The Yersinia Yop Virulon: LcrV Is Required for Extrusion of the Translocators YopB and YopD (original) (raw)
Abstract
LcrV, an essential piece of the Yop virulon, is encoded by the large lcrGVsycDyopBD operon. In spite of repeated efforts, the role of LcrV in the Yop virulon remains elusive. In an attempt to clarify this, we engineered a complete deletion of lcrV in the pYV plasmid of Yersinia enterocolitica E40 and characterized the phenotype of the mutant. Complementation experiments showed that the mutation was not polar with regard to yopB and yopD. Nevertheless the mutation abolished secretion of YopB and YopD, while secretion of the other Yops was unaffected or even increased. Northern blot analysis showed that transcription of yopD was not affected. YopD could be detected inside the bacteria, showing that the lack of its secretion was not due to a lack of translation or to proteolysis. This indicated that LcrV is specifically involved in the process of release of YopB and YopD. We then investigated the possible interactions between LcrV and YopB or YopD. We constructed a glutathione _S_-transferase–LcrV hybrid protein, and we observed that either YopB or YopD could be copurified with it. The same approach showed that LcrV also interacts with LcrG but not with the chaperone SycD. Using deletants of lcrV, we then identified a definite LcrG-binding domain in the C terminus of LcrV.
The capacity of yersiniae (Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica) to resist the immune system of their host depends on the Yop virulon. This system allows extracellular bacteria adhering at the surface of eukaryotic cells to inject bacterial proteins into the cytosol of these cells in order to disarm them or disrupt their communications (for reviews, see references 15, 25, and 52). Translocation of the intracellular effectors (YopE, YopH, YpkA/YopO, YopM) across the eukaryotic cell membrane requires at least two other Yop proteins, namely YopB and YopD (7, 21, 33, 42, 49, 51). Deployment of these translocators at the bacterial surface is triggered by contact with eukaryotic cells and is controlled by proteins of the virulon including YopN, which is supposed to act as a stop valve closing the bacterial secretion channel (7, 18, 42). yopN mutant bacteria are deregulated for Yop secretion in the sense that they release most of their Yop effector load outside eukaryotic cells, but they can nevertheless deliver a portion of these effectors inside eukaryotic cells (7).
Yop proteins are transported outside the bacterial cell by a type III secretion apparatus called Ysc, which consists of proteins YscA through YscU, LcrD, and lipoprotein VirG (1–4, 16, 26, 35, 36, 56). Synthesis of the Yops is subject to feedback inhibition: when the secretion apparatus is closed or defective, transcription of the yop genes is prevented (14, 34). The proper operation of the system also requires the presence in the bacterial cytosol of small individual chaperones called the Syc proteins (for a review see reference 55). Three such chaperones have been described so far: SycE for YopE, SycH for YopH, and SycD (called LcrH in Y. pseudotuberculosis and Y. pestis) for YopD. sycE mutants secrete much less YopE than the wild type does, and they rapidly degrade their pool of intracellular YopE (9, 19, 54). SycE acts by binding the domain of YopE that is recognized by the translocation apparatus (57). SycH acts in a similar way for YopH (53, 57). SycD binds to YopD and is required for secretion of both YopD and YopB (53). Previous work described SycD as a negative regulator of the transcription of yop genes (5, 39). It is not known whether this regulatory effect represents a distinct role of SycD or a consequence of its protective role for YopD.
In vitro, the need for contact with eukaryotic cells can be circumvented by chelating Ca2+ ions with agents such as oxalate or EGTA. Under these conditions, effectors and translocators are released in massive amounts in the bacterial culture medium where they form inert aggregates. The yopN mutants affected in the contact control are also affected in their response to Ca2+ chelation. They secrete Yops, even in the presence of Ca2+ ions, and they are said to be “Ca2+ blind” (18).
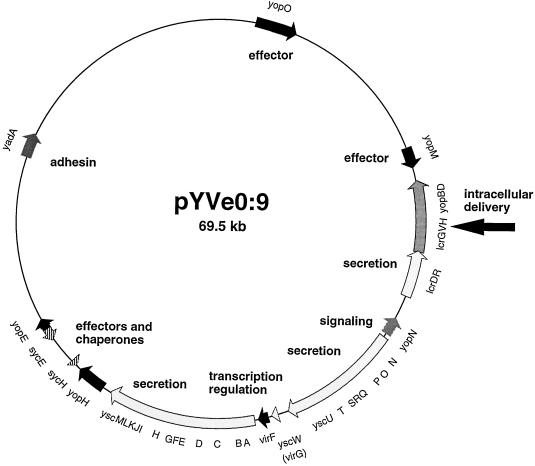
The entire Yop virulon is borne by a 70-kb plasmid called pYV. The genes encoding the Ysc apparatus are arranged as three neighboring operons, while the genes encoding the Yop effectors are scattered around the whole plasmid (Fig. 1). The translocators YopB and YopD as well as LcrG and the chaperone SycD (LcrH) are encoded by the large lcrGVsycDyopBD operon, which also encodes LcrV (5, 29, 38). LcrV is a 41-kDa secreted protein that was described in the mid-1950s as a protective antigen of the plague bacillus Y. pestis (8, 28, 41). LcrV is one of the major Yops, yet its exact role in the virulon is still unclear. As expected, polar mutations in lcrV prevent secretion of LcrV, YopB, and YopD (29, 38). However, nonpolar mutations consisting of small in-frame deletions also impair secretion of other Yops, which suggests that LcrV plays a regulatory role (5, 37, 46). Since the lcrV gene is buried in the operon that encodes the translocators, we considered that LcrV may instead be an element of the translocation apparatus which is required either for the operation or deployment of the latter. In this paper, we present data supporting this view.
FIG. 1.
