anti-Flag monoclonal antibody (Sigma), anti-HA polyclonal antibody (Santa Cruz), anti-His polyclonal antibody (Santa Cruz), anti-ubiquitin polyclonal antibody (Santa Cruz), anti-β-actin monoclonal antibody (Sigma). Cytochrome-conjugated anti-CD4, allophycocyanin-conjugated anti-CD8 (BD Biosciences, San Diego, Calif.), fluorescein isothiocyanate-conjugated anti-TCRβ (H57), and phycoerythrin-conjugated anti-CD45R (B220) monoclonal antibodies were purchased from BD Pharmingen. Secondary anti-rabbit and anti-mouse horseradish peroxidase-conjugated polyclonal antibodies were purchased from Amersham Biosciences. The probes and primers used for real-time PCR of c-IAP2 were as follows: probe, 5′-TTCCCACAGATGACATTGCAGCTCTACCA-3′; c-IAP2 forward primer, 5′-TATTTGTGCAACAGGACATTAGGAGT-3′; and c-IAP2 reverse primer, 5′-TCTTTCCTCCTGGAGTTTCCA-3′. The hypoxanthine phosphoribosyltransferase probe and primers have been previously described (24). The oligonucleotides used in the electrophoretic mobility shift assays (EMSAs) were as follows: NF-κB (5′-GATCAGAGGGGACTTTCCGAG-3′) (11) and AP-1 (8). Mice expressing an NF-κB-regulated luciferase reporter driven by the minimal fos promoter (23) were provided by Mercedes Rincón (University of Vermont, Burlington). XIAP−/− mice were obtained from Craig Thompson (University of Pennsylvania, Philadelphia) and have been backcrossed six times to the C57BL/6 background. TNF-α and interleukin-1 alpha (IL-1α) were purchased from R & D systems. The OPTEIA IL-6 enzyme-linked immunosorbent assay (ELISA) kit was purchased from BD Biosciences.
Gene targeting and generation of c-IAP1−/− mice.
The murine c-IAP1 gene was disrupted in 129 E14 embryonic stem (ES) cells by the use of a targeting vector that was designed to delete the translation initiation codon and first (BIR) domain. Heterozygous ES cell lines containing the mutated c-IAP1 allele were injected into C57BL/6 blastocysts, and male chimeras with germ line transmission were used to generate c-IAP1+/− animals. The mice were backcrossed onto the C57BL/6 background for eight generations and then interbred to produce mice homozygous for the mutated allele. Genotypes were confirmed by Southern blot and immunoblot analysis. Mice used in NF-κB luciferase experiments were backcrossed with NF-κB luciferase reporter transgenic mice on the C57BL/6 background and then interbred to produce c-IAP1−/− × NF-κB luciferase reporter transgenic mice. All animal experimental procedures used in this study were approved by the Animal Care and Use Committee of the National Cancer Institute.
Cell preparation, surface staining, and transient transfection.
Thymus, spleen, and lymph nodes were harvested from wild-type (WT) and c-IAP1−/− mice, and total cell suspensions from each organ were made by disruption of the organ and filtering through nylon mesh (Small Parts Inc., Miami Lakes, Fla.). The distribution of thymocyte and peripheral lymphoid cell populations was determined by cell surface staining and flow cytometry. Murine embryonic fibroblasts (MEFs) were prepared from day 12.5 embryos (1) and were maintained in Alpha modified Eagle's medium supplemented with 15% fetal calf serum purchased from Biofluids, 2 mM l-glutamine (Biofluids), and 50 μM β-mercaptoethanol (Sigma). For transient transfection studies, 293 cells were transfected using Lipofectamine 2000 following the protocol of the manufacturer (Invitrogen). To control for transfection efficiency, cells were transfected with the indicated cDNAs and pEGFP-N1 (Clontech), a plasmid containing green fluorescence protein (GFP) cDNA under the control of the cytomegalovirus promoter (1:10 ratio with experimental plasmid). The percentage of GFP-positive cells was 40 to 60%, and the mean fluorescence intensity of the GFP signal differed by less than twofold between samples. In some experiments, 17 h after transfection 293 cells were incubated with lactacystin (Calbiochem) (25 μM) for an additional 7 h before being lysed.
Southern blotting.
Genomic DNA was isolated from thymocytes harvested from WT, c-IAP1+/−, and c-IAP1−/− littermates by the use of a DNeasy tissue kit (QIAGEN, Valencia, Calif.) and digested with EcoRI and BamHI. The DNA was resolved using a 0.8% agarose gel and denatured for 30 min at 23°C using denaturing solution (Biosource International, Camarillo, Calif.). The denaturing conditions were neutralized for 30 min at 23°C with neutralizing solution (Biosource International), and the DNA was transferred to a Hybond N+ nitrocellulose membrane (Amersham Biosciences) with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and UV-cross-linked to the membrane. The membrane was prehybridized with Hybrisol (Intergen, Purchase, N.Y.) for 1 h at 42°C and then hybridized with the c-IAP1 genomic cDNA probe in Hybrisol for 16 h at 42°C. The c-IAP1 genomic cDNA probe was labeled with [α-32P]dCTP by the use of a random primer kit (Stratagene). The membrane was washed in 3× SSC-0.1% sodium dodecyl sulfate (SDS) for 30 min at 23°C and then twice in 1× SSC-0.1% SDS for 1 h at 50°C. Radioactive DNA duplexes were visualized with a Storm PhosphorImager (Molecular Dynamics).
Immunoprecipitation and immunoblotting.
Total cell lysates from MEF, splenocytes, and thymocytes were prepared using radioimmunoprecipitation buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing 0.5 mM aminoethylbenzene sulfonyl fluoride (AEBSF), leupeptin (20 μg/ml), and aprotinin (20 μg/ml) on ice. For transient transfection studies, transfected 293 cells were harvested and lysed in sample buffer (50 mM Tris [pH 6.8], 10% glycerol, 2% SDS, 2% β-mercaptoethanol, and 0.04% bromophenol blue). For immunoprecipitation, cells were lysed in radioimmunoprecipitation buffer containing 0.5 mM AEBSF, leupeptin (20 μg/ml), and aprotinin (20 μg/ml) on ice and immunoprecipitation was performed with the indicated antibodies. Immunoprecipitated material or cell lysates were denatured at 100°C in sample buffer, resolved by SDS-10% polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose membranes. Membranes were blocked in blocking buffer (150 mM NaCl, 200 mM Tris [pH 8.0], 0.1% Tween, 5% milk), blotted with the appropriate primary antibody in blocking buffer, washed extensively with TBS-T (150 mM NaCl, 200 mM Tris [pH 8.0], 0.1% Tween), blotted with the appropriate secondary horseradish peroxidase-conjugated polyclonal antibody in blocking buffer, and then washed extensively in TBS-T. Immune complexes were detected by enhanced chemiluminescence (Pierce). For stripping, immunoblots were incubated in stripping buffer (2% SDS, 62.5 mM Tris [pH 6.7], 0.75% β-mercaptoethanol) at 55°C for 30 min., washed in TBS-T, blocked, and reimmunoblotted with the appropriate antibodies.
