Chromosome triplication found across the tribe Brassiceae (original) (raw)
Abstract
We have used an ∼8.7-Mb BAC contig of Arabidopsis thaliana Chromosome 4 to trace homeologous chromosome regions in 21 species of the family Brassicaceae. Homeologs of this segment could be identified in all tested species. Painting of pachytene chromosomes of Calepina, Conringia, and Sisymbrium species (2_n_ = 14, 16), traditionally placed in tribe Brassiceae, showed one homeologous copy of the Arabidopsis contig, while the remaining taxa of the tribe (2_n_ = 14–30) revealed three, and three Brassica species (2_n_ = 34, 36, and 38) and Erucastrum gallicum (2_n_ = 30) had six copies corresponding to the 8.7-Mb segment. The multiple homeologous copies corresponded structurally to the Arabidopsis segment or were rearranged by inversions and translocations within the diploidized genomes. These chromosome rearrangements accompanied by chromosome fusions/fissions led to the present-day chromosome number variation within the Brassiceae. Phylogenetic relationships based on the chloroplast 5′-_trn_L (UAA)–_trn_F(GAA) region and estimated divergence times based on sequence data of the chalcone synthase gene are congruent with comparative painting data and place Calepina, Conringia, and Sisymbrium outside the clade of Brassiceae species with triplicated genomes. Most likely, species containing three or six copy pairs descended from a common hexaploid ancestor with basic genomes similar to that of Arabidopsis. The presumed hexaploidization event occurred after the Arabidopsis_–_Brassiceae split, between 7.9 and 14.6 Mya.
The Brassiceae is one of the most morphologically distinct tribes within the Brassicaceae family (Cruciferae). The Brassiceae tribe is a monophyletic group (e.g., Warwick and Black 1997a,b; Anderson and Warwick 1999) comprising ∼240 species in 49–54 genera (Gómez-Campo 1999; Warwick et al. 2000), and includes economically important Brassica crops. Although the basic genome structure of six Brassica crop species was unraveled early (e.g., U 1935), there is a long-lasting debate on the origin and evolution of karyotypes in the genus Brassica and the tribe Brassiceae. The variation in basic chromosome numbers (x = 6–18) (Warwick et al. 2000) makes it difficult to decide whether species with higher basic chromosome numbers are genuine diploids or polyploids. In many Brassiceae genera, species with the lowest chromosome number are considered as diploid. Several hypotheses on ancestral basic numbers (x = 3–7) and karyotype evolution in the Brassiceae have been put forward (for review, see Prakash and Hinata 1980; Truco et al. 1996; Anderson and Warwick 1999; Prakash et al. 1999). However, without knowing phylogenetic relationships and the history of polyploidy of the Brassiceae, unambiguous conclusions cannot be drawn.
Already early studies suggested that diploid Brassica species represent “balanced secondary polyploids” exhibiting internal chromosome homeology and genome duplications (e.g., Catcheside 1934; Röbbelen 1960). Comparative mapping of RFLP probes among Brassica nigra (2_n_ = 16; BB genome), Brassica oleracea (2_n_ = 18; CC), and Brassica rapa (2_n_ = 20; AA) (Lagercrantz and Lydiate 1996), and comparative mapping between the three Brassica species and Arabidopsis thaliana (2_n_ = 10) (e.g., Lagercrantz 1998; Babula et al. 2003) suggested that genomes of the Brassica species are composed of three rearranged variants of an ancestral genome (structurally similar to that of Arabidopsis) and descended from a common hexaploid ancestor (known as the triplication theory). Indeed, syntenic regions corresponding to Arabidopsis chromosome segments could be identified, each in triplicate, within the allopolyploid genome of Brassica napus (Cavell et al. 1998; Parkin et al. 2002, 2003). On the contrary, several whole-genome comparisons between Arabidopsis and “diploid” Brassica species apparently provided only little support for the triplication theory. Although all studies revealed extensive duplications, some Arabidopsis segments were present either in less or more than three copies in Brassica (Truco et al. 1996; Lan et al. 2000; Babula et al. 2003; Li et al. 2003; Lukens et al. 2003). However, at the level of short sequences, less than three copies might be caused by frequent loss in polyploids, and more than three copies might be caused by more remote genome duplications (Bovers et al. 2003) or imbalanced homeologous recombination in allotetraploids (Osborn 2004). FISH mapping of a 431-kb Arabidopsis BAC contig to mitotic chromosomes and DNA fibers of B. rapa (Jackson et al. 2000) and of two Arabidopsis BAC clones (150 kb) to B. oleracea (Ziolkowski and Sadowski 2002) supported an ancestral genome duplication for diploid Brassica species, although it did not clearly prove the genome triplication. Thus, as concluded by Lukens et al. (2004), neither the evidence for an ancestral hexaploid genome nor alternative explanations such as the existence of a tetraploid ancestor and later segmental chromosomal duplications are convincing.
In the present paper we applied comparative chromosome painting using a BAC contig that covers ∼8.7 Mb of the bottom (long) arm of Arabidopsis Chromosome 4 (At4-b contig) to meiotic chromosomes of 21 Brassiceae and related species to explore the level of genome and chromosome homeology between Arabidopsis and Brassiceae species and to look for evidence of genome triplication in the genus Brassica and closely related species. The results were compared with phylogenetic data obtained from sequencing the chloroplast _trn_L intron/_trn_L-F intergenic spacer region in order to gain information on relationships and divergence times within the group.
Results
An Arabidopsis segment of ∼8.7 Mb is found in the genomes of all analyzed species
The At4-b contig spanning ∼8.7 Mb and corresponding ∼7% of the genome or nearly two-thirds of the bottom arm of Arabidopsis Chromosome 4 is comparatively mapped to the syntenic linkage groups G/6 and F/7 of Capsella rubella (Boivin et al. 2004) and Arabidopsis lyrata (Kuittinen et al. 2004), respectively. This contig was arbitrarily divided into three (A–C) differently labeled subcontigs (Fig. 1a) and hybridized to pachytene complements of 21 taxa that were either traditionally placed in the tribe Brassiceae or were considered to be closely related (Table 1). One or more homeologous chromosome regions were detectable in all tested species (Table 1) and showed either the same subcontig structure as in A. thaliana or revealed inversions and/or translocations. Because the _Arabidopsis_-like structure was the most frequent pattern although A. thaliana is rather distantly related to the Brassiceae, we consider this structure of the contig as the ancestral one. In species with one or three homeologous copies of the At4-b segment, the length of FISH signals corresponding to the individual subcontigs was measured for the _Arabidopsis_-like variant as well as for one variant with an inversion (S1, F3, R1 in Figs. 1f,i and 2a). The relative lengths of individual subcontigs in different species were compared with those of the Arabidopsis subcontigs whose lengths, in megabases, are shown in Figure 1a. Although such measurements are likely biased by variation in genome size and/or chromosome condensation, the relative lengths of subcontigs agreed well with those of Arabidopsis. Similar relative subcontig lengths in different species suggest a common origin from an ancestral genome (Table 2).