The pYV plasmid from Y. enterocolitica showing the genes and operons discussed in the text. The arrow indicates the lcrGVHyopBD operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Y. enterocolitica W22703 (nalidixic acid resistant) is a restriction mutant (Res− Mod+) of serotype O:9 strain W227 (11). Y. enterocolitica KNG22703 and MRS40 are the blaA mutants of strains W22703 and E40, respectively, in which the gene encoding β-lactamase A was replaced by the luxAB genes (22, 45). Escherichia coli LK111, received from M. Zabeau (Ghent, Belgium), was used for standard genetic manipulations. E. coli CJ236 was used for site-directed mutagenesis (23). E. coli XL1 Blue (Stratagene, La Jolla, Calif.) was used to produce the glutathione _S_-transferase (GST) fusion proteins. E. coli SM10 lambda pir+ constructed by Miller and Mekalanos (27) was used to deliver the mobilizable plasmids in Y. enterocolitica. This strain allows replication of pir mutants of R6K, and it mobilizes plasmids containing the origin of transfer of RK2. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmids | Relevant characteristics | Reference or source |
|---|---|---|
| pYV plasmids | ||
| pGC153 | pYVe227 yopB-GC153::mini-Mud1 lac ylpA-YL4::Tn_3_ (does not encode YopB and YopD) | 14 |
| pMRS4071 | pYV40 _lcrV_Δ3-324 (pYV40 mutated with pMRS71) | This work |
| pPW2269 | pYVe227 sycD mutant (out-of-frame deletion between nucleotides 111 and 255 of sycD; encodes a 43-residue truncated SycD) | 53 |
| pYV40 | Wild-type PYV plasmid from Y. enterocolitica E40 | 49 |
| pYVe227 | Wild-type pYV plasmid from Y. enterocolitica W227 | 9 |
| Clones and vectors | ||
| pBC18R | 10 | |
| pBC19R | 10 | |
| pCNR21 | pBC19R P_yopE_ yopE | This work |
| pCNR26 | pBC19R P_yopE_ SD T7 _Nde_I yopE | This work |
| pCN29 | pGEX-KG yopD (encodes GST and YopD) | This work |
| pCN40 | pGEX-KG yopB sycD (encodes GST, YopB and SycD) | This work |
| pCNG42 | pGEX-KG gst-lcrV yopB sycD (encodes GST-LcrV, YopB and SycD) | This work |
| pCNG50 | pGEX-KG gst-lcrV sycD (encodes GST-LcrV and SycD) | This work |
| pGEX-KG | p_tac_ _gst ori_pBR322 | 20 |
| pMRS20 | pBluescriptII SK+ and PCR-amplified fragment (by using MIPA271 and MIPA64) containing lcrRGV | 45 |
| pMRS28 | pBC18R and _Eco_RI fragment from pMRS20 (encoding lcrRGV under own promoter) | This work |
| pMRS44 | pT7-7 and PCR-amplified lcrG (by using MIPA354 and MIPA355) from pMRS20 cloned into the _Nde_I-_Sal_I site | This work |
| pMRS46 | pGEX-KG gst-lcrV (encodes GST-LcrV) | 45 |
| pMRS50 | pGEX-KG gst-lcrG yopD (encodes GST-LcrG and YopD) | 45 |
| pMRS52 | pMRS20 _lcrV_Δ224-266 (created by site-directed mutagenesis with MIPA352) | This work |
| pMRS56 | pMRS20 _lcrV_Δ2-32 (created by site-directed mutagenesis with MIPA356) | This work |
| pMRS64 | pBluescript II SK+ and PCR-amplified fragment (by using MIPA384 and MIPA289) from pYV40 containing lcrG, lcrV, and sycD genes | This work |
| pMRS65 | pMRS64, _Nru_I site just before codon 3 of lcrV | This work |
| pMRS67 | pMRS65, _Sma_I site just after codon 324 of lcrV | This work |
| pMRS69 | pMRS67 _lcrV_Δ3-324 | This work |
| pMRS70 | pMRS101, _Sal_I-_Xba_I fragment of pMRS69 | This work |
| pMRS72 | pCNR26 and PCR-amplified fragment (by using MIPA354 and MIPA122) from pYV40 containing lcrG, lcrV, and sycD genes | This work |
| pMRS74 | pMSL56 with cyaA replaced by a PCR-amplified fragment (by using MIPA121 and MIPA317) from pYV40 containing sycD, yopB, and yopD genes | This work |
| pMRS75 | pMRS46 lcrG (encodes GST-LcrV and LcrG) | This work |
| pMRS78 | pMRS75 _lcrV_Δ224-266 (encodes GST-LcrVΔ224-266 and LcrG) | This work |
| pMRS83 | pMRS75 _lcrV_Δ2-32(encodes GST-LcrVΔ2-32 and LcrG) | This work |
| pMRS84 | pMRS50 lcrV (lcrG replaced by lcrV) (encodes GST-LcrV and YopD) | This work |
| pMSL56 | pTM100 P_yopE cyaA_ | M.-P. Sory |
| pPW64 | pBC18R sycD | 53 |
| Suicide vector and mutator | ||
| pMRS101 | _ori_R6K_ori_E1 sacBR oriT_RK2_strAB | 44 |
| pMRS71 | pMRS70 Δ_ori_E1 (encoding _lcrV_Δ3-324) | This work |
Bacteria were routinely grown in tryptic soy broth (Oxoid, Basingstoke, England) and plated on tryptic soy agar. For the induction of the yop regulon, Y. enterocolitica was grown in brain heart infusion broth supplemented with 4 mg of glucose ml−1, 20 mM MgCl2, and 20 mM sodium oxalate. All media were supplemented with the relevant antibiotics. Unless otherwise specified the concentrations were as follows: ampicillin, 200 μg ml−1; kanamycin, 50 μg ml−1; streptomycin, 100 μg ml−1; and nalidixic acid, 35 μg ml−1.
Induction of the yop regulon, SDS-PAGE analysis of Yops, immunoblotting, and genetic conjugation.