In vitro protein binding.
GST and GST-c-IAP1 were expressed in DH5α cells (Invitrogen). pET28, His-TRAF1 (189-416), and His-TRAF2 (272-502) were expressed in BL21(DE3) cells (Invitrogen). Protein expression was induced in log phase cells with 0.05 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 25°C for 4 h. Bacteria expressing GST and GST-c-IAP1 were centrifuged and resuspended in sonication buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100). Bacteria expressing pET28a, His-TRAF1 (189-416), and His-TRAF2 (272-502) were centrifuged and resuspended in GST binding buffer (120 mM NaCl, 10% glycerol, 1% Triton X-100, 50 mM Tris, pH 7.5). All cells were sonicated on ice and centrifuged in a Sorvall SS-34 rotor at 20,000 × g for 10 min, and the supernatants were collected. Recombinant GST proteins were purified using glutathione Sepharose 4B beads (Amersham Biosciences), and expression was confirmed by SDS-PAGE and Coomassie blue staining. Expression of TRAF proteins in the cell lysates was confirmed by SDS-PAGE and Coomassie blue staining. A total of 5 μl of GST- or GST-c-IAP1-coated beads was incubated with pET28a, His-TRAF1 (189-416), or His-TRAF2 (272-502) bacterial cell lysate for 16 h at 4°C. The beads were washed three times in GST binding buffer and incubated with 35S-labeled c-IAP2, which had been translated using the TNT-coupled reticulocyte lysate system (Promega), for 3 h at 4°C. The beads were washed with GST binding buffer four times, and bound material was eluted and resolved by SDS-PAGE. Gels were dried, and the amount of bound 35S-labeled c-IAP2 was visualized with a Storm PhosphorImager.
Northern blotting and real-time PCR.
Total RNA from splenocytes, thymocytes, and murine embryonic fibroblasts was isolated using Ultraspec RNA isolation reagent (Biotecx Laboratory, Houston, Tex.) following the manufacturer's recommended protocol. RNA (15 μg) was examined by Northern blotting as previously described (29). Specific cDNA probes for c-IAP2 mRNA and GAPDH mRNA were labeled with [α-32P]dCTP by the use of a random primer kit (Stratagene). For c-IAP2 mRNA quantitation, total RNA was extracted from splenocytes by the use of Ultraspec RNA isolation reagent (Biotecx) and reverse transcribed. The amount of c-IAP2 cDNA was quantified using a Taqman system (Applied Biosystems, Foster City, Calif.). Values for c-IAP2 cDNA were normalized to the relative quantity of hypoxanthine phosphoribosyltransferase cDNA in each sample.
NF-κB luciferase assays, EMSAs, and measurement of cytokine production.
c-IAP1−/− mice were bred with NF-κB-luciferase reporter transgenic mice to create c-IAP1−/− × NF-κB-luciferase animals. Whole cell extracts were prepared from C57BL/6, NF-κB-luciferase, and c-IAP1−/− × NF-κB-luciferase mice by resuspending 10 × 106 cells in 50 μl of lysis buffer (25 mM Tris [pH 7.8], 2 mM dithiothreitol, 10% glycerol, 1 mM EDTA, 1% Triton X-100). NF-κB luciferase activity was detected using a luminometer (Monolight 2010; BD Biosciences) after the addition of 250 μl of luciferase substrate as described previously (26). Nuclear extracts from MEFs were prepared as described previously (37, 42). Binding reactions for the EMSAs were performed using 2 μg of nuclear protein and [α-32P]dCTP-end-labeled double-stranded oligonucleotides as described previously (28). IL-6 production from MEFs was determined by ELISA per the manufacturer's protocol.
RESULTS
Generation of c-IAP1−/− mice.
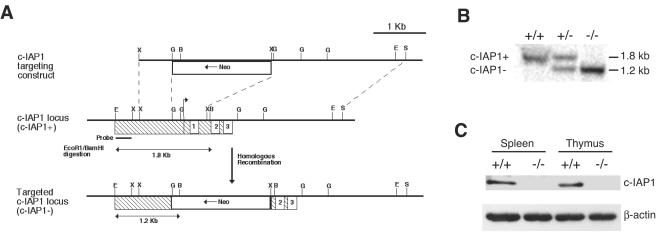
c-IAP1-deficient (c-IAP1−/−) mice were generated by deleting the region that encompasses the translation initiation codon and the first BIR domain (Fig. 1A). The construct p68318S.53 was transfected into mouse embryonic stem (ES) cells. The ES cells were selected for homologous recombination and the presence of the null allele. Stable ES cell transfectants that had incorporated the null allele into genomic DNA were injected into C57BL/6 blastocysts to generate chimeric mice. Heterozygous animals (c-IAP1+/−) with germ line transmission of the null allele were backcrossed for eight generations onto the C57BL/6 background and then intercrossed to generate c-IAP1−/− mice, which were identified by Southern blot analysis of genomic DNA (Fig. 1B). To confirm that incorporation of the null allele disrupted c-IAP1 expression, thymocytes and splenocytes were harvested from wild-type (WT) and c-IAP1−/− mice and the expression of c-IAP1 protein was examined by immunoblot analysis. In results obtained using an antibody against the C-terminal portion of the molecule, c-IAP1 protein was undetectable in cells from c-IAP1−/− mice (Fig. 1C). Thus, deletion of the translation initiation site and a portion of the first exon abolished c-IAP1 expression.
FIG. 1.
Generation of c-IAP1-deficient mice. (A) Strategy for c-IAP1 gene disruption by homologous recombination. The hatched box represents exon 1, the numbered boxes represent the three baculovirus IAP repeats, and the arrow indicates the transcription initiation start codon. Insertion of the neomycin gene (Neo) by homologous recombination replaced the transcription initiation start codon and the first BIR domain. Restriction sites are indicated as follows: E, EcoRI; X, XbaI; G, BglII; B, BamH1; S, SalI. (B) Genomic DNA from wild-type (+/+), c-IAP1 heterozygous (+/−), and c-IAP1 homozygous (−/−) deficient mice was digested with EcoRI and BamHI and probed with the radiolabeled probe indicated in panel A. The upper band (1.8 kb) corresponds to the wild-type c-IAP1 allele (c-IAP1+), and the lower band (1.2 kb) corresponds to the mutant c-IAP1 allele (c-IAP1−). (C) Immunoblot analysis of c-IAP1 expression in spleen and thymus whole-cell lysates from +/+ and −/− mice. Expression of c-IAP1 was determined using a polyclonal antibody that recognizes the C terminus of c-IAP1. β-actin expression was used as a loading control.