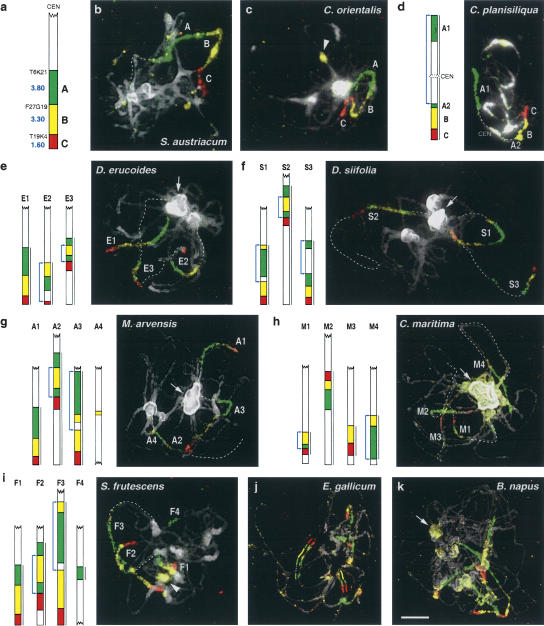
Figure 1.
Comparative chromosome painting using the At4-b Arabidopsis BAC contig. (a) Diagram of the At4-b contig from the bottom arm of A. thaliana Chromosome 4 hybridized to pachytene chromosome complements of species shown in b to k. Starting BAC clones and size (megabases) of subcontigs A, B, and C are shown on the left. Subcontigs A, B, and C were visualized as Alexa 488 (green), Cy3 (yellow), and Texas Red (red) fluorescence, respectively. (b,c) One copy of the At4-b contig in Sisymbrium austriacum and Conringia orientalis. The homeologous segments have the _Arabidopsis_-like structure. (d) One rearranged copy of the At4-b contig in C. planisiliqua. Subcontig A is split into two parts (A1, A2) likely because of a pericentric inversion. (e_–_i) Three copies of the At4-b contig in (e) Diplotaxis erucoides, (f) D. siifolia, (g) Moricandia arvensis, (h) Cakile maritima, and (i) Sinapidendron frutescens. (j) Six copies of the At4-b contig in Erucastrum gallicum. Whereas four copies apparently show the _Arabidopsis_-like structure, two homeologous regions bear an inversion (marked by drawing). (k) Six copies of the At4-b contig in Brassica napus; chromosome clustering does not allow unambiguous tracing of individual fluorescence signals. Chromosomes were counterstained with DAPI. Diagrams were drawn to scale (according to measured regions labeled by a bar on the right) for the corresponding species. Blue arrows in diagrams indicate assumed inversions. Strongly DAPI-stained (peri)centromeric heterochromatin (arrows) and nucleolus organizers (arrowheads) show cross-hybridization with sequences of subcontig B (yellow) in some species. Scale, 5 μm.
Table 1.
Copy number of the At4-b contig, chromosome numbers, origin, and GenBank accession numbers of the sequences used for phylogenetic studies of the investigated species
| Species | At4-bb | 2n | Origin | GenBank accession number of the plastid _trn_L–_trn_F region |
|---|---|---|---|---|
| Arabis pauciflora (Grimm) Garcke [=Fourraea alpina (L.) Greuter & Burdet]a | 14 | Germany, Jena, Hausberg, 300m a.s.l., leg. M. Koch, 1998 | AY751774 | |
| Brassica nigra (L.) W.D.J. Koch | 3 | 16 | Botanic Garden, Hamburg University; Germany, Hafen, Moorburg | AF451578c, AF451579c |
| Brassica oleracea var. capitata L. | 3 | 18 | IPK Gatersleben, GenBank | AF451574c |
| Brassica rapa L. | 3 | 20 | Rapid-cycling B. rapa (Williams and Hill 1986) | AF451571c |
| Brassica carinata A. Braun | 6 | 34 | Rapid-cycling B. carinata (Williams and Hill 1986) | — |
| Brassica juncea (L.) Czern. | 6 | 36 | Rapid-cycling B. juncea (Williams and Hill 1986) | AF451575c |
| Brassica napus L. | 6 | 38 | Rapid-cycling B. napus (Williams and Hill 1986) | — |
| Calepina irregularis (Asso) Thellung | 1 | 14 | Botanic Garden, University of Copenhagen; no. 518 (E4966-0001*A, 219) | AY751760 |
| Cakile maritima Scop. subsp. maritima | 3(4) | 18 | Botanic Garden Berlin-Dahlem; Greece, Sterea Ellas, Athen/Glifada, no. 1416 | AY754817, AY754818 |
| Carrichtera annua (L.) DCa | 16 | Botanic Garden Bordeaux; France, San Vincente de Raspeig, no. 210, AL 0320 | AY751761 | |
| Conringia orientalis (L.) Dumort | 1 | 14 | Botanic Garden Bordeaux; France, Causse Mejean, 48. Loziere, no. 208 (07/98) | — |
| Conringia planisiliqua Fischer & C.A. Meyer | 1 | 16 | India, Ladakh, Zhingchan, leg. A. Pecinka | AY751762 |
| Cordylocarpus muricatus Desf.a | 16 | Botanic Garden Berlin-Dahlem; Marocco, Taza, W Guercif, no. 1436 | AY751759 | |
| Diplotaxis erucoides (L.) DC | 3 | 14 | Botanic Garden Bordeaux; Spain, Vilalba del Arco, no. 211 (05/98) | AY751763 |
| Diplotaxis siifolia Kunze | 3 | 20 | Botanic Garden Bordeaux; Spain, Tarifa, no. 212 (06/99) | AY751764 |
| Eruca sativa Mill. | 3 | 22 | Commercially available seeds | AY751765 |
| Erucastrum gallicum (Willd.) O.E. Schulz | 6 | 30 | Germany, Gatersleben, Seeland lake, leg. M. Lysak | AY751766 |
| Moricandia arvensis (L.) DC | 3 | 28 | Botanic Garden Bordeaux; Spain, Almansa, no. 218 (05/98) | AY751767 |
| Psychine stylosa Desf. | 3 | 30 | Botanic Garden Dijon; no. 312 | AY751768 |
| Raphanus sativus L. var. Saxa | 3 | 18 | Commercially available seeds | AY751770, AF451576c, AF451577c |
| Rapistrum rugosum (L.) All. | 3 | 16 | Madeira, Ponta de São Lourenço, leg. M. Lysak | AY751769 |
| Sinapidendron frutescens (Aiton) Löwe | 3 | 20 | Madeira, mountain chain between Pico do Arieiro Mt. and Pico Ruivo Mt., leg. M. Lysak | AY751771 |
| Sinapis arvensis L. | 3 | 18 | Botanic Garden, J.W. Goethe University, Frankfurt/Main; no. 462 (52) | AY751772 (S. alba: AF451580c, AF451581)c |
| Sisymbrium austriacum Jacq. | 1 | 14 | Botanic Garden, J.W. Goethe University, Frankfurt/Main; no. 464 (52) | (S. altissimum: AY122458c) |
| Vella spinosa Boiss.a | 34 | Botanic Garden Berlin-Dahlem; Spain, Granada, Sierra Nevada, Trevenque, no. 1546 | AY751773 |
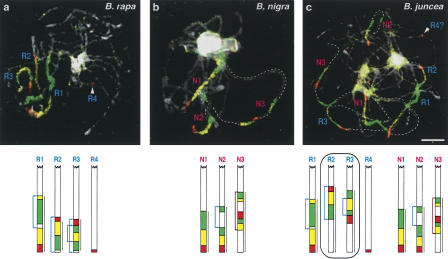
Figure 2.
