Induced Levels of Heat Shock Proteins in a dnaK Mutant of Lactococcus lactis (original) (raw)
Abstract
The bacterial heat shock response is characterized by the elevated expression of a number of chaperone complexes and proteases, including the DnaK-GrpE-DnaJ and the GroELS chaperone complexes. In order to investigate the importance of the DnaK chaperone complex for growth and heat shock response regulation in Lactococcus lactis, we have constructed two dnaK mutants with C-terminal deletions in dnaK. The minor deletion of 65 amino acids in the dnaKΔ2 mutant resulted in a slight temperature-sensitive phenotype. BK6, containing the larger deletion of 174 amino acids (dnaKΔ1), removing the major part of the inferred substrate binding site of the DnaK protein, exhibited a pronounced temperature-sensitive phenotype and showed altered regulation of the heat shock response. The expression of the heat shock proteins was increased at the normal growth temperature, measured as both protein synthesis rates and mRNA levels, indicating that DnaK could be involved in the regulation of the heat shock response in L. lactis. For Bacillus subtilis, it has been found (A. Mogk, G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann, EMBO J. 16:4579–4590, 1997) that the activity of the heat shock repressor HrcA is dependent on the chaperone function of the GroELS complex and that a dnaK insertion mutant has no effect on the expression of the heat shock proteins. The present data from L. lactis suggest that the DnaK protein could be involved in the maturation of the homologous HrcA protein in this bacterium.
DnaK is a bacterial member of the highly conserved, ubiquitous family of 70-kDa heat-shock-induced chaperone proteins (Hsp70 proteins). Genes encoding DnaK have been sequenced from many species, but functional studies have mainly been carried out with eucaryotes and with the gram-negative bacterium Escherichia coli. More recently, such studies have also been conducted with the gram-positive bacterium Bacillus subtilis (34, 43). Studies with E. coli have shown that DnaK functions as a chaperone in collaboration with DnaJ and GrpE and that this chaperone complex plays a significant role in the folding of nascent protein chains during normal growth conditions and in the refolding of proteins after thermal damage (5, 6, 8, 36, 44, 53; for reviews, see references 4 and 17). Furthermore, the DnaK-DnaJ-GrpE chaperone complex participates in ATP-dependent proteolysis in the cell (for a review, see reference 31). The eucaryotic DnaK homolog (Hsp70) was shown to contain an amino-terminal ATP binding domain and a substrate binding domain located immediately after the ATP binding domain. A study of the binding properties of an internal Hsp70 polypeptide covering amino acids Ser-384 to Glu-543, located immediately after the ATPase domain, indicated that the peptide binding domain of Hsp70 is confined within this fragment (51). The C-terminal fragment from amino acid 546 (9) was found to be involved in neither ATP nor substrate binding, yet several studies have indicated that the C terminus is needed for full DnaK activity in eucaryotes (9, 14).
A comparison of the data from E. coli and B. subtilis suggests essential differences between the functions of DnaK in these two bacterial genera. In E. coli, dnaK expression is induced by heat as well as by other stress factors, such as acid stress (19), osmotic stress (33), and carbon starvation (23), indicating that DnaK is involved in the general stress response of E. coli. This finding has been confirmed by the phenotypes of isolated dnaK mutants (5, 7, 33, 45). In B. subtilis, DnaK is induced by heat but not by the addition of salt or by glucose limitation (50), indicating a more limited role in stress response than that found in E. coli. This finding is in accordance with the phenotype displayed by a dnaK insertion mutant (43).
The molecular bases for the regulation of the expression of dnaK and the major heat shock genes are very different in E. coli and B. subtilis. In E. coli, transcription of the dnaKJ operon is stimulated by increased amounts of the heat shock sigma factor ς32 at elevated temperatures. At normal temperatures, ς32 is highly unstable and is degraded by the proteolytic activity of the HflB (FtsH) protease in consort with the DnaK chaperone complex. DnaK is thus a negative factor in the regulation of the heat shock response in E. coli. Elevated growth temperatures prevent the degradation of ς32, presumably by sequestering the DnaK chaperone complex with misfolded proteins, thus inhibiting the degradation of ς32. In accordance with this model, mutations in dnaK, dnaJ, and grpE result in increased expression of the heat shock genes, even in the exponential growth phase, at normal temperatures (46, 48).
In B. subtilis, three classes of heat-induced genes have been found. The dnaK operon, belonging to class I, has been shown to be negatively regulated by a repressor. The repressor is encoded by the hrcA gene, the first gene in the dnaK operon (42, 55). The hrcA gene is followed by grpE, dnaK, dnaJ, and three other genes of unknown function (18, 21, 42). An hrcA deletion mutation results in high levels of constitutive expression of both the dnaK operon and the groESL operon, which is also of class I. The groESL operon encodes another important chaperone complex. Inactivation of dnaK in B. subtilis results in a slight temperature-sensitive phenotype, and no increase in the expression of other genes in the dnaK and the groELS operons has been observed (34). Thus, apparently DnaK is not involved in the regulation of the expression of general heat shock-induced chaperones in B. subtilis. The operator regions in front of the dnaK and groELS operons in B. subtilis contain binding sites for the HrcA repressor. The operator sequences are termed CIRCE, for controlling inverted repeats for chaperone expression, are preserved in many bacterial genera, and are found in the corresponding operons encoding the major heat shock-induced chaperones (18, 38).
The gram-positive bacterium Lactococcus lactis has been found to elicit a heat shock response similar to that of other bacteria (1, 28, 52), and the temporal induction patterns suggest that the heat shock proteins fall in more than one induction class. Apparently, DnaK, GroEL, and GroES fall in the same class, consistent with the finding that both the hrcA (orf1)-grpE-dnaK operon and the groESL operon of L. lactis contain CIRCE-like elements in the promoter regions (13, 29). Also, the dnaJ gene has been shown to contain the CIRCE element (49). Since dnaK mutants of E. coli and B. subtilis have different phenotypes, we wanted to analyze whether the heat shock response is altered in dnaK mutants of L. lactis. For this purpose, two C-terminal deletion mutants were constructed. The most severe mutation removed a major part of the putative substrate binding site of DnaK. Like the E. coli dnaK mutants, this mutant was found to be temperature sensitive for growth and to contain elevated levels of other heat shock proteins at 30°C. In contrast to the E. coli mutants, the lactococcal mutant was able to develop thermotolerance, although not as efficiently as the wild type. Possible implications of these results are discussed.