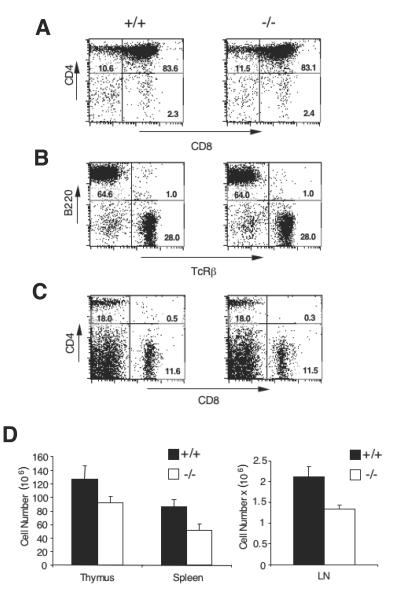
c-IAP1−/− mice were born at the expected Mendelian ratio and had no obvious physical or behavioral abnormalities. c-IAP1−/− mice had life spans of greater than 1 year and did not succumb to disease in our pathogen-free colony. To determine whether c-IAP1 is required for normal lymphoid development, the distribution of thymocyte and peripheral lymphocyte populations was determined by flow cytometry. No difference in the percentages of CD4− CD8− (double-negative), CD4+ CD8+ (double-positive), or single-positive CD4+ and CD8+ thymocytes was observed between WT and c-IAP1−/− mice (Fig. 2A). A normal distribution of splenic B220+ B cells and CD4+ and CD8+ T cells was also found in c-IAP1−/− mice (Fig. 2B and C). Despite this, we consistently observed a modest reduction in the number of lymphocytes in c-IAP1−/− mice (Fig. 2D). This did not appear to be due to an increased sensitivity to apoptotic stimuli, because no quantitative differences in spontaneous, dexamethasone-induced, or TNF-α-induced cell death were observed between WT and c-IAP1−/− cells (data not shown). Furthermore, T-cell receptor-mediated activation of peripheral T cells was normal in c-IAP1−/− mice, as assessed by proliferation and CD25 upregulation (data not shown).
FIG. 2.
T- and B-cell development in wild-type and c-IAP1−/− mice. (A) Thymocytes from wild-type (+/+) and c-IAP1−/− (−/−) mice were isolated, stained with anti-CD4 and anti-CD8 monoclonal antibodies, and analyzed by flow cytometry. (B and C) Splenocytes from +/+ and −/− mice were isolated and stained with anti-TCRβ and anti-CD45R/B220 monoclonal antibodies (B) or anti-CD4 and anti-CD8 monoclonal antibodies (C) and analyzed by flow cytometry. Numbers displayed in the histograms in panels A, B, and C represent the percentages of cells in each quadrant. (D) Cell suspensions were made from thymus (n = 6), spleen (n = 7), and lymph nodes (LN; n = 5) from +/+ and −/− mice. Viable cell recovery was determined by microscopy and trypan blue exclusion. The numbers displayed for lymph nodes reflect the number of cells per lymph node. Error bars represent standard errors of the means.
Increased c-IAP2 levels in c-IAP1-deficient mice.
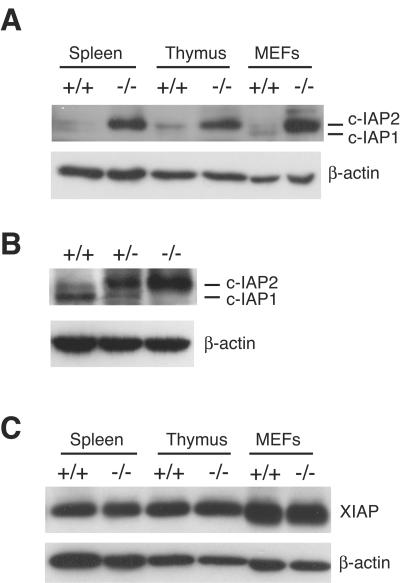
c-IAP1 and c-IAP2 are highly homologous members of the IAP family. We considered the possibility that c-IAP1 loss might be accompanied by changes in expression of other IAP proteins. Splenocytes, thymocytes, and murine embryonic fibroblasts (MEFs) were harvested from WT and c-IAP1−/− mice, and the levels of c-IAP2 were determined by immunoblotting with a rabbit anti-c-IAP antiserum, which was raised against rat c-IAP2 and cross-reacts with mouse c-IAP2 and c-IAP1 (13). Although the ratio of c-IAP1 to c-IAP2 differed among cell types, these proteins were expressed at low levels in all WT cells tested. In contrast, the levels of c-IAP2 were markedly elevated in tissues from c-IAP1−/− animals (Fig. 3A). Comparison of the c-IAP1 and c-IAP2 levels in WT, c-IAP1+/−, and c-IAP1−/− mice showed a clear c-IAP1 gene dose effect on c-IAP2 expression: the levels of c-IAP1 and c-IAP2 in c-IAP1+/− cells were intermediate between those seen with WT and c-IAP1−/− cells (Fig. 3B). Similar results were obtained with thymocytes (data not shown). XIAP is another member of the IAP family that contains a RING domain and has E3 activity (46). In contrast to the elevated levels of c-IAP2, the amounts of XIAP in splenocytes, thymocytes, and primary MEFs from WT and c-IAP1−/− mice were similar (Fig. 3C).
FIG. 3.
Dramatic upregulation of c-IAP2 but not XIAP in c-IAP1−/− mice. (A) Immunoblot analysis of c-IAP expression in spleen, thymus, and murine embryonic fibroblasts (MEFs) using an anti-c-IAP antiserum. (B) Immunoblot analysis of c-IAP expression in splenocytes harvested from +/+, c-IAP1 heterozygous (+/−), and −/− mice. (C) Immunoblot analysis of XIAP expression in spleen, thymus, and MEF lysates from +/+ and −/− mice. β-actin expression was used as a loading control.
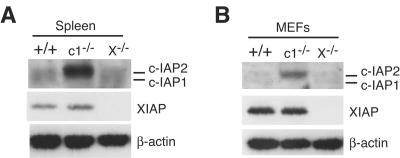
It has been reported that the levels of c-IAP1 and c-IAP2 proteins are elevated in XIAP-deficient (XIAP−/−) MEFs (12), suggesting that there might be mutual regulation between these IAP family members. Given that we didn't find any effect of c-IAP1 deficiency on XIAP levels, we examined the levels of c-IAP1 and c-IAP2 in primary tissues from WT, c-IAP1−/−, and XIAP−/− mice. Unlike the results showing increased levels of c-IAP2 in c-IAP1−/− cells, neither c-IAP1 nor c-IAP2 was elevated in XIAP−/− splenocytes (Fig. 4A). Similar results were obtained with thymocytes (data not shown). To determine whether c-IAP levels might be affected specifically in fibroblasts, primary MEFs were prepared and analyzed for IAP expression. Just as with the other tissues, loss of XIAP had no effect on c-IAP1 or c-IAP2 levels (Fig. 4B). These results were confirmed using two independent MEF preparations, one example of which is shown in Fig. 4B. Therefore, we find no evidence for mutual regulation of XIAP and c-IAP proteins or that the upregulation of c-IAP2 is a specific consequence of the lack of c-IAP1 expression.