Comparative chromosome painting using the At4-b BAC contig in amhidiploid Brassica juncea and its two parental species. (a,b) Three copies of the At4-b contig in pachytene chromosome complements of B. rapa (AA genome, homeologs R1–R4) and B. nigra (BB genome, homeologs N1–N3). (c) Six chromosome regions homeologous to the At4-b contig in B. juncea (AABB genome). Two _B. rapa_-derived homeologs (R2 and R3) showed a structure deviating from that of B. rapa likely because of a translocation (R2) and an inversion (R3), respectively. Chromosomes were counterstained with DAPI. Diagrams were drawn to scale (according to measured regions labeled by a bar on the right) for the corresponding species. Blue arrows in diagrams indicate assumed inversions; brown bracket-like lines depict complex rearrangements. The enumeration of homeologs does not refer to linkage groups of the corresponding species. Scale, 5 μm.
Table 2.
The relative length of subcontigs within the regions homeologous to the At4-b contig based on measurements of FISH signals
| At4-b (_Arabidopsis_-like structure) | At4-b (inverted variant)a | |||||||
|---|---|---|---|---|---|---|---|---|
| Subcontig (%) | ||||||||
| Species | Figure no. | No. of analyzed pachytene complements | A | B | C | A | B | C |
| Arabidopsis thalianab (Columbia) | 1a | — | 43 | 39 | 18 | — | — | — |
| One copy of the At4-b | ||||||||
| Calepina irregularis | Not shown | 3 | 47 | 34 | 19 | — | — | — |
| Sisymbrium austriacum | 1b | 3 | 42 | 41 | 17 | — | — | — |
| Conringia orientalis | 1c | 7 | 45 | 39 | 16 | — | — | — |
| Conringia planisiliqua | 1d | 7 | 47 | 35 | 18 | — | — | — |
| Three copies of the At4-b | ||||||||
| Diplotaxis erucoides | 1e | 7 | 49 | 36 | 15 | — | — | — |
| Moricandia arvensis | 1g | 8 | 53 | 32 | 15 | — | — | — |
| Brassica nigra | 2b | 11 | 46 | 38 | 16 | — | — | — |
| Brassica oleracea | Not shown | 6 | 50 | 32 | 18 | 48 | 36 | 16 |
| Sinapidendron frutescens | 1i | 4 | 33 | 48 | 19 | 44 | 42 | 14 |
| Diplotaxis siifolia | 1f | 8 | — | — | — | 50 | 35 | 15 |
| Brassica rapa | 2a | 5 | — | — | — | 45 | 40 | 15 |
Calepina, Conringia, and Sisymbrium species have one copy of the At4-b contig
One copy of the At4-b contig on one chromosome pair was found in Calepina irregularis, in two Conringia species, and in Sisymbrium austriacum. The contig has the Arabidopsis_-like structure in C. irregularis (data not shown), Conringia orientalis (Fig. 1c), and S. austriacum (Fig. 1b) (all 2_n = 14). In Conringia planisiliqua (2_n_ = 16), however, most of the proximal subcontig A (green) is separated from subcontigs B (yellow) and C (red) by an unlabeled chromosome segment containing the centromere, likely because of a pericentric inversion with breakpoints in the distal part of subcontig A and in a terminal position of the opposite arm (Fig. 1d). The presence of only a single copy of the At4-b contig suggests that these species did not experience the presumed polyploidization after divergence from a common ancestor.
Brassiceae species are paleopolyploid and have multiple copies of the contig
In 13 out of 21 analyzed species, the At4-b contig was found in three copies on three to four chromosome pairs. The homeology patterns were found to be variable and distinct in species with 2_n_ = 14, 16, 18, 20, 22, 28, and 30. Often one of the At4-b homeologous segments showed the Arabidopsis_-like subcontig structure and was positioned distally as in A. thaliana. This is true for Diplotaxis erucoides (2_n = 14) (Fig. 1e), Moricandia arvensis (2_n_ = 28) (Fig. 1g), Sinapidendron frutescens (2_n_ = 20) (Fig. 1i), Sinapis arvensis (2_n_ = 18) (data not shown), B. nigra (2_n_ = 16) (Fig. 2b), and B. oleracea (2_n_ = 18) (data not shown). More often, the corresponding segment was found to be involved in minor or larger rearrangements such as paracentric inversions and reciprocal translocations. Several types of inversions were identified. For example, in Diplotaxis siifolia (2_n_ = 20) and B. rapa (2_n_ = 20), one homeolog pair exhibited a paracentric inversion with breakpoints proximal to subcontig A and within subcontig B (S1 and R1 in Figs. 1f and 2a, respectively). In B. oleracea and S. frutescens (Fig. 1i), one of the three homeologous regions is _Arabidopsis_-like (F1); a second one (F3) revealed a similar inversion as S1 (Fig. 1f) and R1 (Fig. 2a). Another type of paracentric inversion with the breakpoints in subcontig A and at the edge between subcontigs B and C was found in D. erucoides (E3 in Fig. 1e), D. siifolia (S2 in Fig. 1f), M. arvensis (A2 in Fig. 1g), and S. frutescens (F2 in Fig. 1i). The third inversion type with breakpoints within subcontig A and at a proximal position was found in D. siifolia (S3 in Fig. 1f).
In Cakile maritima (2_n_ = 18), B. oleracea, and B. rapa, the entire At4-b contig of one chromosome pair showed an orientation inverse to that of Arabidopsis apparently caused by an inversion with breakpoints proximal to subcontig A and at the very end of the entire At4-b contig (M2 and R2 in Figs. 1h and 2a, respectively).
For the hypothetical reconstruction of more complex rearrangements in B. rapa and B. nigra, the sequence of events is not clear in every case (R3, N3 in Fig. 2a,b). In C. maritima, the fourth homeolog arose probably via a complex rearrangement between M3 and M4 (Fig. 1h). Besides three chromosome pairs containing all three subcontigs, a fourth pair with a minor subcontig-specific signal was found in M. arvensis (A4 in Fig. 1g), S. frutescens (F4 in Fig. 1i), and B. rapa (R4 in Fig. 2a), likely because of one (R4) or two (A4, F4) translocation events. In D. erucoides (E3 in Fig. 1e), D. siifolia (S2 in Fig. 1f), and M. arvensis (A2 in Fig. 1g), one of the chromosomes containing the homeologous segment is flanked on the distal side of subcontig C by an unlabeled region of different size likely because of a translocation with another chromosome at a breakpoint close to the distal border of subcontig C. Additionally, in all three species, the chromosome in question shows an inversion with breakpoints in subcontig A and at the edge between B and C as described above.