Yops were prepared from culture supernatants and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or Tricine-SDS-PAGE and Western blotting as described previously (2, 14, 50). For the analysis of the whole-cell protein extract, 8 × 108 bacteria were applied to SDS–14% PAGE. Immunoblotting was carried out with rat monoclonal antibodies 13A4 (anti-YopD), 7C1 (anti-LcrV), 9B7 (anti-YopB), and 6G1 (anti-YopE) as described by Bodeus et al. (6) and polyclonal antibodies against LcrV and LcrG.
To introduce a plasmid into Y. enterocolitica by conjugation, the plasmid was first introduced in E. coli SM10 lambda pir+ by electroporation. This donor strain and the recipient Y. enterocolitica were mated during a 2- to 3-h period on a plate at 32°C.
Construction of entire lcrV deletion mutant.
A 1,800-bp fragment, containing the entire lcrG, lcrV, and sycD (lcrH) genes, was amplified by PCR with pYV40 as a template. The upstream primer MIPA384 (5′-ATGAATTCATATGAAGTCTTCCCATT-3′) consisted of nucleotides 1 to 16 of lcrG from Y. pestis (38) preceded by an _Eco_RI restriction site (underlined). The downstream primer MIPA289 (5′-CATGGATCCTGGGTTATCAACGCACTCATG-3′) consisted of the nucleotides complementary to the last 20 nucleotides (from 483 to 504) of lcrH from Y. pestis (38) and was preceded by a _Bam_HI restriction site (underlined). The PCR product was then digested with _Eco_RI and _Bam_HI and cloned into the same sites of pBluescriptII SK+, yielding plasmid pMRS64. An _Nru_I restriction site was then introduced just before the third codon of lcrV in pMRS64 by site-directed mutagenesis with oligonucleotide MIPA406 (5′-CAAATTATTTAATATGTCGCGAGCCTACGAACA-3′), generating plasmid pMRS65. Oligonucleotide MIPA407 (5′-CTGCTAGATGACACGCCCGGGAAATGACACGAGGT-3′) was used to introduce a _Sma_I site after codon 324 of lcrV in pMRS65, yielding pMRS67. An in-frame deletion was then generated by digestion of pMRS67 with _Nru_I-_Sma_I followed by religation. This recombinant plasmid containing the mutated allele was called pMRS69. The _Sal_I-_Xba_I fragment of pMRS69 was then cloned into the same sites of the suicide vector precursor pMRS101. This precursor contains two origins of replication: a functional _ori_ColE1 facilitating the production of plasmid DNA and a conditional _ori_R6K that is only functional in E. coli strains producing the π protein (27, 44). The recombinant precursor pMRS70 was then digested by _Not_I and religated to remove _ori_ColE1 and bla. The resulting mutator plasmid, unable to replicate in Y. enterocolitica, was called pMRS71.
To inactivate the lcrV gene on the pYV plasmid, mutator plasmid pMRS71 (_lcrV_Δ3-324) was transferred into Y. enterocolitica MRS40(pYV40) by conjugation, and allelic exchange was selected as described by Kaniga et al. (22) except that in the last step, we added 0.4 mM arsenite to the sucrose-containing plate to select for the pYV plasmid (31). The lcrV mutant pYV plasmid was designated pMRS4071.
In the course of the experiments (see Results) we observed that the deletion was larger than expected, and sequencing revealed that part of sycD was also deleted. This discrepancy presumably resulted from the fact that the design of the oligonucleotides was based on sequence from Y. pestis and not from Y. enterocolitica.
Construction of _lcrV_Δ2-32 and _lcrV_Δ224-266 alleles.
These two alleles were constructed by site-directed mutagenesis as described by Kunkel et al. (23) with single-stranded pMRS20 DNA as a template. A uracil-containing single-stranded pMRS20 was produced from E. coli CJ236 dut ung. The double-stranded DNAs obtained after in vitro synthesis of the second strand were introduced into E. coli LK111, and the mutated plasmids were screened by PCR.
Plasmid pMRS56 containing allele _lcrV_Δ2-32 was constructed by deletion of codons 2 to 32 with MIPA356 (5′-CGAGGGCGCCTTATTTAATATGGAAGAATTGGTTCAGTTAGT-3′), which introduced a _Nar_I restriction site.
Allele _lcrV_Δ224-266 in pMRS52, engineered with MIPA352 (5′-CCTCAAACCACCATTCACGGCGCCACCACCTGC-3′), lost codons 224 to 266 and gained a _Nar_I restriction site.
Construction of pGEX-derived recombinant plasmids.
The lcrG gene, amplified from pMRS44 with MIPA382 (5′-ACGTCGACAAGAAGGAGATATACATATG-3′) and MIPA383 (5′-GATGTCGACTTAAATAATTTGCCCT-3′), was cloned into the _Sal_I site of pMRS46, yielding plasmid pMRS75 (encoding GST-LcrV and LcrG). The _lcrV_Δ2-32 and _lcrV_Δ224-266 alleles were amplified from pMRS52 and pMRS56, respectively (with MIPA364 [5′-CGGAATTCTCATGATTAGAGCCTACG-3′] and MIPA64 [5′-ATGTCGACCTGTCGTCTCTTGTTG-3′], and introduced within _Eco_RI-_Sal_I sites of pMRS75, giving pMRS78 (encoding GST-LcrVΔ2-32 and LcrG) and pMRS83 (encoding GST-LcrVΔ224-266 and LcrG), respectively. The _Bam_HI-_Sal_I fragment, containing the gst-lcrG hybrid gene, from pMRS50 (encoding GST-LcrG and YopD) was then replaced by the _Bam_HI-_Sal_I fragment of pMRS75 to generate plasmid pMRS84 (encoding GST-LcrV and YopD). Plasmid pCN29 (encoding GST and YopD) is a pGEX derivative containing a PCR-amplified _Sal_I fragment of yopD (obtained with MIPA342 [5′-ATGTCGACTCAGACAACACCAAAAGC-3′] and MIPA343 [5′-ATGTCGACAAGAAGGAGATATACATATGAC-3′]) cloned in the _Xho_I site of the vector. Plasmid pCN40 is a pGEX derivative carrying the yopB gene, amplified with MIPA461 (5′-ACGGTCGACCAAAGGAGGATCTAG-3′) and MIPA462 (5′-GGATGAGCTCTTAAACAGTATGGGGTC-3′), cloned in the _Sal_I-_Sac_I sites, and the sycD gene, amplified with MIPA287 (5′-CTCAAGCTTAGCGGTCATGGGTTATCAA-3′) and MIPA 293 (5′-CGCAAGCTTAAGAAGGAGATATACATATGCAAC-3′), cloned in the _Hin_dIII site. Plasmid pCNG42 contains the same genes as pCN40 but also encodes a GST-LcrV fusion protein. The lcrV gene amplified with MIPA364 (5′-CGGAATTCTCATGATTAGAGCCTACG-3′) and MIPA64 (5′-ATGTCGACCTGTCGTCTCTTGTTG-3′) has been cloned in the _Eco_RI-_Sal_I sites. Plasmid pCNG50 contains gst-lcrV as in plasmid pCNG42 and the gene sycD as in plasmid pCN40.