FIG. 4.
Elevated c-IAP2 in c-IAP1-deficient cells but not XIAP-deficient cells. Immunoblot analysis of c-IAP expression in spleen (A) and MEF (B) lysates from wild-type (+/+), c-IAP1−/− (c1−/−), and XIAP−/− (X−/−) mice using anti-c-IAP antiserum. β-actin expression was used as a loading control.
Loss of c-IAP1 and increased levels of c-IAP2 do not affect NF-κB.
Phosphorylation, ubiquitination, and degradation of the cytosolic inhibitory protein IκB induces the nuclear translocation of NF-κB. Nuclear NF-κB binds specific DNA regulatory elements and mediates gene transcription (31). There are a number of reports implicating c-IAPs in activating NF-κB: c-IAP1 can ubiquitinate NEMO/IκB kinase γ (IKKγ), the regulatory subunit of the IKK complex, and activate NF-κB in a TNF-α-dependent manner (41); transient overexpression of c-IAP2 in HeLa cells can induce IκB degradation and subsequent NF-κB activation in a RING-dependent manner (5).
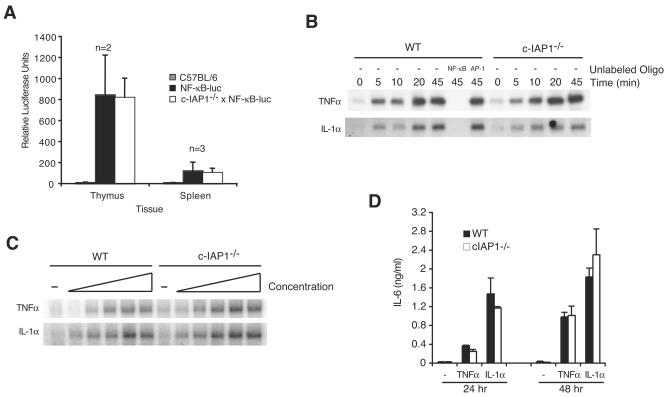
To determine whether the combined effect of c-IAP1 deficiency and increased levels of c-IAP2 led to aberrant activation of NF-κB, c-IAP1−/− mice were crossed with NF-κB-luciferase reporter transgenic (NF-κB-luc) mice (23) and the basal levels of NF-κB-mediated transcription were determined in freshly isolated primary cells. Consistent with previous reports (44), luciferase activity was detected in thymocytes from NF-κB-luc but not C57BL/6 mice (Fig. 5A). Lower but detectable levels were also observed in splenocytes from these animals. Interestingly, the lack of c-IAP1 and dramatically elevated levels of c-IAP2 did not affect the NF-κB-luciferase activity in thymocytes and splenocytes from cIAP1−/− mice (Fig. 5A). Thus, in resting lymphocytes c-IAP1 deficiency and increased c-IAP2 levels do not affect NF-κB activity.
FIG. 5.
Neither c-IAP1 deficiency nor upregulation of c-IAP2 affects NF-κB-mediated transcription. (A) Thymocytes and splenocytes harvested from C57BL/6, NF-κB-luciferase (NF-κB-luc), and c-IAP1−/− × NF-κB-luciferase (c-IAP1−/− × NF-κB-luc) reporter transgenic mice were lysed and assayed for luciferase activity. Values presented are the averages for the indicated numbers (n) of mice. Error bars represent standard deviations. (B) Kinetic analysis of NF-κB DNA binding in stimulated wild-type (WT) and c-IAP1−/− MEFs. Nuclear extracts from unstimulated MEFs or MEFs stimulated with TNF-α (33 ng/ml) or IL-1α (5 ng/ml) for the indicated periods of time were incubated with 32P-end-labeled double-stranded oligonucleotide containing an NF-κB consensus binding site. WT extracts stimulated with TNF-α or IL-1α for 45 min were also incubated with a 32P-end-labeled NF-κB double-stranded oligonucleotide in the absence (−) or presence of a 100-fold excess of unlabeled NF-κB or AP-1 oligonucleotides. (C) Dose-response analysis of NF-κB DNA binding in WT and c-IAP1−/− MEFs by EMSA. Nuclear extracts from unstimulated MEFs or MEFs stimulated with increasing concentrations of TNF-α (0.206, 4.12, 82.5, 1,650, and 33,000 pg/ml) or IL-1α (0.031, 0.625, 12.5, 250, and 5,000 pg/ml) for 25 min were incubated with 32P-end-labeled double-stranded oligonucleotide containing an NF-κB consensus binding site. (D) IL-6 production from WT and c-IAP1−/− MEFs. WT and c-IAP1−/− MEFs were incubated in medium alone or stimulated with TNF-α (33 ng/ml) or IL-1α (5 ng/ml) for 24 and 48 h. The IL-6 concentrations in the supernatants were determined by ELISA. Triplicate samples were assayed, and the error bars represent the standard errors of the means.
TNF-α and IL-1α are cytokines that activate NF-κB (31). To determine whether c-IAP1 is required for TNF-α- or IL-1α-mediated NF-κB activation, we examined NF-κB DNA binding in WT and c-IAP1-deficient cells by EMSA. Nuclear extracts from WT and c-IAP1−/− MEFs stimulated with TNF-α or IL-1α for different amounts of time were incubated with an oligonucleotide containing the consensus DNA sequence recognized by NF-κB (11). Nuclear extracts of WT MEFs caused a gel shift, the NF-κB-binding band increasing in intensity with time of stimulation by TNF-α or IL-1α. The shifted band was eliminated by competing with unlabeled NF-κB, but not AP-1, oligonucleotides (Fig. 5B). Notably, the amounts of NF-κB detected in unstimulated cells and the kinetics of its induction by TNF-α or IL-1α were similar in WT and c-IAP1−/− cells (Fig. 5B). Dose-response analyses yielded similar results: although there was a small degree of variability between experiments, the responses of WT and c-IAP1−/− MEFs to increasing amounts of TNF-α or IL-1α were comparable (Fig. 5C). Similar results were obtained with WT and c-IAP1−/− splenocytes (data not shown).
IL-6 gene expression is induced by TNF-α and IL-1α in an NF-κB-dependent manner (2, 20, 25, 33, 35). To address NF-κB function, WT and c-IAP1−/− MEFs were stimulated with TNF-α or IL-1α and IL-6 production was determined. TNF-α and IL-1α induced easily detectable levels of IL-6 in the cell culture supernatants of WT MEFs at 24 and 48 h (Fig. 5D). Consistent with the above-described results regarding NF-κB activation, c-IAP1−/− MEFs produced similar amounts of IL-6 in response to these stimuli. Taken together, these data indicate that neither the lack of c-IAP1 nor the increased expression of c-IAP2 has a substantial effect on NF-κB activation or function.
Posttranscriptional mechanism for c-IAP2 upregulation in c-IAP1−/− cells.