In the second dnaK mutant, only the extreme C-terminal part of the dnaK gene was deleted. This mutant differed slightly from the wild type with respect to temperature sensitivity.
MATERIALS AND METHODS
Strains and growth conditions.
Table 1 lists the strains used in the present study and how they were constructed. The lactococcal strains used are all derivatives of the plasmid-free prophage-cured strain MG1363 (15). For the growth of L. lactis strains, M17 medium (47) and chemically defined SA medium (24) supplemented with 0.5% glucose (GM17) and 1% glucose, respectively, were used. When required, erythromycin was added at a final concentration of 2 μg/ml. E. coli recombinant strains were grown in LB medium (40) with the addition of ampicillin at 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or construction |
|---|---|---|
| Strains | ||
| Lactococcus lactis subsp. cremoris | ||
| MG1363 | Wild type | 15 |
| BK8 | dnaK+-erm | MG1363 dnaK::pBK101 (integration) |
| BK6 | dnaKΔ1-erm | MG1363 dnaK::pBK103 (integration) |
| BK11 | dnaKΔ2-erm | MG1363 dnaK::pBK105 (integration) |
| Escherichia coli XL1-Blue | endA1 hsdR17 recA | Stratagene |
| Plasmids | ||
| pFI573 | dnaK+ | 13 |
| pBluescriptSKII+ | Apr | Stratagene |
| pBK100 | Apr | _Eco_RI-_Sca_I fragment from pFI573 in pBluescriptSKII+ |
| pBK101 | Apr Emr | erm cassette (_Bam_HI fragment) from pUC7erm in pBK100 |
| pBK102 | Apr | _Hpa_I-_Pst_I fragment from pFI573 in pBluescriptSKII+ |
| pBK103 | Apr Emr | erm cassette (_Bam_HI fragment) from pUC7erm in pBK102 |
| pBK104 | Apr | _Eco_RI-_Hae_II fragment from pFI573 in pBluescriptSKII+ |
| pBK105 | Apr Emr | erm cassette (Bam_HI fragment) from pUC7_erm in pBK104 |
| pUC7_erm_ | Emr | Willem de Vos (10) |
For the determination of growth rates, the Bioscreen C (Labsystems) growth monitoring system was used. The optical density at 600 nm (OD600) was monitored every 15 min during the incubation period. The microtiter plate was shaken before each measurement. Each well contained 400 μl of growth medium and was inoculated with 4 μl of overnight culture. In each experiment, five growth curves were monitored. Growth rates were calculated by linear regression on the linear part of an average growth curve (typically in the OD600 range of 0.3 to 0.8), and growth rates from independent experiments were compared. When MG1363 was grown at 30°C, a series of five experiments gave an average growth rate of 56.8 min with a standard deviation of 3.5. The average growth rate for BK6 was 116.2 min with a standard deviation of 1.2.
Analysis of thermotolerance.
At an OD600 of 0.4, cells from exponentially growing cultures in GM17 were harvested and resuspended in the same volume of GM17 with or without erythromycin in glass tubes with a diameter of 1 cm. After 30 min of incubation at 30 or 40°C, the tubes were placed in a 53°C water bath. The number of viable cells was determined as CFU on GM17 agar plates after 0, 15, 30, 45, and 60 min of incubation at 53°C.
DNA manipulations.
The plasmids constructed in the present study are listed in Table 1. The plasmids were selected in E. coli XL1-Blue (Stratagene). For plasmid construction, standard E. coli techniques were used (40). Southern analysis was carried out as previously described (30) with lactococcal chromosomal DNA prepared as previously described (25).
PCR amplification was carried out either with purified chromosomal DNA or directly as colony PCR with a smear of a bacterial colony as the substrate under standard conditions. For colony PCR with Lactococcus colonies, the cells were treated with 0.1 M NaOH for 30 min at 97°C, vortexed with glass beads, and neutralized with HCl and Tris (pH 7.5) prior to PCR.
Plasmid construction.
Plasmid pBK100 was constructed by inserting a _Sca_I-_Eco_RI fragment from pFI573, containing part of dnaK, into the _Sma_I and _Eco_RI sites of the pBluescriptSKII+ vector. Plasmid pBK102 was constructed by inserting an _Hpa_I-_Pst_I fragment from pFI573 into the _Pst_I and _Hin_cII sites of the pBluescriptSKII+ vector. Plasmid pBK104 was constructed by inserting an _Hae_II-_Eco_RI fragment from pFI573 into the _Sma_I and _Eco_RI sites of the pBluescriptSKII+ vector. The _Hae_II site had been blunted with T4 DNA polymerase before pFI573 was digested with _Eco_RI. Plasmids pBK101, pBK103, and pBK105 were constructed by inserting a _Bam_HI fragment containing a functional erm gene from plasmid pUC7erm (10) into the _Bam_HI sites of plasmids pBK100, pBK102, and pBK104, respectively.
Integration of plasmids into the L. lactis chromosome.
Competent L. lactis cells were transformed by electroporation essentially as previously described (20). One microgram of plasmid DNA was used in each transformation. Selection of erythromycin-resistant transformants was performed on SR plates (20) containing 2 μg of erythromycin per ml.
Confirmation of plasmid integration.
To confirm the site of integration, Southern analysis and PCR analysis were carried out. The chromosomal DNA was digested with _Pst_I; an internal _Eco_RI-_Pst_I fragment from dnaK was used as a probe. For MG1363, a single fragment larger than 10 kb hybridized to the probe. The integrants BK6, BK8, and BK11 all gave two fragments: one of the same size as that found in MG1363 and the additional fragment having a mobility corresponding to the theoretical size of 4,948, 5,276, or 5,074 bp, respectively. PCR analysis was carried out either with chromosomal DNA or directly as colony PCR. The following PCR primers were used: MKP1 (5′-GCA ACT GCT GAA AGC TAC CTT-3′), which corresponds to a sequence in dnaK before the site of integration; ERM1 (5′-CTA TGA GTC GCT TTT GT-3′) and ERM2 (5′-GTT TCC GCC ATT CTT TG-3′), which both correspond to sequences in the erythromycin resistance cassette; and PCK3719 (5′-GTC GCC ATC AAA TGT ATT-3′), which corresponds to a sequence in the C-terminal part of dnaK. When primers MKP1 and ERM1 were used, PCR products corresponding to the theoretical sizes of 1,112 and 1,446 bp were produced from BK6 and BK11, respectively. BK8 gave a PCR product corresponding to the theoretical size of 1,670 bp when primers MKP1 and ERM2 were used.