Construction of vector pCNR26.
pCNR21 is a pBC19R derivative containing the yopE gene and the first 33 codons of sycE. By directed mutagenesis with MIPA395 (5′-TAAATGATGATATTTTCATATGTATTTCCTCCTTTGGCTATTAAAACAAG-3′), the Shine-Dalgarno (SD) sequence of the yopE gene was optimized (AAGGAGG) and a _Nde_I site was introduced, yielding plasmid pCNR26. The ATG within the _Nde_I site is localized nine nucleotides downstream from the SD sequence.
Constructions of the complementing clones.
A DNA fragment containing genes lcrG, lcrV, and sycD was amplified from pYV40 with MIPA354 (5′-CCAAAACCATATGAAGTCTTCCCA-3′) and MIPA122 (5′-CACAAGCTTGACCGACTCCAAT-3′). This _Nde_I-_Hin_dIII fragment was cloned into the same sites of pCNR26, yielding pMRS72 (replacement of yopE by lcrGVsycD).
Plasmid pMRS74 was constructed by cloning a PCR-amplified fragment (using MIPA121 [5′-TTTGGATCCTAATGAATTATCTCAC-3′] and MIPA317 [5′-CGGGGTACCTAAAACTTTCGGTTAATTAA-3′]) containing genes sycD, yopB, and yopD from pYV40 into the _Bam_HI-_Asp7_18 sites of pMSL56, leading to the replacement of cyaA by sycDyopBD.
RNA extraction and Northern blot analysis.
Total RNA of Y. enterocolitica was extracted as described by Lambert de Rouvroit et al. (24), 2 h after induction at 37°C. Electrophoresis, transfer, and hybridization with yop DNA used as probes were done as described by Cornelis et al. (12). The yopD probe was a PCR-amplified fragment of 500 bp obtained with primers MIPA316 (5′-GGAAGATCTCAAATTCTAAACAGTAACA-3′) and MIPA317 (5′-CGGGGTACCTAAAACTTTCGGTTAATTAA-3′).
Production and purification of hybrid GST fusion proteins.
The production and purification of GST fusion proteins were performed as described by Smith and Johnson (47). Briefly, overnight cultures were diluted in 10 ml of broth containing 200 μg of ampicillin/ml to an optical density at 600 nm (OD600) of 0.1. The culture was then incubated vigorously at 37°C until an OD600 of 0.8 to 1.0 was reached. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to a 1 mM final concentration, and the culture was incubated for an additional 3 h at 37°C. A total of 6 × 109 induced bacteria were resuspended and sonicated for 1 min in 1 ml of cold phosphate-buffered saline (PBS). The cell debris were pelleted by a 5-min centrifugation step in a microcentrifuge, and the supernatant was clarified by centrifugation at 10,000 × g for 30 min at 4°C. A total of 800 μl of cleared extract was mixed with 20 μl of glutathione-Sepharose (Pharmacia Biotech) equilibrated with PBS. After 1 to 2 h of incubation with gentle agitation at 4°C, glutathione beads were recovered by centrifugation and washed three times with 100 μl of cold PBS. For SDS-PAGE analysis, proteins were eluted from the glutathione beads by being boiled in sample buffer and were loaded on a gel.
RESULTS
LcrV is necessary for the secretion of YopB and YopD.
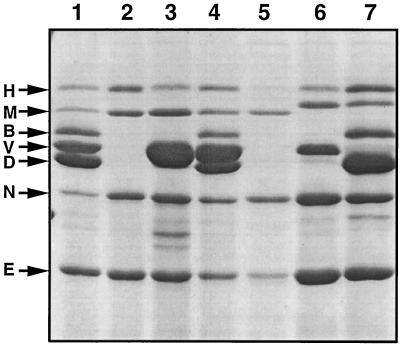
The hypothesis that LcrV is an element of the translocation apparatus implies that LcrV interacts with other Yops. Hence, small in-frame deletions, such as those previously described (37, 46), are likely to give rise to nonfunctional proteins that might interact with other proteins and lead to a biased phenotype. Therefore we engineered a nonpolar null mutation in lcrV by deletion of codons 3 to 324, giving strain MRS40(pMRS4071). The resultant lcrV mutant secreted more YopE, YopH, YopM, and YopN (Fig. 2, lane 2) than the wild type, but secretion of LcrV, YopB, and YopD was completely abolished. Surprisingly, the secretion defects for YopB and YopD could not be complemented by the introduction of plasmid pMRS28 (Fig. 2, lane 3) containing lcrRGV, which suggested that the mutation in lcrV could have a polar effect on the downstream sycDyopByopD genes. To address this question, we introduced plasmid pMRS72, containing lcrGVsycD, or plasmid pPW64, containing only sycD, into the lcrV mutant strain MRS40(pMRS4071). Plasmid pMRS72 restored the secretion of LcrV, YopB, and YopD but plasmid pPW64 could not (Fig. 2, lanes 4 and 5). The sycD gene in pPW64 was, however, functional because it could complement a previously characterized sycD mutation (Fig. 2, lanes 6 and 7). These results indicated that the lcrV mutation had an impact on sycD but not on yopB and yopD. They also showed that the lack of secretion of YopB and YopD was due not only to sycD deficiency but also to the loss of the lcrV gene. In order to understand the unexpected effect of our mutation on sycD, we sequenced the allele _lcrV_Δ3-324 in pMRS69. We found that the deletion extended not to codon 324 of lcrV but to nucleotide 109 of sycD and thus removed the whole lcrV gene and a part of sycD. This abnormal extension of the deletion could be explained by the fact that we designed our oligonucleotides based on the sequence of lcrGVHyopBD from Y. pestis and not from Y. enterocolitica. Hence, the genotype of pMRS4071 was quoted as lcrVsycD.
