Use of Thioredoxin as a Reporter To Identify a Subset of Escherichia coli Signal Sequences That Promote Signal Recognition Particle-Dependent Translocation (original) (raw)
Abstract
We have previously reported that the DsbA signal sequence promotes efficient, cotranslational translocation of the cytoplasmic protein thioredoxin-1 via the bacterial signal recognition particle (SRP) pathway. However, two commonly used signal sequences, those of PhoA and MalE, which promote export by a posttranslational mechanism, do not export thioredoxin. We proposed that this difference in efficiency of export was due to the rapid folding of thioredoxin in the cytoplasm; cotranslational export by the DsbA signal sequence avoids the problem of cytoplasmic folding (C. F. Schierle, M. Berkmen, D. Huber, C. Kumamoto, D. Boyd, and J. Beckwith, J. Bacteriol. 185 **:**5706-5713, 2003). Here, we use thioredoxin as a reporter to distinguish SRP-dependent from non-SRP-dependent cleavable signal sequences. We screened signal sequences exhibiting a range of hydrophobicity values based on a method that estimates hydrophobicity. Successive iterations of screening and refining the method defined a threshold hydrophobicity required for SRP recognition. While all of the SRP-dependent signal sequences identified were above this threshold, there were also a few signal sequences above the threshold that did not utilize the SRP pathway. These results suggest that a simple measure of the hydrophobicity of a signal sequence is an important but not a sufficient indicator for SRP recognition. In addition, by fusing a number of both classes of signal sequences to DsbA, we found that DsbA utilizes an SRP-dependent signal sequence to achieve efficient export to the periplasm. Our results suggest that those proteins found to be exported by SRP-dependent signal sequences may require this mode of export because of their tendency to fold rapidly in the cytoplasm.
Several pathways have been described for the export of proteins to the periplasm of the bacterium Escherichia coli (13). Of these, the most widely used and conserved system is the general secretory (Sec) pathway, which passes proteins through the SecYEG membrane-embedded translocon (13). Periplasmic proteins, outer membrane proteins, and at least one inner membrane protein (31) are recognized and targeted to the Sec pathway by short, cleavable, N-terminal signal sequences.
Josefsson and Randall showed that, among several periplasmic proteins that use the Sec pathway, all are exported posttranslationally (19, 34). That is, the translocation of these proteins across the cytoplasmic membrane begins only after a substantial amount of the polypeptide chain has been synthesized. As a result of these studies, it has traditionally been assumed that all proteins targeted for translocation by a cleavable signal sequence are exported posttranslationally in E. coli.
Since folded proteins cannot pass through the SecYEG translocon, features of the cytoplasm ensure that precursor proteins destined to be exported do not fold stably into their final conformations (14, 39). Two periplasmic proteins, alkaline phosphatase (PhoA) and maltose binding protein (MalE), illustrate this principle. PhoA requires disulfide bonds to achieve a stable conformation. These covalent modifications do not take place in the cytoplasm and can only occur in the periplasm catalyzed by the oxidoreductase, DsbA (21). MalE is maintained in an unfolded conformation by the cytoplasmic chaperone SecB (14). The signal sequence of MalE also plays a role in SecB recognition by retarding folding enough to allow an interaction with SecB (40).
In addition to the posttranslational export mechanism, there is a pathway for cotranslational targeting of proteins to the SecYEG translocon. This latter pathway is mediated by the signal recognition particle (SRP), a ribonucleoprotein whose components, the 54-kDa protein homologue (Ffh) and 4.5S RNA, are widely conserved across all domains of life (28). It is currently thought that proteins exported by the SRP pathway are recognized as substrates for this pathway based on their highly hydrophobic signal sequences (25). In fact, it appeared initially that the function of this pathway was restricted to the assembly of the highly hydrophobic membrane proteins into the cytoplasmic membrane. Recently, however, evidence has been presented that some proteins with cleavable signal sequences are cotranslationally exported to the periplasm. One native E. coli protein, DsbA, is a substrate for the SRP pathway (37), and artificially increasing the hydrophobicity of a number of other cleavable signal sequences will target them to this pathway (4, 25). In addition, the Hbp signal sequence appears to target proteins to the SRP pathway (38). However, this signal sequence contains a long N-terminal extension that may affect its targeting in unknown ways.
In contrast to proteins destined to be exported, cytoplasmic proteins presumably fold rapidly in the cytoplasm. Thus, attempts to export proteins that are normally located in the cytoplasm by attaching a cleavable signal sequence to them has generally met with, at best, only partial success. In the case of the cytoplasmic protein, thioredoxin-1, only a small percentage of the protein is exported when it is fused to the MalE or PhoA signal sequences. However, thioredoxin is exported with high efficiency when fused to the DsbA signal sequence (18, 37). We have attributed this difference to the ability of the DsbA signal sequence to bypass cytoplasmic folding of thioredoxin by promoting cotranslational translocation via the SRP pathway (37). We will present direct evidence in a subsequent paper for the role of thioredoxin folding in its ability to be exported (D. Huber, M. Cha, M. L. Tasayco, A. G. Planson, L. Debarbieux, A. Chaffotte, and J. Beckwith, unpublished data).