PCK3719 and MKP1 were used to confirm the absence of intact dnaK in BK6 and BK11. In these experiments, BK8 and MG1363 were used as controls and gave the expected 1,495-bp PCR product, while no PCR product was obtained from BK6 and BK11.
Curing of L. lactis strains for the integrated plasmids.
Liquid GM17 was inoculated with integrants grown on a GM17 agar plate containing erythromycin, and the culture was incubated overnight. A 1% dilution of the overnight culture in GM17 was subsequently incubated overnight. Cured strains were obtained from this culture by screening for erythromycin-sensitive colonies.
Northern blotting.
At an OD600 of 0.4, cells from GM17 cultures were harvested either directly or after incubation for 15 min at 43°C. RNA isolation and Northern blotting were carried out as previously described (1). The following probes were used for hybridization: a 1,704-bp (dnaK) _Dra_I fragment from pFI573 (13), a 576-bp (orf1) _Hin_dIII-_Bst_EII fragment from pFI573, a 1,392-bp (ftsH) _Hin_dIII fragment from pLN32 (35), and a 726-bp (dnaJ) _Hin_dIII fragment from pKS2 (1).
Western blotting.
Total cell proteins were extracted essentially as described previously (28) from cells harvested at an OD600 of 0.4. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) was carried out on a 12% acrylamide gel (29:1 acrylamide-bisacrylamide; Bio-Rad). The gel was transferred by semidry electroblotting to an Immobilon-P membrane (Millipore) as recommended by the supplier. Chemiluminescence detection of DnaK with rabbit antibodies against DnaK from B. subtilis and horseradish peroxidase-conjugated secondary antibodies was performed with an Amersham ECL Western blotting analysis system.
2D gel analysis.
Cells were grown in SA medium with reduced amounts of unlabelled methionine. Labelling with [35S]methionine, harvesting, extraction, and two-dimensional (2D) gel analysis were performed as described previously (28).
RESULTS
Construction of dnaK mutants.
The dnaK operon from L. lactis, consisting of hrcA (orf1)-grpE-dnaK, was cloned and sequenced by Eaton et al. (13). Downstream of dnaK a putative transcriptional terminator structure was identified; the structure was followed by an open reading frame (orf4) with no similarity to known protein genes. Transcriptional analysis previously showed that orf4 expression was not induced by heat shock, in contrast to the expression of the dnaK operon, arguing that transcription does not proceed past the terminator structure (1). Therefore, since dnaK is the last gene of the operon, it should be possible to delete different parts of the dnaK gene in L. lactis by insertion mutagenesis without creating polar effects on downstream genes in the operon. We used this strategy to construct two different dnaK deletion mutants of MG1363 by homologous recombination of nonreplicating plasmids carrying internal fragments of the dnaK operon into the chromosome.
Mutant BK6 was obtained by integration of plasmid pBK103, which contains an 873-bp _Hpa_I-_Pst_I internal fragment of dnaK. The resulting C-terminal deletion (dnaKΔ1) from the _Pst_I site in dnaK is shown in Fig. 1B. Similarly, mutant BK11 (dnaKΔ2) was obtained by integration of pBK105, which contains a 999-bp _Eco_RI-_Hae_II fragment of dnaK. The resulting C-terminal deletion from the _Hae_II site in dnaK is shown in Fig. 1B. In order to obtain an erythromycin-resistant control strain for the BK6 and BK11 mutants, we integrated plasmid pBK101 into strain MG1363. Plasmid pBK101 contains a 1,201-bp _Eco_RI-_Sca_I DNA fragment with most of the dnaK gene, including the entire C-terminal sequence. The integration event resulted in strain BK8, which carried no gene disruptions. The correct integration of the plasmids was verified by both PCR and Southern blotting as described in Materials and Methods (data not shown).
FIG. 1.
Physical map of the dnaK region in wild-type strain MG1363 and dnaK mutants and Western blot analysis of mutant proteins. (A and B) Gray boxes represent structural genes, and the gene designation is indicated. (A) Physical map of the hrcA-grpE-dnaK operon in MG1363. (B) Extent of dnaK sequences in the hrcA-grpE-dnaK operon in mutant strains BK6 and BK11, shown by lines below a partial restriction map of the dnaK gene. The mutants were constructed by homologous recombination into the chromosome of various plasmids (see text for details). (C) Functional domains recognized for the DnaK protein from E. coli and the eucaryotic DnaK homologs Hsp70. (D) Western blot analysis of mutant proteins with antibodies against DnaK from B. subtilis. Proteins were extracted from exponentially growing cells and separated by SDS-PAGE. Following transfer of the protein bands to an Immobilon-P membrane, the DnaK protein was detected by chemiluminescence with an Amersham ECL Western blotting analysis system and antibodies against DnaK from B. subtilis. The approximate sizes of the proteins are shown above the bands.
In the dnaKΔ1 mutant BK6, 522 bp of the 3′ end of the dnaK gene is deleted, specifying 174 amino acid residues, while the vector sequence adds 84 bp to the reading frame before a stop codon is encountered. Thus, a truncated DnaK protein of 49.4 kDa should be synthesized. In the dnaKΔ2 mutant BK11, 188 bp (65 amino acids) is deleted, and a truncated DnaK protein of 58.8 kDa should be synthesized when the addition of five vector-specified codons is taken into account. The full-length DnaK protein has a calculated molecular mass of 65.0 kDa. We tested if it was possible to identify the smaller DnaK proteins in the mutant strains by means of Western blotting with antibodies raised against the purified DnaK protein from B. subtilis (43). As shown in Fig. 1D, it was possible to detect mutant proteins with apparent sizes of 50 and 65 kDa (DnaKΔ1 and DnaKΔ2, respectively). The wild-type DnaK protein had an apparent molecular mass of 70 kDa, as determined previously (28).