FIG. 2.
SDS-PAGE analysis of the Yops (identified at left) secreted by wild-type Y. enterocolitica MRS40(pYV40) (lane 1), by MRS40(pMRS4071), an lcrVsycD mutant (lane 2), by MRS40(pMRS4071)(pMRS28), an lcrVsycD mutant complemented with lcrRGV (lane 3; note the overproduction of LcrV), by MRS40(pMRS4071)(pMRS72), an lcrVsycD mutant complemented with lcrGVsycD (lane 4), by MRS40(pMRS4071)(pPW64), an lcrVsycD mutant complemented with sycD (lane 5), by KNG22703(pPW2269), a nonpolar sycD mutant (lane 6; note that YopD and YopB do not appear in the absence of SycD [53]), and by KNG22703(pPW2269)(pPW64), an sycD mutant complemented with sycD (lane 7) (Note that YopB and YopD are present. LcrV [V] cannot be distinguished clearly. However, overexpression of SycD does not affect LcrV production and secretion [not shown].) Note that the size of YopM varies between strain E40 and strain W22703.
To confirm that the lack of secretion of YopB and YopD did not result from a polar effect of the lcrVsycD mutation on the downstream yopB and yopD genes, we constructed a plasmid that contains sycDyopByopD genes cloned behind the strong yopE promoter. We introduced this plasmid, called pMRS74, into the lcrVsycD mutant strain MRS40(pMRS4071) and into the yopByopD double-mutant strain W22703(pGC153) (13). We observed that YopB and YopD secretion was restored in strain W22703(pGC153) (not shown), but it was not restored in strain MRS40(pMRS4071) (not shown). If we assume that complementing the nonpolar lcrVsycD mutation with a multicopy plasmid carrying sycD did not deeply modify the system, all these results indicate that LcrV is a factor specifically required for secretion of YopB and YopD.
LcrV is not required for transcription of yop genes.
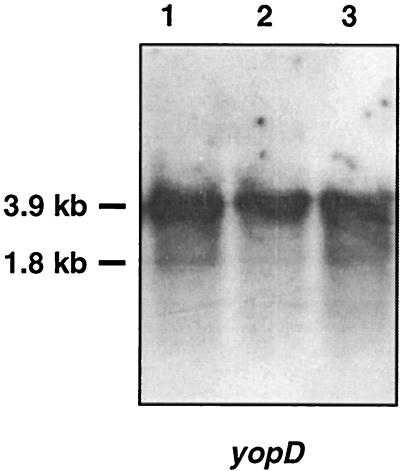
Since previous observations suggested that lcrV could be a regulator, we analyzed the transcription of yop genes in our lcrVsycD mutant. We carried out a Northern blot analysis on RNA extracted from the wild-type strain [MRS40(pYV40)], the lcrVsycD mutant strain [MRS40(pMRS4071)], and the lcrVsycD mutant complemented by sycD [MRS40(pMRS4071)(pPW64)] by using the yopD gene as a probe. As seen in Fig. 3, with wild-type bacteria, we observed a major yopD transcript of approximately 3,900 nucleotides and a minor transcript of about 1,800 nucleotides (5). The intensities of the transcripts did not differ among the wild type, the lcrV mutant, and the lcrV mutant complemented by sycD, indicating that LcrV is not required for transcription of yopD.
FIG. 3.
Northern blot analysis of RNA extracted from wild-type Y. enterocolitica MRS40(pYV40) (lane 1), from the lcrVsycD mutant MRS40(pMRS4071) (lane 2), and from the lcrVsycD mutant complemented by sycD, MRS40(pMRS4071)(pPW64) (lane 3). The probe was a PCR-amplified fragment internal to yopD.
LcrV is specifically required for the secretion step.
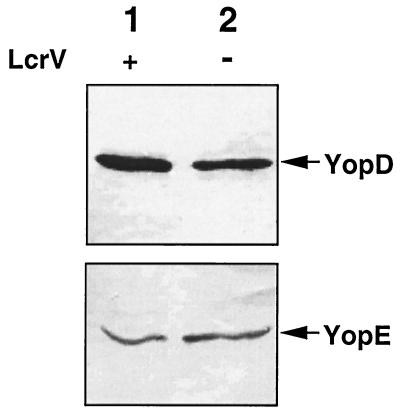
To determine at what level LcrV is required for secretion of YopB and YopD, we monitored the intracellular amounts of YopD and YopE (taken as a control) in wild-type MRS40(pYV40) and in the lcrVsycD mutant bacteria complemented by sycD [MRS40(pMRS4071)(pPW64)]. After temperature induction of Yop synthesis, total-cell lysates were separated by SDS-PAGE, and YopD and YopE were monitored by immunoblotting with rat monoclonal antibodies. As shown in Fig. 4, equivalent amounts of YopD and YopE could be detected in the total-cell extracts from the lcrV mutant MRS40(pMRS4071)(pPW64) and wild-type MRS40. From this result we concluded that the main effect of LcrV on YopD is to promote its secretion.
FIG. 4.
Western blot analysis of YopD and YopE in total-cell proteins using anti-YopD and anti-YopE monoclonal antibodies. Lane 1, wild-type Y. enterocolitica MRS40(pYV40); lane 2, MRS40(pMRS4071)(pPW64), an lcrVsycD mutant complemented with the sycD gene.