Six copies of the At4-b contig confirmed the neopolyploid origin of three amphidiploid Brassica species (B. carinata, B. juncea, B. napus) and of E. gallicum (2_n_ = 30). We identified the copies of the contig derived from B. rapa (2_n_ = 20; AA) and from B. nigra (2_n_ = 16; BB) in the amphidiploid B. juncea (2_n_ = 36; AABB) (Fig. 2; see U 1935). Three and one of the homeologs of B. nigra and B. rapa, respectively, could be identified in B. juncea. Two of the _B. rapa_-derived homeologous segments in B. juncea deviate from the B. rapa copies structurally and by occupying an interstitial position (Fig. 2c). In E. gallicum, four of the six chromosomes with painting signals show the _Arabidopsis_-like structure and two bear inversions (Fig. 1j).
In some species, three or six copies were clearly discernible, but their structure could not unambiguously be analyzed because of obscure pachytene configurations (e.g., B. napus) (Fig. 1k) and/or insufficient FISH signals. This is true for Eruca sativa, Psychine stylosa, Raphanus sativus, Rapistrum rugosum, S. arvensis (all having three copies of the At4-b), B. carinata, and B. napus (both with six copies of the At4-b) (data not shown).
Phylogenetic relationships indicate the branchpoint for the genome triplication and define the tribe Brassiceae
Phylogenetic reconstructions using parsimony analysis resulted in 50% majority-rule consensus trees, which are totally in congruence with those trees obtained with the maximum-likelihood approach. The tree parameters obtained from parsimony analysis were 84.3%, 84.8%, 191 steps (with C. maritima containing a sequence within the _trn_L-F intergenic spacer that was not detectable for other taxa) and 84.1%, 85.2%, and 182 steps (without C. maritima) for consistency index, retention index, and tree length, respectively. However, a lower tree resolution is obtained when the strict consensus tree has been generated instead of the 50% majority rule consensus tree (data not shown). Although statistics support as indicated by bootstrap values is low for some taxa, reliable phylogenetic relationships can be inferred. The comparison of parsimony and maximum likelihood methods implies that the presented phylogenetic hypothesis is highly significant.
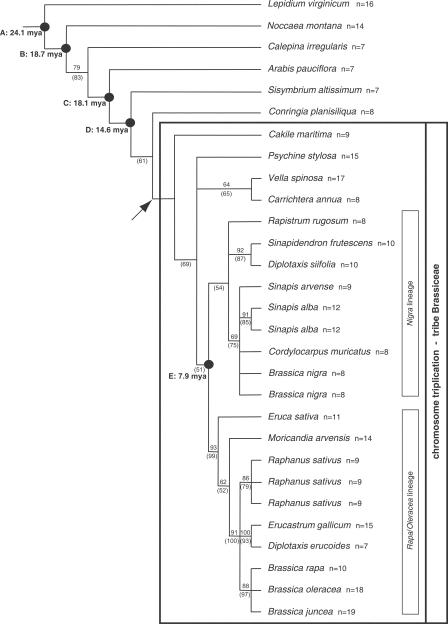
The phylogenetic analysis based on sequences of the chloroplast _trn_L intron/_trn_L-F intergenic spacer region placed the taxa with the triplicated At4-b chromosome region in a single major clade, the Brassiceae lineage, divided into three main groups (Fig. 3). The largest group comprising 15 species was divided into the Nigra and the Rapa/Oleracea lineages (Warwick and Black 1991). Vella spinosa, Carrichtera annua, and Psychine stylosa were placed outside these lineages and C. maritima was sister taxon to all remaining species within the Brassiceae lineage. Our data indicate that the Brassiceae represent a monophyletic group and suggest that all extant species analyzed have descended from a common hexaploid ancestor. Species with a single copy of the At4-b contig, C. irregularis, Sisymbrium altissimum and C. planisiliqua, as well as Arabis pauciflora, were placed outside the Brassiceae clade.
Figure 3.
Phylogenetic hypothesis based on DNA sequences of the plastid trnL(UAA)–trnF(GAA) region of taxa from tribe Brassiceae and several outgroups using a maximum-likelihood (ML) approach as implemented in PAUP 4.0 (for details, see Methods). Bootstrap values are indicated above (ML analysis without Cakile) and below branches (ML analysis with Cakile), respectively. Divergence time estimates (nodes A–E) are given in million years ago (Mya) (see Table 3). The presumed polyploidization event is indicated by the arrow. Intratribal classification (Nigra and Rapa/Oleracea lineages) follows Warwick and Black (1991). Note that both Brassica and Diplotaxis are polyphyletic taxa.
Specific homeology patterns for the At4-b segment do not correspond with phylogenetic relationships within the Brassiceae lineage, that is, species in the Nigra and the Rapa/Oleracea lineages show the comparable variation in the At4-b contig. Similarly, no obvious correlation between phylogenetic relationships and chromosome number variation was found (Fig. 3).
In order to estimate the timing of the presumed polyploidization event in the Brassiceae lineage, the synonymous substitution rates and divergence time estimates of several species pairs have been adopted from Koch et al. (2001) and are summarized in Table 3 and Figure 3. Because the genus Lepidium represents a basal member of the evolutionary lineage that also contains the genus Arabidopsis (Koch et al. 2005), the present analysis indicates that A. thaliana and the genus Brassica/the Brassiceae lineage diverged ∼24 million years ago (Mya). In spite of the uncertain relationship between Noccaea (Thlaspi) and A. pauciflora lineages caused by different results obtained from plastid and nuclear gene sequences (Koch et al. 2001), divergence time estimates do not differ significantly from each other (Fig. 3, nodes B and C: 18.7 and 18.1 Mya; see Table 3). However, radiation of multiple lineages is possible with nearly identical divergence time estimates. As a consequence, phylogenetic trees are not able to resolve these particular relationships in a dichotomous way. The genus Sisymbrium has been separated from the Brassiceae lineage and from the genus Conringia ∼14.6 Mya, and the Nigra and Rapa/Oleracea clades within the Brassiceae lineage diverged ∼7.9 Mya. Hence, the genome triplication should have occurred between 7.9 and 14.6 Mya (Table 3; Fig. 3).
Table 3.
Pairwise synonymous substitutions (Ks) (lower triangle) and calculated divergence times (Mya, upper triangle)a
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| 1 | Sinapis alba | 7.9 | 14.3 | 18.6 | 20.0 | 17.3 |
| 2 | Raphanus sativus | 0.221 | 14.8 | 17.4 | 28.0 | 21.1 |
| 3 | Sisymbrium irio | 0.401 | 0.413 | 23.3 | 24.6 | 16.1 |
| 4 | Noccaea/Thlaspi | 0.521 | 0.486 | 0.652 | 22.8 | 15.6 |
| 5 | Lepidium campestre | 0.673 | 0.784 | 0.690 | 0.637 | 25.0 |
| 6 | Arabis pauciflora | 0.485 | 0.590 | 0.450 | 0.438 | 0.699 |
Discussion
The ∼8.7-Mb region is the largest Arabidopsis chromosome segment cross-hybridized to Brassica species and the first Arabidopsis probe hybridized to chromosomes of other Brassiceae species so far. The triplication of the At4-b BAC contig together with the comparative genetic mapping that revealed six copies of A. thaliana sequences in Brassica species (Lagercrantz and Lydiate 1996; Cavell et al. 1998; Lagercrantz 1998; Parkin et al. 2002, 2003) and less comprehensive previous FISH data (Jackson et al. 2000; Ziolkowski and Sadowski 2002) suggest a hexaploid ancestor for the Brassiceae tribe. The presumed hexaploidization should not be confused with the genome duplications preceding the origin of Brassicaceae and evident in the A. thaliana genome (Bovers et al. 2003) that are no longer detectable by cytogenetic means (Lysak et al. 2001). Homeologous recombination in an allotetraploid, followed by imbalanced segregation of the products (Osborn 2004), should yield two instead of three labeled regions on pachytene chromosome complements. Translocations between heterologous chromosomes followed by imbalanced segregation could also be tolerated in tetraploids and cannot be excluded as a reason for segment triplication but would require independent events for each triplication.