Physiological characterization of mutants.
Heat sensitivity is encountered for all dnaK mutants of both E. coli and B. subtilis. We therefore tested the heat sensitivity of the lactococcal dnaK mutants by their growth on GM17 agar at temperatures of 10 to 37°C. At 30°C, which is the standard growth temperature for L. lactis, colonies with a 1-mm diameter were formed after 1 day of incubation for both dnaK+ strains MG1363 and BK8 and the dnaKΔ2 mutant BK11, while the dnaKΔ1 mutant BK6 required 2 days at 30°C (Table 2). An equal reduction in the growth rate of BK6, compared to the other strains, was observed when the strains were grown in liquid GM17 at 30°C (Table 2). When the growth rates of MG1363 and BK8 were compared, a small negative effect of the addition of erythromycin was seen. It was also evident that the dnaKΔ2 mutation in BK11 caused a growth rate slightly lower than that of BK8 containing the wild-type dnaK allele.
TABLE 2.
Growth of dnaK mutants at various temperatures
| Strain | Relevant genotype | Doubling time at 30°C on GM17 (min) | Incubation time (days) needed to obtain colonies of 1 mm on GM17 agar plates at the following temp (in °C): | ||||
|---|---|---|---|---|---|---|---|
| 30 | 33 | 35 | 37 | 10 | |||
| MG1363 | Wild type | 57 | 1 | 1 | 1 | 2 | 6 |
| BK8 | dnaK+-erm | 62 | 1 | 1 | 1 | 2 | 6 |
| BK11 | dnaKΔ2-erm | 69 | 1 | 1 | 1 | NG | 6 |
| BK6 | dnaKΔ1-erm | 120 | 2 | NGa | NG | NG | 10–11 |
At higher temperatures (Table 2), the growth of BK6 decreased with increasing temperature until growth stopped at 35°C. Concomitant with the lower growth rates, the sizes of individual colonies became much more variable and a smaller fraction of cells survived the challenge. MG1363, BK8, and BK11 all grew as well at 33 and 35°C as at 30°C, but at 37°C the dnaK+ strains showed approximately half the growth rate found at 30°C while BK11 grew considerately more slowly and with variable colony sizes.
The ability of the mutants to grow at low temperatures was tested by plating at 10°C. All strains showed growth rates approximately 15 to 20% the growth rate found at 30°C. It took MG1363, BK8, and BK11 6 days to form 1-mm colonies at 10°C (Table 2) and 1 day to do so at 30°C (Table 2), and it took BK6 10 to 11 days to form colonies at 10°C and 2 days to do so at 30°C. Thus, neither of the dnaK mutants showed increased cold sensitivity.
In order to confirm that the observed heat-sensitive phenotype of strain BK6 containing the dnaKΔ1 mutation was due to the deletion in dnaK, the strain was cured of the inserted plasmid by growth in the absence of erythromycin and screening for erythromycin-sensitive revertants as described in Materials and Methods. These revertants, verified by PCR for the loss of the plasmid, were able to plate at 35 and 37°C, like MG1363, and showed the same growth rate as MG1363 at 30°C in GM17 (data not shown). Thus, the temperature-sensitive phenotype of BK6 was indeed due to the dnaKΔ1 mutation present in this strain.
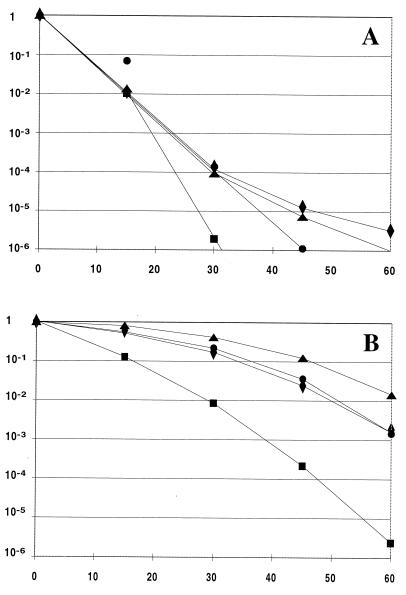
The heat sensitivity of the dnaK mutants was further analyzed by challenging exponentially growing cells at 30°C to the lethal temperature of 53°C for 60 min and monitoring the fraction of surviving cells by measuring CFU; the results are shown in Fig. 2A. The dnaKΔ1 mutant BK6 appeared to be more heat sensitive than the control strain, while no heat-sensitive phenotype could be attributed to the dnaKΔ2 mutant BK11.
FIG. 2.
Survival of dnaK mutants at 53°C. Exponentially growing cultures in GM17 were harvested and resuspended in fresh medium. (A) The washed cells were incubated at 30°C for 30 min and then at 53°C for the indicated times (minutes) (x axis). The fractions of surviving cells were determined as CFU at 30°C on GM17 agar plates and are given relative to those for untreated cultures (y axis). (B) The effects of preincubating the cultures at 40°C were determined as described above, except that cells were incubated for 30 min at 40°C instead of at 30°C before subjection to 53°C. Symbols: diamonds, MG1363; triangles, BK8 (wild type); circles, BK11 (dnaKΔ2); squares, BK6 (dnaKΔ1).
Induction of thermotolerance has been found to be optimal when L. lactis is preheated for 30 min at 40°C before incubation at 53°C (3). These conditions were therefore used for testing the dnaK mutants for thermotolerance (for further details of the performance of the experiment, see Materials and Methods). The data shown in Fig. 2B indicate that the acquired thermotolerance of the dnaKΔ1 mutant BK6 was significantly lower than that of the control strains MG1363 and BK8. From the data in Fig. 2A, however, it is evident that the mutant strain showed significantly better survival after pretreatment at 40°C, so the acquisition of thermotolerance by exposure to sublethal temperatures is possible, even with a severely damaged DnaK protein. The induction of thermotolerance in the dnaKΔ2 mutant BK11 was not significantly different from that in the control strains.