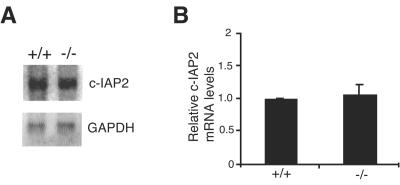
Although c-IAP2 gene expression can be upregulated by NF-κB (5, 17), the analyses described above argue against its upregulation in c-IAP1−/− cells being a consequence of enhanced NF-κB activity. Furthermore, c-IAP2 protein was undetectable in c-IAP1-deficient testes (data not shown), a tissue in which c-IAP1 but not c-IAP2 mRNA is normally expressed (21). To directly determine whether c-IAP2 upregulation in the absence of c-IAP1 was or was not at the transcriptional level, mRNA was isolated from cells from WT and c-IAP1−/− mice and the levels of c-IAP2 mRNA were determined by Northern blotting. Comparable levels of c-IAP2 mRNA were detected in WT and c-IAP1−/− splenocytes (Fig. 6A). Although the absolute amount of c-IAP2 mRNA was lower in thymocytes and MEFs than in spleen, the levels of c-IAP2 mRNA in these tissues between WT and c-IAP1−/− mice were also similar (data not shown). To obtain better quantitation, c-IAP2 mRNA levels in WT and c-IAP1−/− cells were determined by quantitative real-time PCR. The levels of c-IAP2 mRNA in splenocytes (Fig. 6B), thymocytes, and MEFs from WT and cIAP1−/− mice were similar (data not shown). Therefore, the upregulation of c-IAP2 protein in c-IAP1−/− cells is not due to increased c-IAP2 gene expression.
FIG. 6.
c-IAP1 deficiency does not affect c-IAP2 gene expression. (A) c-IAP2 mRNA expression in splenocytes harvested from wild-type (+/+) and c-IAP1−/− (−/−) mice was determined by Northern blot analysis. Expression of GAPDH was used as a loading control. (B) c-IAP2 mRNA expression in splenocytes harvested from +/+ and −/− mice was determined by real-time PCR. Values for c-IAP2 mRNA expression in −/− splenocytes (n = 3) are expressed relative to the levels of c-IAP2 mRNA in +/+ splenocytes (n = 3). Error bars represent standard errors of the means.
c-IAP1 regulates c-IAP2 levels in a ubiquitin protein ligase-dependent manner.
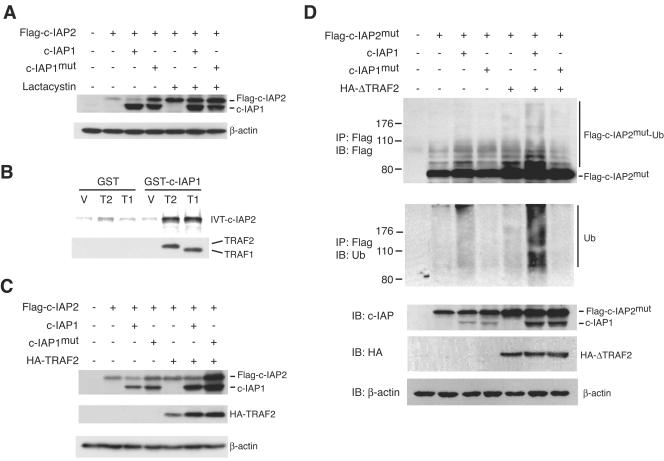
The E3 activity of c-IAP1 is mediated by the carboxy-terminal RING domain (46). Deletion of the RING domain or mutation of its Zn2+-coordinating histidine residue dramatically reduces auto- and _trans_-ubiquitination activity (19, 46). Given the substantial upregulation of c-IAP2 protein without a corresponding increase in mRNA in c-IAP1−/− cells, we considered the possibility that c-IAP1 might normally regulate c-IAP2 protein by promoting its degradation in proteasomes. To test this, Flag-tagged c-IAP2 (Flag-c-IAP2) was transiently expressed in 293 cells alone or in combination with wild-type c-IAP1 or a point mutant of c-IAP1 (c-IAP1mut) with an alanine substitution for the critical histidine residue in the RING domain. c-IAP1mut behaves as a dominant negative in cells expressing endogenous wild-type c-IAP1, such as 293 cells (19). After 17 h the transfected cells were incubated for an additional 7 h with or without the proteasome inhibitor lactacystin, and the levels of Flag-c-IAP2 and c-IAP1 were determined by immunoblotting with the anti-c-IAP antiserum. Ectopically expressed Flag-c-IAP2 was detected in 293 cells (Fig. 7A). Cotransfection with c-IAP1 caused a small reduction in the level of Flag-c-IAP2, and coexpression of the E3-inactive c-IAP1mut caused an increase, consistent with the dominant-negative nature of the mutant. Incubation with lactacystin significantly increased the level of Flag-c-IAP2 (compare lanes 5 and 2 from the left of the panel). Importantly, in the presence of the proteasome inhibitor, coexpression of c-IAP1 did not reduce Flag-c-IAP2 levels and was there no substantial increase in Flag-c-IAP2 levels when c-IAP1mut was coexpressed. Similar results were observed using an E3-defective c-IAP2 mutant (data not shown). Therefore, the regulation of c-IAP2 by c-IAP1 requires proteasome catalytic activity.
FIG. 7.
c-IAP1 ubiquitinates and downregulates c-IAP2 in an E3-dependent manner. (A) Immunoblot analysis of 293 cells transiently transfected with Flag-tagged-c-IAP2 in combination with vector (−), c-IAP1, or c-IAP1mut. Cells were transfected for 17 h and then treated with or without lactacystin (25 μM) for an additional 7 h. The amounts of Flag-c-IAP2, c-IAP1, and c-IAP1mut were determined with an anti-c-IAP antiserum. β-actin expression was used as a loading control. (B) c-IAP1 binds c-IAP2 only in the presence of TRAF1 or TRAF2. Bacterially expressed and purified GST- or GST-c-IAP1-coated beads were incubated with the lysate of bacteria transformed with vector (V) or His-tagged TRAF1 (189-416) (T1) or His-tagged TRAF2 (272-501) (T2) cDNAs. The beads were washed and incubated with in vitro-translated and 35S-labeled c-IAP2. After extensive washing, the bound material was eluted and resolved by SDS-PAGE. The presence of His-TRAF1 (189-416) and His-TRAF2 (272-501) in the protein complexes was determined by immunoblotting. (C) 293 cells were transiently transfected with Flag-tagged-c-IAP2 (Flag-c-IAP2) in combination with vector (−), c-IAP1, c-IAP1mut, or HA-tagged TRAF2 (HA-TRAF2). The amounts of Flag-c-IAP2, c-IAP1, c-IAP1mut, and HA-TRAF2 were determined by immunoblot analysis. Flag-c-IAP2, c-IAP1, and c-IAP1mut expression was determined using anti-c-IAP antiserum. (D) 293 cells were transfected with Flag-tagged-c-IAP2mut alone or in combination with the indicated cDNAs. After overnight culture, the cells were treated with lactacystin for an additional 7 h and then harvested and lysed. Flag-c-IAP2mut was immunoprecipitated (IP) and immunoblotted (IB) with anti-Flag to detect the native molecule as well as higher-molecular-weight species representing ubiquitinated c-IAP2mut (Flag-c-IAP2mut-Ub). The membrane was stripped and reblotted with anti-ubiquitin polyclonal antibody to directly detect the presence of ubiquitinated Flag-c-IAP2mut. Expression of Flag-c-IAP2mut, c-IAP1, c-IAP1mut, and HA-ΔTRAF2 in cell lysates was detected by immunoblotting.