The At4-b BAC contig was frequently found to be rearranged in Brassiceae species with three copies of the contig. Some specific rearrangements involving the At4-b segment have been found in several Brassiceae taxa (Figs. 1 and 2). Because an independent origin of similar rearrangements in several species is less parsimonious, the different rearrangements were probably already present in the ancestral genomes. In none of the species with three copies were the same structural rearrangements found. This observation might indicate an allopolyploid nature of the putative hexaploid ancestor, as has been proposed on the basis of comparative genetic mapping between Arabidopsis and Brassica species (Lagercrantz 1998; Babula et al. 2003). The most frequent chromosome rearrangements involving the At4-b contig are inversions. Similarly, by comparative genetic mapping, Babula et al. (2003) found that 43% of the investigated regions with homeology to Arabidopsis chromosomes involved inversions. The six homeologous copies on the bottom arm of Arabidopsis Chromosome 5 were found to be inverted, whereas an 8-Mb region from the top arm of Chromosome 5 was found in six collinear copies in the amphidiploid genome of B. napus (2_n_ = 38, AACC) (U 1935; Parkin et al. 2002). The 7.5-Mb chromosome region from the bottom arm of Arabidopsis Chromosome 4, partially overlapping with the At4-b segment, was genetically mapped as six homeologous copies in B. napus (Cavell et al. 1998). Two homeologs revealed an identical large paracentric inversion compared with Arabidopsis Chromosome 4, suggesting, together with the present data, that the bottom arm region of A. thaliana Chromosome 4 is prone to chromosome rearrangements. Rearrangements (such as translocations or inversions) might reduce/prevent undesirable pairing and recombination between homeologous chromosomes/chromosome regions (Rieseberg 2001) and lead to reproductive isolation between populations eventually contributing to speciation processes.
Although the karyotype of A. thaliana is a highly derived one (Boivin et al. 2004; Kuittinen et al. 2004; Koch and Kiefer 2005), the _Arabidopsis_-like structure of the ∼8.7-Mb region was found in species with a single copy of the At4-b contig (except in C. planisiliqua). Hence, this region remained apparently unchanged during at least 18 Mya since the divergence of Arabidopsis and Calepina and probably longer when considering that the Arabidopsis lineage was separated together with Lepidium (∼24 Mya) (Fig. 3). The same region remained stable also in seven species within the Arabidopsis lineage as shown by comparative chromosome painting (Lysak et al. 2003). Extended regions, often comprising entire chromosome arms, were found to be collinear to Arabidopsis chromosomes in Brassica genomes (Cavell et al. 1998; Parkin et al. 2002, 2003; Babula et al. 2003). The presumed pericentric inversion found in C. planisiliqua and the occurrence of Brassiceae species with only rearranged versions of the triplicated segment suggest that these rearrangements within the At4-b region have no deleterious effects.
The importance of the Arabidopsis genome structure (n = 5) for the polyploid evolution within the Brassiceae should be evaluated with precaution. Although our data strongly support the view that the structure of the ancestral genome prior to the presumed polyploidization event was similar to that of A. thaliana (Lagercrantz and Lydiate 1996; Lagercrantz 1998), the Arabidopsis karyotype is evolutionarily derived from an ancestral _Capsella_-like genome with eight linkage groups (Boivin et al. 2004; Kuittinen et al. 2004; Koch and Kiefer 2005). Thus, assuming an ancestral genome with n = 5 and yielding a hexaploid one with n = 15 chromosomes is not substantiated by the available data.
There are conflicting genetic data as to the karyotype stability of B. rapa (AA) and B. nigra (BB) genomes in the amphidiploid B. juncea (AABB). According to Axelsson et al. (2000), parental genomes are stably maintained in natural as well as in synthetic B. juncea. On the contrary, Song et al. (1995) observed rapid changes in synthetic Brassica allopolyploids with the highest rate of rearrangements in B. juncea. Similarly, chromosome rearrangements correlated with the phenotypic variation have been reported in resynthesized B. napus (for review, see Osborn 2004; Pires et al. 2004). Our data obtained with the At4-b contig show conservation of homeologous regions derived from B. nigra and an altered structure for two of the three B. rapa homeologs in the genome of a natural B. juncea. However, since the actual parental genotypes of the analyzed B. juncea are not known, it remains elusive whether the rearrangements within the B. rapa homeologs occurred before or after the emergence of B. juncea. Therefore, it would be challenging to monitor the stability of particular chromosome regions by comparative chromosome painting in _F_1 and subsequent generations of newly synthesized Brassica allopolyploids.
Our phylogenetic analysis of the chloroplast _trn_L intron/_trn_L-F intergenic spacer region is congruent with results obtained from the chloroplast gene maturase K (_mat_K) (Koch et al. 2001) and shows that species bearing the triplicated At4-b region represent a monophyletic group (Fig. 3) as was postulated for Brassiceae previously (Warwick and Black 1997a,b; Warwick and Sauder 2005). Internal branching patterns elucidating relationships among taxa without the genome triplication, Conringia, Calepina, A. pauciflora, and Sisymbrium, are not well supported by high bootstrap values. Other markers such as the internal transcribed spacer (ITS) of nuclear rDNA suggest, for instance, monophyly of Conringia and Calepina (Warwick and Sauder 2005). However, “concerted evolution” (Zimmer et al. 1980; Dover 1982) of the ITS marker sequences has been documented in Brassicaceae (Koch et al. 2003a). Therefore, it remains elusive whether ITS-based trees reflect the actual phylogeny or rather the reticulate pattern of ITS evolution (Álvarez and Wendel 2003). C. irregularis and the Conringia species do not possess segmented (heteroarthrocarpous) fruits and/or conduplicate cotyledons characteristic for the Brassiceae (Appel and Al-Shehbaz 2003). Whereas the monotypic genus Calepina was traditionally placed outside the tribe, the genus Conringia was regarded as a member of the Brassiceae (Gómez-Campo 1999). Our comparative painting and phylogenetic analysis suggest the exclusion of Calepina and Conringia from the tribe Brassiceae as postulated by Anderson and Warwick (1999) on the basis of the absence in both genera of duplicated Pgm-2 and Tpi-1 isozyme loci, which occur in the core Brassiceae taxa. Because of the close relationships between Sisymbrium and the tribe Brassiceae (Warwick et al. 2002), we have analyzed S. austriacum for the genome triplication and S. altissimum for phylogenetic position. S. austriacum does not bear the genome triplication and, thus, should be placed together with S. altissimum outside the tribe Brassiceae (Fig. 3) in accordance with previously published phylogenetic data (Warwick et al. 2002). However, it was shown that the genus Sisymbrium is polyphyletic and two other species have been grouped in the Brassiceae clade (Warwick et al. 2002). It would be intriguing to analyze these two species for the presence of the genome triplication.