It was previously shown that the heat shock-induced chaperones DnaK, GroEL, and GroES are induced during salt stress (28), suggesting a function of the chaperones under this condition. Therefore, we wanted to determine whether a functional DnaK protein is required for growth at high salt concentrations. The addition of 2.5 to 4% NaCl reduced the growth rate of MG1363 in GM17 (data not shown). The growth rate was reduced to the same degree for both the dnaK mutants BK11 and BK6 and the wild-type control strain BK8. Thus, neither of the mutants showed altered salt sensitivity.
Induction of heat shock proteins by the dnaKΔ1 mutation.
In E. coli, dnaK mutants show increased synthesis of heat shock-regulated genes, compared to the wild-type strain, when grown at the normal growth temperature (17, 46, 48). This was not found to be the case for a dnaK mutant of B. subtilis (43). It was therefore of considerable interest to test the rate of synthesis of heat shock-regulated proteins in the dnaKΔ1 mutant of L. lactis. The synthesis of both mRNA and proteins was monitored.
By Northern blotting, we determined the levels of heat shock mRNAs in the two dnaK mutants. As shown in Table 3, _hrcA_-, _dnaK_-, and _dnaJ_-specific mRNA levels were greatly elevated in the dnaKΔ1 mutant BK6 (14-, 7.3-, and 2.3-fold for the three mRNA species, respectively). In the dnaKΔ2 mutant BK11, the mRNA levels were also elevated; however, in this strain they were only slightly elevated (3.1-, 3.2-, and 1.2-fold, respectively). The HflB protease, encoded by the hflB gene (35), was previously inferred to be involved in heat shock regulation in L. lactis (12). The level of _hflB_-specific mRNA was not, however, elevated in either BK6 or BK11 (Table 3).
TABLE 3.
Relative heat shock mRNA levelsa
| Strain | Level of the following specific mRNAb: | |||
|---|---|---|---|---|
| hrcA | dnaK | dnaJ | hflB | |
| BK8 (dnaK+-erm) | 1.0 | 1.0 | 1.0 | 1.0 |
| BK11 (dnaKΔ2) | 3.0 | 3.2 | 1.2 | 1.1 |
| BK6 (dnaKΔ1) | 14 | 7.3 | 2.3 | 1.4 |
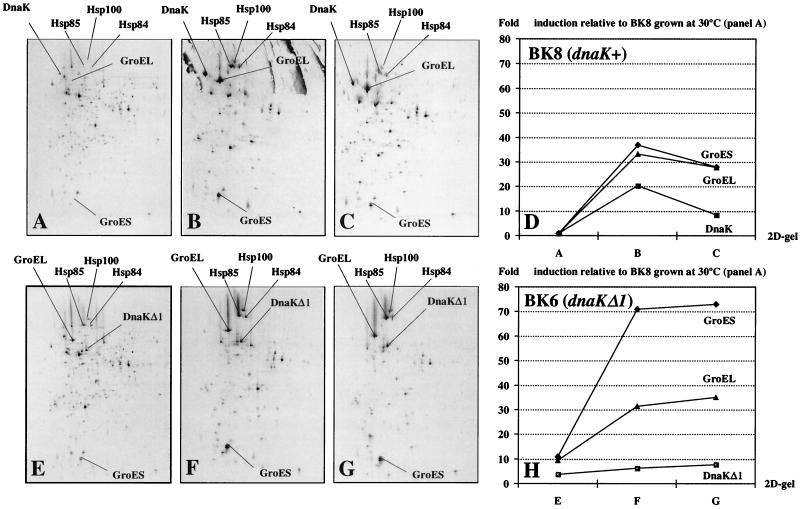
At the protein level, the dnaKΔ1 deletion resulted in the induction of heat shock proteins when the synthesis rate was visualized by [35S]methionine incorporation (Fig. 3). When the pattern of proteins synthesized in BK8 (dnaK+-erm) during a 15-min period at 30°C (Fig. 3A) was compared to the pattern in BK6 grown and labelled under identical conditions (Fig. 3E), most proteins were labelled at the same relative intensities in the two strains, indicating that their rates of synthesis were comparable. Some proteins, however, were much more intensely labelled in BK6, showing that they were induced in the dnaK mutant. Many of these induced proteins (GroEL, GroES, Hsp84, Hsp85, and Hsp100) were identical to the heat stress-induced proteins which were seen when BK8 was subjected to 43°C for 5 min (Fig. 3B) or 30 min (Fig. 3C), followed by [35S]methionine labelling of the newly synthesized proteins for 15 min at 43°C. The presence of the dnaKΔ1 mutation resulted in a 10-fold increase in both GroEL and GroES synthesis rates at 30°C (Fig. 3H). The DnaK protein which was visible in BK8 at both 30 and 43°C was clearly absent in BK6. However, the truncated DnaKΔ1 protein which was detected as a 50-kDa protein by Western blotting in Fig. 1D most likely corresponded to the intensely labelled spot which was located at the position indicated in Fig. 3E, F, and G and which had no counterpart in Fig. 3A, B, and C. This position had the coordinates (7 and 80) in the L. lactis reference gel described elsewhere (28) from which an apparent molecular mass of 49 kDa and an isoelectric point of 4.8 could be calculated. These values are in agreement with the values predicted from the DNA sequence of the dnaKΔ1 gene (49.4 kDa; pI, 4.78). The synthesis rate for this protein was increased fivefold compared to the intensity of the DnaK spot in Fig. 3A (Fig. 3H).
FIG. 3.
Analysis of protein synthesis rates in the dnaKΔ1 mutant BK6 by 2D PAGE. Bacterial cultures were grown exponentially at 30°C to an OD450 of 0.4, followed by a shift to 43°C. At the indicated times after the temperature shift, [35S]methionine was added and growth was continued for 15 min. Following extraction, the labelled proteins were separated by 2D PAGE. The erythromycin-resistant control strain BK8 (dnaK+-erm; A to C) and the dnaK mutant BK6 (dnaKΔ1; E to G) were analyzed. Growth temperatures and labelling periods were as follows: 30°C, −15 to 0 min (A and E); 43°C, 5 to 20 min (B and F); and 43°C, 30 to 45 min (C and G). (D and H) Relative protein synthesis rates for DnaK, GroEL, and GroES. The amounts of radioactivity in the protein spots were measured with a Packard Instant Imager. The values are given relative to the total amount of radioactivity on the gels and are normalized to the values from panel A, so that they represent the fold induction compared to that in the dnaK+ strain grown at 30°C. (D) Values from the gels shown in panels A to C. (H) Values from the gels shown in panels E to G. The pictures were scanned at 300 dpi with a Scan Jet 4c/T (Hewlett-Packard Co.) and DeskScan II version 2.3 software. The TIF file was imported into Top Draw version 3.1 for the addition of text.