c-IAP1 and c-IAP2 do not bind one another directly but independently bind the C-terminal TRAF domains of TRAF2 and TRAF1 (30, 43), which can homo- and heterodimerize (40). c-IAP1-induced degradation of c-IAP2 suggests that c-IAP1 and c-IAP2 coexist in a multimeric complex. To explore the possibility that such a complex was formed in the presence of the adaptor TRAF proteins, in vitro binding studies were performed using recombinant GST-conjugated c-IAP1, His-tagged mutants of TRAF1 [His-TRAF1 (189-416)] and TRAF2 [His-TRAF2 (272-501)] containing the IAP-binding TRAF domains, and in vitro-translated and radiolabeled c-IAP2. Beads coated with GST or GST-c-IAP1 and control bacterial lysate bound weakly to c-IAP2 (Fig. 7B, lanes 1 and 4 from the left of the panel), and incubation of GST-coated beads with bacterial lysates containing either His-TRAF1 (189-416) or His-TRAF2 (272-501) did not increase this binding. However, the combination of GST-c-IAP1 beads and His-TRAF2 (272-501) or His-TRAF1 (189-416) lysate resulted in markedly increased c-IAP2 binding (Fig. 7B, lanes 5 and 6). Consistent with this, immunoblotting of the eluted proteins showed that His-TRAF2 (272-501) and His-TRAF1 (189-416) bound specifically to GST-c-IAP1 (lanes 5 and 6) and not GST (lanes 2 and 3). Therefore, c-IAP1 and c-IAP2 can coexist in a multimeric complex through their interactions with the TRAF domains of TRAF1 and TRAF2.
Because TRAFs form complexes containing both c-IAP1 and c-IAP2, we asked whether the level of endogenous TRAF in 293 cells might limit the dominant-negative effect of c-IAP1mut on c-IAP2 expression. 293 cells were transfected with HA-tagged TRAF2 (HA-TRAF2), Flag-c-IAP2, and either c-IAP1 or c-IAP1mut. As described for Fig. 7A, coexpression of c-IAP1 or c-IAP1mut caused a modest decrease or increase, respectively, in Flag-c-IAP2 levels (Fig. 7C). Expression of HA-TRAF2 reproducibly caused a small increase in the level of Flag-c-IAP2. Coexpression of c-IAP1 in the presence of HA-TRAF2 had little if any effect on Flag-c-IAP2, which may have been due to the generation of c-IAP1/TRAF and c-IAP2/TRAF complexes lacking the reciprocal IAP. Notably, however, coexpression of HA-TRAF2 greatly potentiated the dominant-negative effect of c-IAP1mut (Fig. 7C, lane 7).
These results show that c-IAP2 and c-IAP1 coexist in a multimeric complex with TRAF1 and TRAF2 and that proteasome-mediated c-IAP2 degradation requires the E3 activity of c-IAP1. The most straightforward explanation for these observations would be that TRAF1 and TRAF2 function as adaptors to bring the two IAP molecules together, allowing c-IAP1 to ubiquitinate c-IAP2 in vivo. To examine this, 293 cells were transfected with a Flag-tagged E3-inactive mutant of c-IAP2 (c-IAP2mut) to prevent autoubiquitination and different combinations of c-IAP1, c-IAP1mut, and an HA-tagged TRAF2 mutant lacking the N-terminal RING domain (HA-ΔTRAF2). The latter retains its C-terminal IAP-binding region but precludes the possibility that TRAF2 might directly contribute to IAP ubiquitination via its RING domain. The transfected cells were treated with lactacystin for 7 h prior to harvest and immunoprecipitation of Flag-c-IAP2mut. A small amount of ubiquitinated Flag-c-IAP2mut was detected in cells transfected with Flag-c-IAP2 alone. This amount of ubiquitinated c-IAP2 increased to a small extent when c-IAP1, but not the E3-defective c-IAP1mut, was coexpressed (Fig. 7D, lanes 3 and 4). In contrast, coexpression of c-IAP1 and ΔTRAF2 caused a substantial increase in the amount of ubiquitinated c-IAP2mut (lane 6). This increase was not observed when the E3-defective c-IAP1mut was expressed (lane 7). These results demonstrate that c-IAP1 directly ubiquitinates c-IAP2 when the two are brought together in a TRAF-containing complex.
DISCUSSION
Apoptosis is required for normal embryonic development, growth, and homeostasis. IAP proteins can inhibit cell death, either by binding to and inhibiting caspases (32) or by ubiquitinating the proapoptotic mitochondrial protein Smac/DIABLO (14). Note, however, that a physiologic anti-apoptotic role for mammalian IAPs, and for c-IAP1 and c-IAP2 in particular, has been inferred almost entirely from studies in which these proteins were overexpressed (7, 14, 19, 32, 36). Mice deficient in XIAP were initially shown to have no obvious abnormalities and, in particular, no defects in apoptosis (12). Recent studies have shown that cells derived from these mice are more sensitive to nerve growth factor withdrawal and cytochrome _c_-induced sympathetic neuronal apoptosis (27) and have defects in copper homeostasis (4). As shown in the present report, the lack of a major cell apoptosis phenotype cannot be explained, as was originally suggested, by compensatory changes in c-IAP1 or c-IAP2 levels. It is perhaps noteworthy that the highly homologous c-IAP1 and c-IAP2 were not identified by function but rather were discovered as components of the TNFR2 signaling complex (30). A number of reports have shown that c-IAPs can actually promote apoptosis. One study found that c-IAP1 is cleaved during apoptosis and that the resulting C-terminal RING-containing fragment induces apoptosis in transient transfection assays (6). We have shown that in response to TNFR2 signaling, c-IAP1 ubiquitinates and causes the proteasome-mediated degradation of TRAF2 and that overexpression of E3-inactive c-IAP1 prevents TRAF2 degradation and apoptosis (19). The present study demonstrates that in vivo, the loss of c-IAP1 does not have an obvious effect on cell survival, although it is possible that the upregulation of c-IAP2 might have a mitigating effect on this phenotype. Studies with animals deficient in both IAPs will be required to explore this further.