Based on the data presented here, we propose a taxonomic circumscription of the tribe Brassiceae without the genera Calepina, Conringia, and without S. austriacum, S. altissimum, and A. pauciflora (Fourraea alpina). We have shown that the chromosome triplication serves as an apomorphic character defining the tribe Brassiceae and providing an explanation for the reticulate evolutionary patterns and incongruent evolutionary hypothesis derived from nuclear versus plastid sequence data (Warwick and Sauder 2005).
According to our data the tribe Brassiceae evolved quite recently. Divergence time estimates demonstrate that the presumed genome triplication and initial diversification of the tribe must have occurred between 7.9 and 14.6 Mya (Table 3; Fig. 3). In contrary, the evolutionary split separating the Brassiceae and the A. thaliana lineage occurred ∼20 Mya (24 Mya calculated herein; <20.4 Mya calculated by Yang et al. 1999). Taking into account the age of the Brassicaceae as a whole with the deepest known intrafamilar split of ∼50 Mya (Koch et al. 2000, 2001; Davies et al. 2004), the Brassiceae can be regarded as a young monophyletic assemblage of taxa. Subsequently, at least one major evolutionary event gave rise to two lineages within the tribe Brassiceae, Nigra and Rapa/Oleracea lineages according to Warwick and Black (1991). The maternally inherited plastome allows one to estimate the divergence times because it is not or rarely subjected to recombination and biparental inheritance. Our estimate for the divergence time of this split (7.9–14.6 Mya) is in a good agreement with previously reported estimates (5–10 Mya) (Uyenoyama 1995).
In conclusion, we propose that an ancestral Brassiceae genome became triplicated via allohexaploidy 7.9–14.6 Mya, which might be confirmed by further regions to be tested. Subsequently, various chromosome rearrangements have accumulated leading to genome diploidization. Chromosome fusions and fissions played a role in karyotype evolution of the Brassiceae and resulted in the present-day chromosome number variation. The actual chromosome number of the common ancestor remains uncertain and needs more detailed investigation.
Methods
Plant material
Plants were grown from seeds in a greenhouse of IPK, Gatersleben, or collected in the field. The origin of investigated species is given in Table 1. Leaves were dried in silica gel for subsequent DNA extraction. Entire inflorescences were fixed in ethanol/chloroform/acetic acid (6:3:1) fixative. Spreads of nuclei in meiotic stages (pachytene) were done as described (Lysak et al. 2001, 2003) and treated with pepsin (100 μg/mL 0.01 HCl) prior to FISH.
DNA probes and multicolor FISH
A BAC contig of ∼8.7 Mb of the bottom (long) arm of A. thaliana Chromosome 4 was divided into three subcontigs: (A) BAC T6K21 (AL021889)–T24A18 (AL035680), (B) F27G19 (AL078467)–F4B14 (AL031986), (C) T19K4 (AL022373)–T5J17 (AL035708). Subcontigs A–C cover the same chromosome regions as contigs V and VI in C. rubella, I–III in C. bursa-pastoris, and IV–VIII in Arabis alpina used by Lysak et al. (2003). For selection of individual BAC clones in each subcontig, see Lysak et al. (2001). DNA isolated by a standard alkaline extraction from individual BAC clones was labeled by biotin-dUTP, digoxigenin-dUTP, and Cy3-dUTP, respectively, via nick translation. Labeled BAC clones were mixed and precipitated prior to FISH. Probes and chromosome preparations were denatured together on slides at 80°C for 2 min, hybridized at 37°C for ∼60 h, and subsequently washed in 20% formamide in 2× SSC at 42°C. Biotin-labeled BAC clones were detected using avidin∼Texas Red (Vector Laboratories) and signal amplification by anti-avidin∼biotin (Vector Laboratories) and avidin∼Texas Red. The digoxigenin-labeled subcontig was detected by mouse anti-digoxigenin (Jackson ImmunoResearch) and by goat anti-mouse∼Alexa 488 (Molecular Probes). Slides were evaluated using a Zeiss Axioplan 2 fluorescence microscope. Images were captured by a Spot CCD camera and processed using Photoshop 6.0 imaging software. Fluorescence signals were measured using the ImageJ software (Wayne Rasband, National Institute of Mental Health).
DNA isolation, PCR, and DNA sequencing
Total genomic DNA was obtained from dried leaf tissue from single individuals. DNA extraction followed the procedure of Doyle and Doyle (1987) with some modifications: grinding of only 50–75 mg of dried leaf tissue using a Retsch swing mill (MM 200), addition of two units of ribonuclease per extraction to the isolation buffer, and washing of the DNA pellet twice with 70% ethanol. DNA was finally dissolved in 50 μL of TE buffer for long-term storage.
The 50-μL PCR reactions were performed in a master mix containing 1× PCR buffer (10 mM TRIS/50 mM KCl buffer at pH 8.0), 3 mM MgCl, 0.4 μM each primer, 0.2 mM each dNTP, one unit of Taq DNA polymerase (Schott-Eppendorf), and ∼5 ng of template DNA using a PTC200 (Biozym Diagnostics) thermal cycler. Thermal cycling started with a 5-min denaturation step at 95°C followed by 35 cycles each comprising 60 sec of denaturation at 95°C, 45 sec of annealing at 38°C (_trn_L intron)/45°C trnL/F intergenic spacer, and 60 sec of elongation at 72°C, an elongation phase at 72°C for 10 min, and a final hold at 4°C. The _trn_L intron was amplified using the forward primer 5′-CGAAATCGGTAGACGCTACG-3′ and the reverse primer 5′-GGGGATAGAGGGACTTGAAC-3′ (primers c and d according to Taberlet et al. 1991), which anneal in the first and second exon of the _trn_L gene, respectively. Sequences comprised the complete intron and the second exon of the _trn_L gene. For amplification of the _trn_L/F IGS primers, 5′-GGTTCAAGTCCCTCTATCCC-3′ (primer e according to Taberlet et al. 1991) and 5′-GATT TTCAGTCCTCTGCTCTAC-3′ (designed in this study) annealing in the second exon of the _trn_L gene and the _trn_F gene, respectively, were used. PCR products were checked for length and concentration on a 1.5% agarose gel. No purification of PCR products was necessary for subsequent sequence reactions. Cycle sequencing was performed using the TaqDyeDeoxy Terminator Cycle Sequencing Kit (ABI Applied Biosystems, Inc.) and the original amplification primers. However, the reverse _trn_L/F IGS primer was modified by adding an additional cytosine to its 3′-end as described (Dobeš et al. 2004). Products were analyzed on an ABI 377XL automated sequencer. Cycle sequencing was performed on both strands; in the majority of cases each reaction spanned the complete sequence.
Data analysis: Phylogenetic reconstructions and divergence time estimates
In all, 29 samples were analyzed for sequence variation of the plastid _trn_L intron/_trn_L-F intergenic spacer region. One outgroup sequence from Noccaea montana (Thlaspi montanum) was obtained from Koch and Al-Shehbaz (2004). Additional sequences for Lepidium virginicum as the second outgroup and some other ingroup accessions (B. nigra, R. sativus, and Sinapis alba) were obtained from Yang et al. (2002). The S. altissimum sequence was published by Hall et al. (2002). Sources and GenBank accession numbers are listed in Table 1.