When the temporal synthesis rates of GroEL and GroES after the 43°C heat shock were compared for strains BK8 and BK6 (Fig. 3D and H), it appeared that the maximal levels of expression were approximately the same for the two strains, with GroEL reaching a plateau at between 15 and 19% the total protein synthesis rate after heat shock. The maximal levels of the GroES synthesis rate were about 4% the total synthesis rate for BK8 and 7% that for BK6. When the elevated levels of expression at 30°C for the GroEL and GroES proteins in BK6 were taken into account, the 35-fold induction seen in the dnaK+ strain after transfer of the cells to 43°C (Fig. 3D) was reduced to 4- to 7-fold induction in the dnaKΔ1 mutant (Fig. 3H). In the dnaK+ strain, the induction of DnaK exhibited a peak of synthesis at a 20-fold-elevated level between 5 and 20 min after subjection to 43°C (Fig. 3D), similar to the heat shock response in wild-type strain MG1363 (28). In contrast, the induction of the truncated DnaKΔ1 protein showed a progressive increase with time (Fig. 3H), resulting in a modest twofold-elevated level, but the end-point synthesis level reached for either DnaK species was about 5% the total synthesis rate. The values for the incorporation of radioactive methionine in the two proteins could be directly compared as synthesis rates, since only 1 of the 11 methionine residues present in DnaK was missing from DnaKΔ1. The possibility that the DnaKΔ1 protein is unstable cannot be ruled out and would have the effect of underestimating its synthesis rate in Fig. 3H. Whether the lack of a synthesis peak between 5 and 20 min after heat shock could be the result of increased degradation during this period is also not known.
Cell morphology and phage sensitivity of dnaK mutants.
For further comparison with the phenotypes of dnaK mutants of E. coli, the lactococcal dnaK mutants were tested for filamentation and propagation of phages. In E. coli, several dnaK mutants have shown filamentous morphology due to defects in cell division (5, 32, 36). Therefore, the cell morphology of MG1363, BK8, BK6, and BK11 was studied after growth at 30°C in GM17 with or without the addition of erythromycin. In exponentially growing (4-h) and overnight (24-h) cultures, MG1363, BK8, and BK11 typically appeared in short chains comprised of 4 to 10 cells in the chain, but in less than 1% of the chains, the chain length was as high as 20 cells. However, in BK6, chains comprised of more than 16 to 20 cells were dominant, and some chains had up to 40 to 60 cells (data not shown). The shape of the individual cells in the chain was not changed.
A dnaK mutant was originally isolated from E. coli as a mutant showing increased phage resistance. It was later shown that DnaK was necessary for the replication of the E. coli phages λ and P1 (54, 56) and for late transcription of the E. coli phage Mu (41). We therefore tested the dnaKΔ1 mutant strain BK6 for sensitivity to lactococcal phages. The lactococcal phages have been divided into 11 species based on DNA-DNA hybridization (22). We tested lactococcal phages from the two most common groups of phages, namely, the small isometric phages (three phages) and the prolate phages (one phage), for their ability to form plaques on BK6. In the plaque assays, we did not see any difference in plaquing ability or plaque morphology between the mutant strain BK6 and the wild-type strain MG1363 for the small isometric phages sk1 (37), p2 (26), and φjj50 (27) or the prolate phage c2 (37).
DISCUSSION
In the present study, we constructed two dnaK deletion mutants of L. lactis MG1363. The smallest deletion is found in the dnaKΔ2 mutant BK11, which has a deletion of 63 C-terminal amino acids. BK11 differs only slightly from the wild-type strain, but it has one mutant phenotype; a slight temperature-sensitive phenotype was observed when cells were plated at 37°C (Table 2). The chain length of growing BK11 cells is not significantly different from that of the wild-type cells, indicating that the level of excreted lysozyme is unaltered. Thermotolerance develops as efficiently in BK11 as in the wild-type strain, and the mutant was not found to be more susceptible to thermal killing. This result indicates that the function of DnaK is only slightly impaired by the dnaKΔ2 mutation. However, the existence of a phenotype for this mutant indicates a function of the distal end of the DnaK protein. A similar deletion mutant of E. coli showed no phenotype (8).
For the eucaryotic Hsp70 proteins, the amino acids from Ser-384 to Glu-543 are proposed to constitute the peptide binding domain (51). Based on alignments, this region corresponds to amino acids 367 to 510 in the lactococcal DnaK sequence (data not shown). In the dnaKΔ1 mutant BK6, the region from amino acid 432 is deleted. We therefore presume that interactions with substrates are severely affected in this mutant, but we cannot exclude the possibility that some residual DnaK activity exists, especially since the truncated DnaK peptide can be detected, as shown by Western blotting (Fig. 1D). However, in E. coli it has been shown that mutants lacking the substrate region have the same phenotype as mutants lacking the entire protein (36).
BK6 (dnaKΔ1) is heat sensitive for growth, a phenotype which is also displayed by both E. coli and B. subtilis dnaK null mutants. In the E. coli dnaK52 null mutant, elevated temperatures are detrimental, since cells incubated for 2 h at 42°C were found to be incapable of colony formation at 30°C (36) and at 50°C the rate of killing was found to be much higher for the E. coli dnaK null mutant than for the wild type (11). The increased thermosensitivity of the lactococcal dnaKΔ1 mutant (BK6) is in agreement with the data for the E. coli mutant. BK6 is, however, capable of developing thermotolerance when pretreated at 40°C, although not as efficiently as the wild-type strain (compare BK6 to BK8 in Fig. 2). This acquired thermotolerance was not observed in the E. coli dnaK52 null mutant, which is deficient in the development of both heat- and starvation-induced thermotolerance (11, 39).