Although c-IAP1 and c-IAP2 are widely expressed, their abundance is generally low. The present results suggest that the cause of low c-IAP2 expression is constitutive ubiquitination by c-IAP1 and its resultant degradation. While there could be indirect mechanisms as well, strong evidence for a direct effect comes from the in vitro binding studies and the coexpression of c-IAP1, c-IAP2, and TRAF2, which demonstrates that the IAPs and TRAF1/2 form a complex. Whereas the expression of c-IAP1 alone increased c-IAP2 ubiquitination, this effect was markedly enhanced by coexpression of ΔTRAF2, indicating that IAP-binding TRAFs promote c-IAP1-mediated ubiquitination by acting as a scaffold or adaptor protein. This, combined with the finding that c-IAP2 mRNA was not elevated in c-IAP1−/− cells, suggests that physiologic levels of c-IAP1 regulate c-IAP2 posttranscriptionally via ubiquitination and subsequent degradation.
It has been reported that XIAP−/− MEFs have increased levels of c-IAP1 and c-IAP2 (12), suggesting that XIAP might normally suppress the expression of these other IAP family members. We found in contrast that c-IAP1 and c-IAP2 are expressed at comparable levels in fresh splenocytes, thymocytes, and primary MEFs from WT and XIAP−/− mice. We believe it is likely that the difference between our and the previous results is that the latter were generated with immortalized MEF lines (Colin Duckett, personal communication), which may have differentiated during long-term culture or become oligoclonal. Given that c-IAP1 and c-IAP2, but not XIAP, are found together in the TNFR signaling complex, it seems reasonable that c-IAP2 could be ubiquitinated or otherwise directly regulated by c-IAP1. It will be of interest to learn whether c-IAP1 is also a substrate for c-IAP2 and therefore might accumulate in c-IAP2−/− mice.
Following the degradation of IκB, NF-κB translocates to the nucleus, where it binds to DNA and induces gene transcription. TNF-α and IL-1α activate NF-κB (38) and lead to the upregulation of anti-apoptotic genes, among them ciap2 (5, 45). c-IAP1 can ubiquitinate NEMO/IKKγ, the regulatory subunit of the IKK complex, resulting in the activation of NF-κB (41). In addition, transient expression of c-IAP2 in cell lines has been reported to cause the degradation of IκB and subsequent activation of NF-κB, suggesting that c-IAP2 might potentiate NF-κB-mediated gene expression by a feed-forward mechanism (5). The present results demonstrate that NF-κB in primary cells is unaffected by c-IAP1 deficiency. Furthermore, the levels of XIAP and the production of IL-6, which are upregulated by NF-κB (20, 39), are similar in WT and c-IAP1−/− cells. Therefore, although c-IAPs can affect NF-κB activation when manipulated in cell lines, they do not seem to play a critical role in vivo.
c-IAP1−/− mice have revealed an unexpected facet of IAP posttranslational regulation. Analysis of c-IAP1 regulation of c-IAP2 expression also underscores the role of RING-dependent ubiquitination, as opposed to BIR-dependent inactivation of caspases, as a mechanism for IAP function. It will be of interest to revisit these issues in c-IAP2−/− mice and ask whether posttranscriptional regulation of c-IAPs is reciprocal.
Acknowledgments
We thank Bei Dong for expert technical assistance with c-IAP1 and c-IAP2 mutagenesis and subcloning, Peter Liston for supplying the c-IAP1 and c-IAP2 expression plasmids and the anti-c-IAP antiserum, Chuanjin Wu for supplying the HA-tagged ΔTRAF2, and Mercedes Rincón for providing the NF-κB-luciferase reporter transgenic mice.
REFERENCES
- 1.Abbondanzo, S. J., I. Gadi, and C. L. Stewart. 1993. Derivation of embryonic stem cell lines. Methods Enzymol. 225**:**803-823. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., and T. Kishimoto. 1992. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol. Rev. 127**:**25-50. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds. J. Mol. Biol. 295**:**1103-1112. [DOI] [PubMed] [Google Scholar]
- 4.Burstein, E., L. Ganesh, R. D. Dick, B. van De Sluis, J. C. Wilkinson, L. W. Klomp, C. Wijmenga, G. J. Brewer, G. J. Nabel, and C. S. Duckett. 2004. A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO J. 23**:**244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, Z. L., T. A. McKinsey, L. Liu, J. J. Gentry, M. H. Malim, and D. W. Ballard. 1997. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc. Natl. Acad. Sci. USA 94**:**10057-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clem, R. J., T. T. Sheu, B. W. Richter, W. W. He, N. A. Thornberry, C. S. Duckett, and J. M. Hardwick. 2001. c-IAP1 is cleaved by caspases to produce a proapoptotic C-terminal fragment. J. Biol. Chem. 276**:**7602-7608. [DOI] [PubMed] [Google Scholar]
- 7.Conte, D., P. Liston, J. W. Wong, K. E. Wright, and R. G. Korneluk. 2001. Thymocyte-targeted overexpression of xiap transgene disrupts T lymphoid apoptosis and maturation. Proc. Natl. Acad. Sci. USA 98**:**5049-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conze, D., T. Krahl, N. Kennedy, L. Weiss, J. Lumsden, P. Hess, R. A. Flavell, G. G. Le, R. J. Davis, and M. Rincon. 2002. c-Jun NH(2)-terminal kinase (JNK)1 and JNK2 have distinct roles in CD8(+) T cell activation. J. Exp. Med. 195**:**811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67**:**2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai, Z., W. G. Zhu, C. D. Morrison, R. M. Brena, D. J. Smiraglia, A. Raval, Y. Z. Wu, L. J. Rush, P. Ross, J. R. Molina, G. A. Otterson, and C. Plass. 2003. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum. Mol. Genet. 12**:**791-801. [DOI] [PubMed] [Google Scholar]
- 11.Guerder, S., M. Rincon, and A. M. Schmitt-Verhulst. 2001. Regulation of activator protein-1 and NF-kappa B in CD8+ T cells exposed to peripheral self-antigens. J. Immunol. 166**:**4399-4407. [DOI] [PubMed] [Google Scholar]
- 12.Harlin, H., S. B. Reffey, C. S. Duckett, T. Lindsten, and C. B. Thompson. 2001. Characterization of XIAP-deficient mice. Mol. Cell. Biol. 21**:**3604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holcik, M., C. A. Lefebvre, K. Hicks, and R. G. Korneluk. 2002. Cloning and characterization of the rat homologues of the inhibitor of apoptosis protein 1, 2, and 3 genes. BMC Genomics 3**:**5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, S., and X. Yang. 2003. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 278**:**10055-10060. [DOI] [PubMed] [Google Scholar]
- 15.Huang, H., C. A. Joazeiro, E. Bonfoco, S. Kamada, J. D. Leverson, and T. Hunter. 2000. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275**:**26661-26664. [DOI] [PubMed] [Google Scholar]