LcrV binds to YopB and YopD.
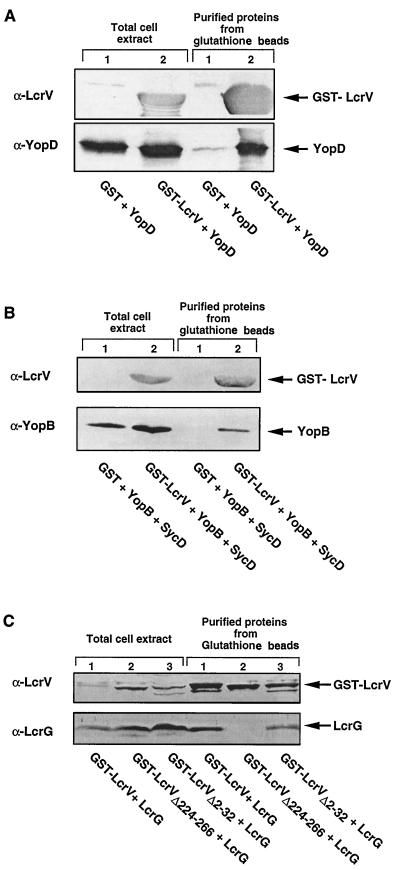
Since LcrV is involved in secretion of YopB and YopD, we tested whether it could interact with these proteins. In this attempt, we took advantage of the GST fusion protein expression and purification system (47). We constructed plasmid pMRS84, which encodes a GST-LcrV hybrid protein and YopD, in order to overproduce the two proteins simultaneously. To serve as a control, we constructed pCN29, which encodes GST and YopD separately. After production in E. coli XL1 Blue, GST-LcrV hybrid proteins were purified from the crude extracts with glutathione-Sepharose beads and were analyzed for the recovery of a second protein. As shown in Fig. 5A, when YopD was overproduced together with GST-LcrV, it copurified with GST-LcrV on the glutathione beads. In contrast, when YopD was overproduced together with GST instead of GST-LcrV, no copurification occurred, which indicated that YopD specifically binds to LcrV. In order to investigate the binding of YopB to GST-LcrV, we then constructed plasmid pCNG42, which encodes the fusion protein GST-LcrV and YopB but also SycD in order to avoid degradation of YopB (30). To serve as a control, we also constructed plasmid pCN40, which encodes GST and YopB separately as well as encoding SycD. After overproduction in E. coli XL1 Blue, the soluble extracts were incubated with glutathione-Sepharose beads. After the beads were washed, the purified proteins were analyzed by immunoblotting (Fig. 5B). As shown in Fig. 5B, YopB copurified with GST-LcrV but not with GST alone. To rule out any positive role of SycD in the binding of LcrV to YopB, we tested the capacity of LcrV to bind SycD. We cloned sycD downstream of gst-lcrV, giving plasmid pCNG50. The GST-LcrV hybrid protein was purified from the crude extract of E. coli carrying pCNG50 with glutathione-Sepharose beads and was analyzed for the recovery of SycD by immunoblotting. We could not copurify SycD with GST-LcrV (data not shown), which indicated that SycD does not bind to LcrV, a result that is in good agreement with the observation of Fields et al. (17). From these experiments, we conclude that LcrV binds to YopB and that SycD does not prevent this binding.
FIG. 5.
Binding of YopD, YopB, and LcrG to a GST-LcrV hybrid. (A) Western blot analysis with anti-LcrV and anti-YopD monoclonal antibodies of total-cell extracts and of the soluble extracts purified on glutathione-Sepharose beads from E. coli XL1(pCN29) overproducing GST and YopD (lanes 1) and XL1(pMRS84) overproducing GST-LcrV and YopD (lanes 2). (B) Western blot analysis with polyclonal anti-LcrV and monoclonal anti-YopB antibodies of the total-cell extracts and of the soluble extracts purified on glutathione-Sepharose beads from XL1(pCN40) overproducing GST, YopB, and SycD (lanes 1) and XL1(pCNG42) overproducing GST-LcrV, YopB, and SycD (lanes 2). (C) Western blot analysis with anti-LcrV and anti-LcrG polyclonal antibodies of the total-cell extracts and of the soluble extracts purified on glutathione-Sepharose beads from XL1(pMRS75) producing GST-LcrV and LcrG (lanes 1), XL1(pMRS78) producing GST-LcrVΔ224-266 and LcrG (lanes 2), and XL1(pMRS83) producing GST-LcrVΔ2-32 and LcrG (lanes 3).
LcrV binds to LcrG.
Since LcrG is encoded by the same large lcrGVHyopBD operon, we tested whether it could also interact with LcrV. To demonstrate such an association, we cloned the lcrG gene downstream of gst-lcrV, giving plasmid pMRS75. A soluble extract from E. coli carrying plasmid pMRS75 was mixed with glutathione-Sepharose beads, and proteins absorbed on the beads were analyzed by Western blotting. As shown in Fig. 5C, LcrG copurified with GST-LcrV. We have previously shown that binding between YopE and SycE and binding between YopH and SycH occur at definite domains of the Yop proteins (57). Therefore, we tested whether the association that we observed between LcrV and LcrG would also involve a specific domain of LcrV. To analyze this, we engineered two in-frame deletion mutations in lcrV, namely _lcrV_Δ2-32 and _lcrV_Δ224-266, and we substituted these deletants for lcrV in pMRS75, giving plasmids pMRS83 and pMRS78, respectively. Total-cell extracts from E. coli carrying either pMRS83 or pMRS78 were mixed with glutathione-Sepharose beads, and proteins absorbed on the beads were analyzed by immunoblotting. As shown in Fig. 5C, LcrG copurified with GST-LcrVΔ2-32 but not with GST-LcrVΔ224-266, suggesting that there is a unique LcrG-binding site situated in the carboxy-terminal domain of LcrV.