The outgroup choice was based on previous family-wide phylogenetic reconstructions (Koch et al. 2000, 2001, 2003b; Koch 2003). From these studies it is obvious that species relationships are as follows: {Lepidium {Noccaea {ingroup taxa}}}. The distant phylogenetic position of Lepidium has been substantiated recently by the occurrence of plastid pseudogenes (Koch et al. 2005). From this study it is also obvious that Lepidium represents a basal member of the evolutionary lineage comprising the genus Arabidopsis (Koch et al. 2005). In the case of C. maritima, we obtained a sequence from the _trn_L-F intergenic spacer, which was not alignable with all remaining sequences. Consequently, we analyzed two different data sets (with and without the C. maritima data). In both cases the maximum-likelihood method retained identical tree topologies, and we present the results from both analyses to demonstrate which degree of confidence values (bootstrap support) is affected (Fig. 3). The corresponding alignment is available on request.
For phylogenetic reconstructions, insertions and deletions were treated as missing data. Characters and character states were weighted equally (Fitch parsimony). All phylogenetic analyses were run using the PAUP 4.0* beta10 version (Swofford 2002). Parsimony was performed under the following conditions: HEURISTIC, TBR, STEEPEST DESCENT. The bootstrap option of PAUP (100 replicates) was used to assess relative support in the unweighted analysis. The maximum-likelihood option was also used for phylogenetic reconstructions under the PAUP standard settings assuming constant mutational rates among lineages and empirical base frequencies of A = 0.324, C = 0.184, G = 0.168, and T = 0.322.
The plastid _trn_L intron/_trn_L-F intergenic spacer region did not evolve constantly as revealed by relative rate tests (data not shown), and, consequently, the data set violated any assumptions of a constant molecular clock. However, within the Brassicaceae family the _trn_L-F spacer region is highly subjected to trn_F(GAA) pseudogene integration, and further research is needed before molecular clock assumptions can be applied correctly (Koch et al. 2005). Therefore, divergence time estimates were calculated according to the previously published sequence set for the nuclear encoded chalcone synthase gene (chs) (Koch et al. 2001). For the chs we calculated a synonymous mutation rate of 1.4 × 10-8 mutations/site/year (Koch et al. 2001). Divergence time estimates could be easily estimated using the equation: KS/2_T = synonymous mutation rate. The advantages of using the chalcone synthase data set are: (1) Constancy of mutational rates has been demonstrated over the entire family, and therefore, a molecular clock can be applied. (2) The chs data set comprises several taxa included in the present study, which allows a calculation of simple means of divergence time estimates.
Acknowledgments
We are thankful to T. Ksiazczyk (University of Silesia, Katowice, Poland) for providing seeds of rapid-cycling Brassica species and M. Kühne for technical assistance. This work was supported by a grant of the German Research Foundation (DFG) to M.A.L. (LY 19/1-1).
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: T. Ksiazczyk.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3531105\. Article published online before print in March 2005.
References
- Álvarez, I. and Wendel, J.F. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phyl. Evol. 29**:** 417-434. [DOI] [PubMed] [Google Scholar]
- Anderson, J.K. and Warwick, S.I. 1999. Chromosome number evolution in the tribe Brassiceae (Brassicaceae): Evidence from isozyme number. Pl. Syst. Evol. 215**:** 255-285. [Google Scholar]
- Appel, O. and Al-Shehbaz, I.A. 2003. Cruciferae. In The families and genera of vascular plants (eds. K. Kubitzki and C. Bayer), pp. 75-174. Springer-Verlag, Berlin, Heidelberg.
- Axelsson, T., Bowman, C.M., Sharpe, A.G., Lydiate, D.J., and Lagercrantz, U. 2000. Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 43**:** 679-688. [PubMed] [Google Scholar]
- Babula, D., Kaczmarek, M., Barakat, A., Delseny, M., Quiros, C.F., and Sadowski, J. 2003. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: Complexity of the comparative map. Mol. Gen. Genomics 268**:** 656-665. [DOI] [PubMed] [Google Scholar]
- Boivin, K., Acarkan, A., Mbulu, R.-S., Clarenz, O., and Schmidt, R. 2004. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135**:** 735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422**:** 433-438. [DOI] [PubMed] [Google Scholar]
- Catcheside, D.G. 1934. The chromosomal relationships in the Swede and turnip groups of Brassica. Ann. Bot. 48**:** 601-633. [Google Scholar]
- Cavell, A.C., Lydiate, D.J., Parkin, I.A.P., Dean, C., and Trick, M. 1998. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41**:** 62-69. [PubMed] [Google Scholar]
- Davies, T.J., Barraclough, T.G., Chase, M.W., Soltis, P.S., Soltis, D.E., and Savolainen, V. 2004. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proc. Natl. Acad. Sci. 101**:** 1904-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobeš, C., Mitchell-Olds, T., and Koch, M. 2004. Phylogeographic analysis of extensively sympatric and highly diverse chloroplast haplotypes (trnL intron–trnF IGS) in North American _Arabis drummondii, A._x divaricarpa, and A. holboellii (Brassicaceae). Mol. Ecol. 13**:** 349-370. [DOI] [PubMed] [Google Scholar]
- Dover, G.A. 1982. Molecular drive: A cohesive mode of species evolution. Nature 299**:** 111-117. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 19**:** 11-15. [Google Scholar]
- Gómez-Campo, C. 1999. Taxonomy. In Biology of Brassica coenospecies (ed. C. Gómez-Campo), pp. 3-32. Elsevier, Amsterdam.