- 16.Imoto, I., Z. Q. Yang, A. Pimkhaokham, H. Tsuda, Y. Shimada, M. Imamura, M. Ohki, and J. Inazawa. 2001. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 61**:**6629-6634. [PubMed] [Google Scholar]
- 17.Lamb, J. A., J. J. Ventura, P. Hess, R. A. Flavell, and R. J. Davis. 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell 11**:**1479-1489. [DOI] [PubMed] [Google Scholar]
- 18.LeBlanc, A. C. 2003. Natural cellular inhibitors of caspases. Prog. Neuro-psychopharmacol. Biol. Psychiatry 27**:**215-229. [DOI] [PubMed] [Google Scholar]
- 19.Li, X., Y. Yang, and J. D. Ashwell. 2002. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416**:**345-347. [DOI] [PubMed] [Google Scholar]
- 20.Libermann, T. A., and D. Baltimore. 1990. Activation of interleukin-6 gene expression through the NF-κB transcription factor. Mol. Cell. Biol. 10**:**2327-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liston, P., C. Lefebvre, W. G. Fong, J. Y. Xuan, and R. G. Korneluk. 1997. Genomic characterization of the mouse inhibitor of apoptosis protein 1 and 2 genes. Genomics 46**:**495-503. [DOI] [PubMed] [Google Scholar]
- 22.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96**:**11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millet, I., R. J. Phillips, R. S. Sherwin, S. Ghosh, R. E. Voll, R. A. Flavell, A. Vignery, and M. Rincon. 2000. Inhibition of NF-kappaB activity and enhancement of apoptosis by the neuropeptide calcitonin gene-related peptide. J. Biol. Chem. 275**:**15114-15121. [DOI] [PubMed] [Google Scholar]
- 24.Mittelstadt, P. R., and J. D. Ashwell. 2003. Disruption of glucocorticoid receptor exon 2 yields a ligand-responsive C-terminal fragment that regulates gene expression. Mol. Endocrinol. 17**:**1534-1542. [DOI] [PubMed] [Google Scholar]
- 25.Okazaki, T., S. Sakon, T. Sasazuki, H. Sakurai, T. Doi, H. Yagita, K. Okumura, and H. Nakano. 2003. Phosphorylation of serine 276 is essential for p65 NF-kappaB subunit-dependent cellular responses. Biochem. Biophys. Res. Commun. 300**:**807-812. [DOI] [PubMed] [Google Scholar]
- 26.Petrak, D., S. A. Memon, M. J. Birrer, J. D. Ashwell, and C. M. Zacharchuk. 1994. Dominant negative mutant of c-Jun inhibits NF-AT transcriptional activity and prevents IL-2 gene transcription. J. Immunol. 153**:**2046-2051. [PubMed] [Google Scholar]
- 27.Potts, P. R., S. Singh, M. Knezek, C. B. Thompson, and M. Deshmukh. 2003. Critical function of endogenous XIAP in regulating caspase activation during sympathetic neuronal apoptosis. J. Cell Biol. 163**:**789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rincon, M., B. Derijard, C. W. Chow, R. J. Davis, and R. A. Flavell. 1997. Reprogramming the signalling requirement for AP-1 (activator protein-1) activation during differentiation of precursor CD4+ T-cells into effector Th1 and Th2 cells. Genes Funct. 1**:**51-68. [DOI] [PubMed] [Google Scholar]
- 29.Rincon, M., A. Tugores, A. Lopez-Rivas, A. Silva, M. Alonso, M. O. De Landazuri, and M. Lopez-Botet. 1988. Prostaglandin E2 and the increase of intracellular cAMP inhibit the expression of interleukin 2 receptors in human T cells. Eur. J. Immunol. 18**:**1791-1796. [DOI] [PubMed] [Google Scholar]
- 30.Rothe, M., M. G. Pan, W. J. Henzel, T. M. Ayres, and D. V. Goeddel. 1995. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 83**:**1243-1252. [DOI] [PubMed] [Google Scholar]
- 31.Rothwarf, D. M., and M. Karin. 26. October 1999, posting date. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci. STKE 1999**:**RE1. [Online.] [DOI] [PubMed] [Google Scholar]
- 32.Roy, N., Q. L. Deveraux, R. Takahashi, G. S. Salvesen, and J. C. Reed. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 16**:**6914-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudolph, D., W. C. Yeh, A. Wakeham, B. Rudolph, D. Nallainathan, J. Potter, A. J. Elia, and T. W. Mak. 2000. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 14**:**854-862. [PMC free article] [PubMed] [Google Scholar]
- 34.Salvesen, G. S., and C. S. Duckett. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3**:**401-410. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt-Supprian, M., W. Bloch, G. Courtois, K. Addicks, A. Israel, K. Rajewsky, and M. Pasparakis. 2000. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol. Cell 5**:**981-992. [DOI] [PubMed] [Google Scholar]
- 36.Schoemaker, M. H., J. E. Ros, M. Homan, C. Trautwein, P. Liston, K. Poelstra, G. H. van, P. L. Jansen, and H. Moshage. 2002. Cytokine regulation of pro- and anti-apoptotic genes in rat hepatocytes: NF-kappaB-regulated inhibitor of apoptosis protein 2 (cIAP2) prevents apoptosis. J. Hepatol. 36**:**742-750. [DOI] [PubMed] [Google Scholar]
- 37.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17**:**6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seitz, C., P. Muller, R. C. Krieg, D. N. Mannel, and T. Hehlgans. 2001. A novel p75TNF receptor isoform mediating NFkappa B activation. J. Biol. Chem. 276**:**19390-19395. [DOI] [PubMed] [Google Scholar]
- 39.Stehlik, C., R. de Martin, I. Kumabashiri, J. A. Schmid, B. R. Binder, and J. Lipp. 1998. Nuclear factor (NF)-kappaB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha-induced apoptosis. J. Exp. Med. 188**:**211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi, M., M. Rothe, and D. V. Goeddel. 1996. Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J. Biol. Chem. 271**:**19935-19942. [DOI] [PubMed] [Google Scholar]
- 41.Tang, E. D., C. Y. Wang, Y. Xiong, and K. L. Guan. 2003. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J. Biol. Chem. 278**:**37297-37305. [DOI] [PubMed] [Google Scholar]
- 42.Tugores, A., M. A. Alonso, F. Sanchez-Madrid, and M. O. de Landazuri. 1992. Human T cell activation through the activation-inducer molecule/CD69 enhances the activity of transcription factor AP-1. J. Immunol. 148**:**2300-2306. [PubMed] [Google Scholar]
- 43.Uren, A. G., M. Pakusch, C. J. Hawkins, K. L. Puls, and D. L. Vaux. 1996. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc. Natl. Acad. Sci. USA 93**:**4974-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voll, R. E., E. Jimi, R. J. Phillips, D. F. Barber, M. Rincon, A. C. Hayday, R. A. Flavell, and S. Ghosh. 2000. NF-kappa B activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity 13**:**677-689. [DOI] [PubMed] [Google Scholar]
- 45.Wang, C. Y., M. W. Mayo, R. G. Korneluk, D. V. Goeddel, and A. S. J. Baldwin. 1998. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281**:**1680-1683. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Y., S. Fang, J. P. Jensen, A. M. Weissman, and J. D. Ashwell. 2000. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288**:**874-877. [DOI] [PubMed] [Google Scholar]