DISCUSSION
In this study, we attempted to construct a complete nonpolar deletion of lcrV in the pYV plasmid. Complementation and sequence analysis showed that the deletion also encompassed part of sycD. Nevertheless, the mutation did not prevent transcription of the distal yopB and yopD genes. The mutation abolished secretion of LcrV, YopB, and YopD. When lcrV and sycD were supplied in trans, secretion of YopB and YopD was restored. When sycD only was reintroduced in this mutant, secretion of YopB and YopD did not resume but YopD could be detected inside the bacteria, showing that the lack of secretion was not due to a lack of transcription or translation or to proteolysis. These observations strongly suggest that LcrV could be specifically involved in the process of release of YopB and YopD. If this is true, one would expect some interactions between LcrV and YopB or YopD. Using GST-LcrV fusion proteins, we observed that YopB or YopD could indeed be copurified with GST-LcrV. The same approach showed that the region spanning residues 224 to 266 of LcrV also interacts with LcrG. This result confirmed the previous findings of Nilles et al. (32) and suggests that LcrG is another piece of the translocation apparatus. In good agreement with results reported by Fields et al. (17), we did not observe any interaction between GST-LcrV and SycD, indicating that SycD does not act as a chaperone for LcrV as it does for YopD (53). We conclude from all these results that LcrV interacts with YopB and YopD to promote their release from the bacterium. How could this occur? One should point out first that, so far, the secretion domain has been clearly identified only for effector Yops but not for the translocator Yops (see reference 15 for a review). Thus, one cannot exclude the possibility that the Ysc secretion apparatus only recognizes LcrV or a complex involving LcrV, YopB, and YopD rather than YopB and YopD individually. Further work is required to clarify this.
Our results also show that when YopB and YopD are not secreted they do not obstruct the secretion channel. Indeed the lcrV mutant does not secrete YopB and YopD but secretes the effectors. This indicates that YopB and YopD without LcrV do not obstruct the secretion channel. In this respect, it would be worthwhile trying to localize YopB and YopD in the bacterium in the presence and in the absence of LcrV.
Finally, does LcrV only play a role in YopB and YopD secretion or does it also play a structural role in the translocation apparatus? We would like to suggest that LcrV is not only required for proper placement (localization) of YopB and YopD but that it also forms some kind of a short pilus below YopB and YopD. However, at this stage, this remains pure speculation.
According to our hypothesis, LcrV is expected to be essential for virulence because it is needed for the deployment of the translocation apparatus. This last conclusion is in perfect agreement with the conclusion drawn by Skrzypek and Straley (46), who showed that LcrV is essential for the virulence of Y. pestis.
Finally, our results rule out the idea issued from previous studies that LcrV is a regulator: yopD transcription was not affected in a mutant completely lacking LcrV and the YopD protein was clearly present in the extract from these mutant bacteria. Such observations may seem to be contradictory to those of Bergman et al. (5), who observed that a nonpolar deletion of Y. pseudotuberculosis lcrV, leaving a truncated gene of 750 bp, is severely downregulated in transcription of the lcrGVHyopBD operon and of yopE. We think that the discrepancy could be explained by the presence of a truncated LcrV in these previous studies. Indeed, if a truncated LcrV protein associates with YopB and YopD, such a complex might obstruct the secretion channel, which could result in turning on the feedback regulatory mechanism which prevents transcription of yop genes when Yops release is compromised (14, 34, 40, 51a). Experiments using Y. pestis by Price et al. (37) and by Skrzypek and Straley (46) also led to the conclusion that LcrV plays a regulatory role. However, in Y. pestis, this type of analysis is hampered by the fact that the Yops are attacked by the plasminogen activator protease Pla (43, 48) and Yops are detected by immunoblotting. The analysis of the lcrV mutants of Y. pestis focused on the secretion of YopM rather than YopB and YopD, which may have been misleading. However, in their more recent work showing the interaction between LcrV and LcrG, Nilles et al. (32) suggest that LcrV could function in the control of secretion. Thus, we think that the data presented in this paper can be conciliated with previous observations and promote a reevaluation of the role of LcrV in Yop secretion.
ACKNOWLEDGMENTS
We acknowledge D. Desnoeck and I. Lambermont for excellent technical assistance and Scott Mills for a critical reading and discussions. We are grateful to Patrick Gildemeester for participating in the formation of some of the genetic constructs.
M.R.S. was the recipient of a fellowship from ICP. C.N and I.S. were recipients of fellowships from the Belgian “Fonds pour la Formation à la Recherche dans l’Industrie et l’Agriculture.” This work was supported by the Belgian “Fonds National de la Recherche Scientifique Médicale” (Convention 3.4595.97), the “Direction Générale de la Recherche Scientifique-Communauté Française de Belgique” (Action de Recherche Concertée” 94/99-172), and the “Interuniversity Poles of Attraction Program—Belgian State, Prime Minister’s Office, Federal Office for Scientific, Technical and Cultural Affairs” (PAI 4/03).