At all temperatures tested (from 10 to 33°C), dnaKΔ1 BK6 showed reduced growth rates compared to the wild-type strain. This result demonstrates that a functional DnaK protein is needed during normal growth in L. lactis, as has been found for E. coli mutants. In conclusion, the phenotype of the lactococcal dnaKΔ1 mutant resembles the severe effects in the E. coli mutants more than the modest temperature-sensitive phenotype of the dnaK mutant isolated from B. subtilis (43).
A major point of interest in this study was to test how a defective DnaK protein would affect the expression of heat shock-induced chaperones. In E. coli, the DnaK chaperone complex is known to be the sensor of denatured proteins, and the availability of the complex determines the rate of proteolysis of the heat shock sigma factor (16, 46). A dnaK mutant has impaired proteolysis of the sigma factor and therefore contains increased levels of heat shock proteins at the normal growth temperature and shows greatly diminished shutoff of protein synthesis after heat shock. If the availability of the DnaK chaperone complex could also function as a sensor of denatured proteins in HrcA-regulated organisms, then the HrcA heat shock repressor would be expected to be dependent upon the DnaK complex for activity. During heat shock, when the complex is sequestered by denatured proteins, the repressor might not mature properly, leading to increased expression from CIRCE-regulated promoters. Recently, however, it was reported that the GroELS chaperone complex, and not the DnaK complex, is the sensor of denatured proteins in B. subtilis (34). Also, in Bradyrhizobium japonicum, a functional groEL gene product seems to be necessary for the repression of the CIRCE-regulated _groELS_4 operon at the lower growth temperature (2). For B. subtilis the conclusion was partly based upon the fact that a dnaK null mutation does not result in increased expression of CIRCE-regulated genes, as it should if DnaK were needed for repressor activity.
The orf1 gene in the dnaK operon of L. lactis has high similarity to hrcA of B. subtilis. Since L. lactis has an HrcA-like protein and CIRCE regulatory elements just like B. subtilis, we expected that BK6 would not overexpress chaperones, but much to our surprise the defect in the lactococcal dnaKΔ1 mutant resulted in increased expression of all of the known CIRCE-containing operons: dnaJ (Table 3), groELS (Fig. 3), and the dnaK operon itself (Table 3 and Fig. 3). It therefore appears that the dnaKΔ1 mutation does influence HrcA activity in L. lactis. Yet, the activity of HrcA is not likely to be solely dependent upon the DnaK chaperone complex, because BK6 can still acquire thermotolerance and can still elicit a limited heat shock response. The induction of the heat shock proteins in BK6 is due to the dnaKΔ1 mutation and is not an indirect effect of the slow growth rate of this mutant, since reduced growth rates in purine- and pyrimidine-requiring mutants do not induce heat shock proteins (27a). It is also unlikely that the effect is due to the presence of the integrated plasmid, e.g., by the production of an antisense RNA from a plasmid promoter which could prevent hrcA, grpE, or dnaK translation. We did not observe any such RNA species in a Northern analysis in which a double-stranded DNA fragment was used as probe (Table 3). Accordingly, no promoter is known to be present in the plasmid immediately downstream of dnaK.
Concerning the role of the DnaK chaperone complex in HrcA stabilization, it is noteworthy that the dnaK mutant of B. subtilis is polar for the expression of the downstream genes. The downstream genes are, however, constitutively expressed from a vegetative promoter immediately upstream of dnaJ, resulting in the expression of dnaJ at a reduced level (21, 43). Also, the dnaKΔ52 mutant of E. coli is polar and DnaJ is expressed constitutively at a lower level from a similar internal promoter. For the latter, it has been shown that the phenotype of the dnaKΔ52 mutant can be complemented by a wild-type dnaK gene provided by a lambda phage and is thus not due to the reduced level of DnaJ (5). The same may well be true for the dnaK mutant of B. subtilis. It would, however, be important in light of the role of DnaK in L. lactis to observe the phenotype of a nonpolar dnaK mutant of B. subtilis; in addition, it would be most interesting to obtain results from nonpolar dnaK mutants of other CIRCE-containing organisms.
ACKNOWLEDGMENTS
The outstanding technical assistance of Tim Evison and Kristina Brandborg Jensen is gratefully acknowledged. We gratefully acknowledge the gift of the antibody against DnaK from B. subtilis from W. Schumann as well as the dnaK plasmid, pFI573, from M. Gasson and the ftsH plasmid, pLN32, from D. Nilsson.
This work was financed by MFF (Danish Dairy Board Research Foundation) and by the FØTEK Program through the Center for Advanced Food Studies.