- Hall, J.C., Sytsma, K.J., and Iltis, H.H. 2002. Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Am. J. Botany 89**:** 1826-1842. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Cheng, Z.K., Wang, M.L., Goodman, H.M., and Jiang, J.M. 2000. Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics 156**:** 833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, M. 2003. Molecular phylogenetics, evolution and population biology in the Brassicaceae. In Plant genome: Biodiversity and evolution. Vol. 1: Phanaerogams (eds. A.K. Sharma and A. Sharma), pp. 1-35. Science Publishers, Enfield, NH. [Google Scholar]
- Koch, M. and Al-Shehbaz, I.A. 2004. Taxonomic and phylogenetic evaluation of the American “_Thlaspi_” species: Identity and relationship to the Eurasian genus Noccaea (Brassicaceae). System. Botany 29**:** 375-384. [Google Scholar]
- Koch, M. and Kiefer, M. 2005. Genome evolution among cruciferous plants—A lecture from the comparison of the genetic maps of three diploid species: Capsella rubella, Arabidopsis lyrata ssp. petraea and Arabidopsis thaliana. Am. J. Botany (in press). [DOI] [PubMed]
- Koch, M., Haubold, B., and Mitchell-Olds, T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17**:** 1483-1498. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Botany 88**:** 534-544. [PubMed] [Google Scholar]
- Koch, M., Dobeš, C., and Mitchell-Olds, T. 2003a. Multiple hybrid formation in natural populations: Concerted evolution of the internal transcribed spacer of nuclear ribosomal DNA (ITS) in North American Arabis divaricarpa (Brassicaceae). Mol. Biol. Evol. 20**:** 338-350. [DOI] [PubMed] [Google Scholar]
- Koch, M., Al-Shehbaz, I., and Mummenhoff, K. 2003b. Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae). Ann. Missouri Botan. Garden 90**:** 151-171. [Google Scholar]
- Koch, M.A., Dobeš, C., Matschinger, M., Bleeker, W., Vogel, J., Kiefer, M., and Mitchell-Olds, T. 2005. Evolution of the trnF(GAA) gene in Arabidopsis relatives and the Brassicaceae family: Monophyletic origin and subsequent diversification of a plastidic pseudogene. Mol. Biol. Evol. (in press). [DOI] [PubMed]
- Kuittinen, H., de Haan, A.A., Vogl, C., Oikarinen, S., Leppälä, J., Koch, M., Mitchell-Olds, T., Langley, C.H., and Savolainen, O. 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168**:** 1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U. 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150**:** 1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U. and Lydiate, D. 1996. Comparative genome mapping in Brassica. Genetics 144**:** 1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, T.-H., DelMonte, T.A., Reischmann, K.P., Hyman, J., Kowalski, S.P., McFerson, J., Kresovich, J., and Paterson, A.H. 2000. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 10**:** 776-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Gao, M., Yang, B., and Quiros, C.F. 2003. Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor. Appl. Genet. 107**:** 168-180. [DOI] [PubMed] [Google Scholar]
- Lukens, L., Zou, F., Lydiate, D., Parkin, I., and Osborn, T. 2003. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164**:** 359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, L.N., Quijada, P.A., Udall, J., Pires, J.C., Schranz, M.E., and Osborn, T.C. 2004. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linnean Soc. 82**:** 665-674. [Google Scholar]
- Lysak, M.A., Fransz, P.F., Ali, H.B.M, and Schubert, I. 2001. Chromosome painting in Arabidopsis thaliana. Plant J. 28**:** 689-697. [DOI] [PubMed] [Google Scholar]
- Lysak, M.A., Pecinka, A., and Schubert I. 2003. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 11**:** 195-204. [DOI] [PubMed] [Google Scholar]
- Osborn, T.C. 2004. The contribution of polyploidy to variation in Brassica species. Physiol. Plant. 121**:** 531-536. [Google Scholar]
- Parkin, I.A.P., Lydiate, D.J., and Trick, M. 2002. Assessing the level of collinearity between Arabidopsis thaliana and Brassica napus for A. thaliana chromosome 5. Genome 45**:** 356-366. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P., Sharpe, A.G., and Lydiate, D.J. 2003. Patterns of genome duplication within the Brassica napus genome. Genome 46**:** 291-303. [DOI] [PubMed] [Google Scholar]
- Pires, J.C., Zhao, J., Schranz, M.E., Leon, E.J., Quijada, P.A., Lukens, L.N., and Osborn, T.C. 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linnean Soc. 82**:** 675-688. [Google Scholar]
- Prakash, S. and Hinata, K. 1980. Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot. 55**:** 1-57. [Google Scholar]
- Prakash, S., Takahata, Y., Kirti, P.B., and Chopra, V.L. 1999. Cytogenetics. In Biology of Brassica coenospecies (ed. C. Gómez-Campo), pp. 59-106. Elsevier, Amsterdam.
- Rieseberg, L.H. 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16**:** 351-358. [DOI] [PubMed] [Google Scholar]
- Röbbelen, G. 1960. Beiträge zur Analyse des _Brassica_-Genoms. Chromosoma 11**:** 205-228. [DOI] [PubMed] [Google Scholar]
- Song, K., Lu, P., Tang, K., and Osborn, T.C. 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. 92**:** 7719-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. 2002. Phylogenetic analysis using parsimony (and other methods). Version 4. Sinauer, Sunderlands, MA.
- Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17**:** 1105-1109. [DOI] [PubMed] [Google Scholar]
- Truco, M.J., Hu, J., Sadowski, J., and Quiros, C.F. 1996. Inter- and infra-genomic homology of the Brassica genomes: Implications for their origin and evolution. Theor. Appl. Genet. 93**:** 1225-1233. [DOI] [PubMed] [Google Scholar]
- U, N. 1935. Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan J. Bot. 7**:** 389-452. [Google Scholar]
- Uyenoyama, M.K. 1995. A generalized least-square estimate for the origin of sporophytic self-incompatibility. Genetics 139**:** 975-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick, S.I. and Black, L.D. 1991. Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae)—Chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82**:** 81-92. [DOI] [PubMed] [Google Scholar]
- ———. 1997a. Molecular phylogenies from theory to application in Brassica and allies (Tribe Brassiceae, Brassicaceae). Opera Bot. 132**:** 159-168. [Google Scholar]
- ———. 1997b. Phylogenetic implications of chloroplast DNA restriction site variation in subtribes Raphaninae and Cakilinae (Brassicaceae, tribe Brassiceae). Can. J. Bot. 75**:** 960-973. [Google Scholar]
- Warwick, S.I. and Sauder, C. 2005. Phylogeny of tribe Brassiceae (Brassicaceae) based on chloroplast restriction site polymorphisms and nuclear ribosomal internal transcribed spacer (ITS) and chloroplast _trn_L intron sequences. Can. J. Bot. (in press).
- Warwick, S.I., Francis, A., and La Fleche, J. 2000. Guide to the wild germplasm of Brassica and allied crops (tribe Brassiceae, Brassicaceae), II. Chromosome numbers, 2nd ed., pp. 1-49. Agriculture and Agri-Food Canada, Eastern Cereal and Oilseed Research Centre, Ottawa, Ontario, Canada.
- Warwick, S.I., Al-Shehbaz, I.A., Price, R.A., and Sauder, C. 2002. Phylogeny of Sisymbrium (Brassicaceae) based on ITS sequences of nuclear ribosomal DNA. Can. J. Bot. 80**:** 1002-1017. [Google Scholar]
- Williams, P.H. and Hill, C.B. 1986. Rapid-cycling populations of Brassica. Science 232**:** 1385-1389. [DOI] [PubMed] [Google Scholar]
- Yang, Y.W., Lai, K.N., Tai, P.Y., and Li, W.H. 1999. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48**:** 597-604. [DOI] [PubMed] [Google Scholar]
- Yang, Y.W., Tai, P.Y., Chen, Y., and Li, W.H. 2002. A study of the phylogeny of Brassica rapa, B. nigra, Raphanus sativus, and their related genera using noncoding regions of chloroplast DNA. Mol. Phylogenet. Evol. 23**:** 268-275. [DOI] [PubMed] [Google Scholar]
- Zimmer, E.A., Martin, S.L., Beverley, S.M., Kan, Y.M., and Wilson, A.C. 1980. Rapid duplication and loss of genes-coding for the α-chains of hemoglobin. Proc. Natl. Acad. Sci. 77**:** 2158-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski, P.A. and Sadowski, J. 2002. FISH-mapping of rDNAs and Arabidopsis BACs on pachytene complements of selected Brassicas. Genome 45**:** 189-197. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.arabidopsis.org; The Arabidopsis Information Resource.