REFERENCES
- 1.Allaoui A, Scheen R, Lambert de Rouvroit C, Cornelis G R. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to ExsB of Pseudomonas aeruginosa. J Bacteriol. 1995;177:4230–4237. doi: 10.1128/jb.177.15.4230-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui A, Woestyn S, Sluiters C, Cornelis G R. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergman T, Erickson K, Galyov E, Persson C, Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman T, Hakansson S, Forsberg A, Norlander L, Macellaro A, Backman A, Bolin I, Wolf-Watz H. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J Bacteriol. 1991;173:1607–1616. doi: 10.1128/jb.173.5.1607-1616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodeus M, Sory M-P, Fang J C, Janssens M, Delferriere N, Cornelis G, Wauters G, Burtonboy G. Production of rat hybridomas directed against Yersinia enterocolitica. In: Bazin H, editor. Rat hybridoma and rat monoclonal antibodies. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 335–338. [Google Scholar]
- 7.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows T W, Bacon G A. The basis of virulence in Pasteurella pestis: an antigen determining virulence. Br J Exp Pathol. 1956;37:481–493. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 10.China B, Michiels T, Cornelis G R. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol Microbiol. 1990;4:1585–1593. doi: 10.1111/j.1365-2958.1990.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis G R, Colson C. Restriction of DNA in Yersinia enterocolitica detected by recipient ability for a derepressed R factor from Escherichia coli. J Gen Microbiol. 1975;87:285–291. doi: 10.1099/00221287-87-2-285. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis G R, Sluiters C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis G R, Sory M P, Laroche Y, Derclaye I. Genetic analysis of the plasmid region controlling virulence in Yersinia enterocolitica 0:9 by mini-Mu insertions and lac gene fusions. Microb Pathog. 1986;2:367–379. doi: 10.1016/0882-4010(86)90067-7. [DOI] [PubMed] [Google Scholar]
- 14.Cornelis G R, Vanooteghem J C, Sluiters C. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb Pathog. 1987;2:367–379. doi: 10.1016/0882-4010(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 15.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 16.Fields K A, Plano G V, Straley S C. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields K A, Williams A W, Straley S C. Failure to detect binding of LcrH to the V antigen of Yersinia pestis. Infect Immun. 1997;65:3954–3957. doi: 10.1128/iai.65.9.3954-3957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg A, Viitanen A M, Skurnik M, Wolf-Watz H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 19.Frithz-Lindsten E, Rosqvist R, Johansson L, Forsberg A. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensable for targeting to the secretion loci. Mol Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- 20.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 21.Håkansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Lambert de Rouvroit C, Sluiters C, Cornelis G R. Role of the transcriptional activator VirF in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol Microbiol. 1992;6:395–409. [PubMed] [Google Scholar]
- 25.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells. Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 26.Michiels T, Vanooteghem J C, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis G R. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motin V L, Nakajima R, Smirnov G B, Brubaker R R. Passive immunity to yersiniae mediated by anti-recombinant V antigen and protein A-V antigen fusion peptide. Infect Immun. 1994;62:4192–4201. doi: 10.1128/iai.62.10.4192-4201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulder B, Michiels T, Simonet M, Sory M P, Cornelis G R. Identification of additional virulence determinants on the pYV plasmid of Yersinia enterocolitica W227. Infect Immun. 1989;57:2534–2541. doi: 10.1128/iai.57.8.2534-2541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neyt, C., and G. R. Cornelis. Unpublished data.
- 31.Neyt C, Iriarte M, Ha Thi V, Cornelis G R. Virulence and arsenic resistance in yersiniae. J Bacteriol. 1997;179:612–619. doi: 10.1128/jb.179.3.612-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilles M L, Williams A W, Skrzypek E, Straley S C. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson C, Nordfelth R, Holmström A, Hakansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 34.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 35.Plano G V, Straley S C. Multiple effects of lcrD mutations in Yersinia pestis. J Bacteriol. 1993;175:3536–3545. doi: 10.1128/jb.175.11.3536-3545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plano G V, Straley S C. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J Bacteriol. 1995;177:3843–3854. doi: 10.1128/jb.177.13.3843-3854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price S B, Cowan C, Perry R D, Straley S C. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2+-dependent growth and maximal expression of low-Ca2+ response virulence genes. J Bacteriol. 1991;173:2649–2657. doi: 10.1128/jb.173.8.2649-2657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price S B, Leung K Y, Barve S S, Straley S C. Molecular analysis of lcrGVH, the V antigen operon of Yersinia pestis. J Bacteriol. 1989;171:5646–5653. doi: 10.1128/jb.171.10.5646-5653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price S B, Straley S C. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun. 1989;57:1491–1498. doi: 10.1128/iai.57.5.1491-1498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rimpiläinen M, Forsberg A, Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J Bacteriol. 1992;174:3355–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roggenkamp A, Geiger A M, Leitritz L, Kessler A, Heesemann J. Passive immunity to infection with Yersinia spp. mediated by anti-recombinant V antigen is dependent on polymorphism of V antigen. Infect Immun. 1997;65:446–451. doi: 10.1128/iai.65.2.446-451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosqvist R, Magnusson K E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sample A K, Brubaker R R. Post-translational regulation of Lcr plasmid-mediated peptides in pesticinogenic Yersinia pestis. Microb Pathog. 1987;3:239–248. doi: 10.1016/0882-4010(87)90057-x. [DOI] [PubMed] [Google Scholar]
- 44.Sarker M R, Cornelis G R. An improved version of suicide vector pKNG101 for gene replacement in gram-negative bacteria. Mol Microbiol. 1997;23:410–411. doi: 10.1046/j.1365-2958.1997.t01-1-00190.x. [DOI] [PubMed] [Google Scholar]
- 45.Sarker, M. R., M.-P. Sory, A. P. Boyd, M. Iriarte, and G. R. Cornelis. Unpublished data.
- 46.Skrzypek E, Straley S C. Differential effects of deletions in lcrV on the secretion of V antigen, regulation of the low-Ca2+ response, and virulence of Yersinia pestis. J Bacteriol. 1995;177:2530–2542. doi: 10.1128/jb.177.9.2530-2542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 48.Sodeinde O A, Sample A K, Brubaker R R, Goguen J D. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect Immun. 1988;56:2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sory M-P, Boland A, Lambermont I, Cornelis G R. Identification of YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sory M-P, Cornelis G R. Yersinia enterocolitica O:9 as a potential live oral carrier for protective antigens. Microb Pathog. 1988;4:431–442. doi: 10.1016/0882-4010(88)90028-9. [DOI] [PubMed] [Google Scholar]
- 51.Sory M-P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 51a.Stainier I, Iriarte M, Cornelis G R. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol. 1997;26:833–843. doi: 10.1046/j.1365-2958.1997.6281995.x. [DOI] [PubMed] [Google Scholar]
- 52.Straley S C, Perry R D. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995;3:310–317. doi: 10.1016/s0966-842x(00)88960-x. [DOI] [PubMed] [Google Scholar]
- 53.Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 55.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 56.Woestyn S, Allaoui A, Wattiau P, Cornelis G R. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woestyn S, Sory M-P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]