REFERENCES
- 1.Arnau J, Sørensen K I, Appel K, Vogensen F K, Hammer K. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology. 1996;142:1685–1691. doi: 10.1099/13500872-142-7-1685. [DOI] [PubMed] [Google Scholar]
- 2.Babst M, Hennecke H, Fischer H-M. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 3.Boutibonnes P, Gillot B, Auffray Y, Thammavongs B. Heat shock induced thermotolerance and inhibition of lysis in a lysogenic strain of Lactococcus lactis. Int J Food Microbiol. 1991;14:1–10. doi: 10.1016/0168-1605(91)90031-j. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 5.Bukau B, Walker G. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau B, Walker G. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkholder W F, Panagiotidis C A, Silverstein S J, Cegielska A, Gottesmann M E, Gaitanaris G A. Isolation and characterization of an Escherichia coli DnaK mutant with impaired ATPase activity. J Mol Biol. 1994;242:364–377. doi: 10.1006/jmbi.1994.1587. [DOI] [PubMed] [Google Scholar]
- 8.Cegielska A, Georgopoulos C. Functional domains of the Escherichia coli DnaK heat shock protein as revealed by mutational analysis. J Biol Chem. 1989;264:21122–21130. [PubMed] [Google Scholar]
- 9.Chappell T G, Konforti B B, Schmid S L, Rothman J E. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 10.Christiansen B, Johnsen M G, Stenby E, Vogensen F K, Hammer K. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J Bacteriol. 1994;176:1069–1076. doi: 10.1128/jb.176.4.1069-1076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaney J M. Requirement of the Escherichia coli dnaK gene for thermotolerance and protection against H2O2. J Gen Microbiol. 1990;136:2113–2118. doi: 10.1099/00221287-136-10-2113. [DOI] [PubMed] [Google Scholar]
- 12.Duwat P, Ehrlich S D, Gruss A. The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol Microbiol. 1995;17:1121–1131. doi: 10.1111/j.1365-2958.1995.mmi_17061121.x. [DOI] [PubMed] [Google Scholar]
- 13.Eaton T, Shearman C, Gasson M. Cloning and sequence analysis of the dnaK gene region of Lactococcus lactis subsp. lactis. J Gen Microbiol. 1993;139:3253–3264. doi: 10.1099/00221287-139-12-3253. [DOI] [PubMed] [Google Scholar]
- 14.Freeman B C, Myers M P, Schumacher R, Morimoto R I. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasson M J. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, et al., editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–251. [Google Scholar]
- 17.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 18.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 19.Heyde M, Portalier R. Acid shock proteins of Escherichia coli. FEMS Microbiol Lett. 1990;69:19–26. doi: 10.1016/0378-1097(90)90406-g. [DOI] [PubMed] [Google Scholar]
- 20.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis A W, Fitzgerald G F, Mata M, Mercenier A, Neve H, Powell I B, Ronda C, Saxelin M, Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32:2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins D E, Auger E A, Martin A. Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J Bacteriol. 1991;173:1992–1996. doi: 10.1128/jb.173.6.1992-1996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen P R, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen E, Kibenich A. Characterization of Leuconostoc isolates from commercial mixed strain starter cultures. J Dairy Sci. 1992;75:1186–1191. [Google Scholar]
- 26.Josephsen J, Klaenhammer T. Stacking of three different restriction and modification systems in Lactococcus lactis by cotransformation. Plasmid. 1990;23:71–75. doi: 10.1016/0147-619x(90)90046-f. [DOI] [PubMed] [Google Scholar]
- 27.Josephsen J, Vogensen F K. Identification of three different plasmid-encoded restriction/modification systems in Streptococcus lactis subsp. cremoris W56. FEMS Microbiol Lett. 1989;59:161–166. [Google Scholar]
- 27a.Kilstrup, M. Unpublished data.
- 28.Kilstrup M, Jacobsen S, Hammer K, Vogensen F K. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol. 1997;63:1826–1837. doi: 10.1128/aem.63.5.1826-1837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S G, Batt C A. Cloning and sequencing of the Lactococcus lactis subsp. lactis groEL operon. Gene. 1993;127:121–126. doi: 10.1016/0378-1119(93)90626-e. [DOI] [PubMed] [Google Scholar]
- 30.Koch B M, Sibbesen O, Halkier B A, Svendsen I, Møller B L. The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalysing the conversion of l-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- 31.Mayhew M, Hartl F-U. Molecular chaperone proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 922–937. [Google Scholar]
- 32.McCarty J S, Walker G C. DnaK mutants defective in ATPase activity are defective in negative regulation of the heat shock response: expression of mutant DnaK proteins results in filamentation. J Bacteriol. 1994;176:764–780. doi: 10.1128/jb.176.3.764-780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meury J, Kohiyama M. Role of heat shock protein DnaK in osmotic adaptation of Escherichia coli. J Bacteriol. 1991;173:4404–4410. doi: 10.1128/jb.173.14.4404-4410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson D, Lauridsen A A, Tomoyasu T, Ogura T. A Lactococcus lactis gene encodes a membrane protein with putative ATPase activity that is homologous to the essential Escherichia coli ftsH gene product. Microbiology. 1994;140:2601–2610. doi: 10.1099/00221287-140-10-2601. [DOI] [PubMed] [Google Scholar]
- 36.Paek K-H, Walker G C. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pillidge C J, Jarvis A W. DNA restriction maps and classification of the lactococcal bacteriophages c2 and sk1. N Z J Dairy Sci Technol. 1988;23:411–416. [Google Scholar]
- 38.Roberts R C, Toochinda C, Avedissian M, Baldini R L, Gomes S L, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rockabrand D, Arthur T, Korinek G, Livers K, Blum P. An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reduction division. J Bacteriol. 1995;177:3695–3703. doi: 10.1128/jb.177.13.3695-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sand O, Desmet L, Toussaint A, Pato M. The Escherichia coli DnaK chaperone machine and bacteriophage Mu late transcription. Mol Microbiol. 1995;15:977–984. doi: 10.1111/j.1365-2958.1995.tb02366.x. [DOI] [PubMed] [Google Scholar]
- 42.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz A, Tzschaschel B, Schumann W. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol Microbiol. 1995;15:421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x. [DOI] [PubMed] [Google Scholar]
- 44.Skowyra D, Georgopoulos C, Zylicz M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990;62:939–944. doi: 10.1016/0092-8674(90)90268-j. [DOI] [PubMed] [Google Scholar]
- 45.Spence J, Cegielska A, Georgopoulos C. Role of Escherichia coli heat shock proteins DnaK and HtpG (C62.5) in response to nutritional deprivation. J Bacteriol. 1990;172:7157–7166. doi: 10.1128/jb.172.12.7157-7166.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 47.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The DnaK protein modulates the heat shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- 49.van Asseldonk M, Simons A, Visser H, de Vos W M, Simons G. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J Bacteriol. 1993;175:1637–1644. doi: 10.1128/jb.175.6.1637-1644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 51.Wang T-F, Chang J H, Wang C. Identification of the peptide binding domain of hsc70 18-kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J Biol Chem. 1993;268:26049–26051. [PubMed] [Google Scholar]
- 52.Whitaker R D, Batt C A. Characterization of the heat shock response in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1991;57:1408–1412. doi: 10.1128/aem.57.5.1408-1412.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickner S, Hoskins J, McKenney K. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature. 1991;350:165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- 54.Wickner S H. Three Escherichia coli heat shock proteins are required for P1 plasmid DNA replication: formation of an active complex between E. coli DnaJ protein and the P1 initiator protein. Proc Natl Acad Sci USA. 1990;87:2690–2694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan G, Wong S-L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of λ DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]