These findings raise two questions. What features of cleavable signal sequences target some to the SRP-dependent, cotranslational pathway and others to the posttranslational pathway? To answer this question, we screened for native E. coli signal sequences that promote export of thioredoxin and those that do not. By analyzing them, we have found hydrophobicity scales that allow us to predict a minimum threshold for SRP recognition. However, results presented in this paper also suggest that features other than hydrophobicity may influence the choice of export pathways. Why do some proteins, such as DsbA, utilize cleavable signal sequences that interact with the SRP pathway while most of them do not? We have constructed fusions of signal sequences to the mature portion of DsbA that promote co- or posttranslational translocation. Our results suggest that, in the case of DsbA, it may be exported cotranslationally to avoid protein folding in the cytoplasm which would inhibit its export.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids are listed in Table 1. Cells were generally grown at 37°C in NZ medium, described previously (35). For pulse-labeling, cells were grown in minimal glycerol medium (M63 salts with 0.4% glycerol, 1 mg/ml of vitamin B1, 1 mM MgSO4, 50 mg/ml of 18 amino acids [methionine and cysteine were not included in the amino acid solution]). Antibiotic selection was maintained for selected markers at the following concentrations: ampicillin, 200 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 40 μg/ml; tetracycline, 15 μg/ml. Induction of lac promoter constructs was accomplished by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 10 μM.
TABLE 1.
Strains and Plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| MC1000 | F−araD139 Δ(ara-leu)7697 galU galK ΔlacX74 rpsL thi | 9 |
| MC1061 | F−hsdR2 mcrA mcrB1 araD139 Δ(ara-leu)7696 ΔlacX74 galE15 galU galK16 rpsL thi | 6 |
| MC4100 | F−araD139 Δ(argF-lac)U169 prsL150 relA1 deoC1 rbsR fthD5301 fruA25 | 42 |
| DHB3 | MC1000 Δ_malF3_ Δ(_phoA_[_PvuII/_]) phoR | 16 |
| DHB4 | DHB3/F′ _lac-pro lacl_q | 16 |
| MB68 | DHB4 Δ_dsbA_ | Laboratory stock |
| JAH143 | MC1061 Δ_malE444_/F′ _lacI_q Kanr | 37 |
| DRH223 | JAH143 Δ_trxA_ | This study |
| CFS456 | MC4100 Δ_ara714_ Δ_trxA_ | 37 |
| CFS459 | CFS456 ffh77 | This study |
| Plasmids | ||
| pTRC99a | IPTG inducible, high expression | Promega |
| pDSW204 | pTRC99a, attenuated promoter | 43 |
| pDSW206 | pTRC99a, attenuated promoter | 43 |
| pCFS119 | pDSW204 + dsbAss | 37 |
| pCFS122 | pDSW204 + phoAss | 37 |
| pCFS123 | pCFS119 + trxA | 37 |
| pCFS126 | pCFS122 + trxA | 37 |
| pMO1 | pTRC99a + trxA with 5′ NheI site | This study |
| pMO2 | pDSW204 + trxA with 5′ NheI site | This study |
| pMO3 | pDSW206 + trxA with 5′ NheI site | This study |
| pMO15 | pMO2 + torTs | This study |
| pMO16 | pMO2 + appAss | This study |
| pDHSS0 | pMO2 + a_smAss_ | This study |
| pDHSS1 | pMO2 + sfmCss | This study |
| pDHSS5 | pMO2 + fecBss | This study |
| pDHSS9 | pMO2 + treAss | This study |
| pDHSS11 | pMO2 + papJss | This study |
| pDHSS13 | pMO2 + pcoEss | This study |
| pDHSS14 | pMO2 + tolBss | This study |
| pDHSS17 | pMO2 + flgAss | This study |
| pDHSS18 | pMO2 + yraPss | This study |
| pDHSS19 | pMO2 + artIss | This study |
| pDHSS20 | pMO2 + yraIss | This study |
| pDHSS21 | pMO2 + ccmHss | This study |
| pDHSS22 | pMO2 + focCss | This study |
| pDHSS24 | pMO2 + nikAss | This study |
| pDHSS25 | pMO2 + ycfSss | This study |
| pDHSS29 | pMO2 + livJss | This study |
| pDHSS34 | pMO2 + agpss | This study |
| pDHSS35 | pMO2 + traUss | This study |
| pDHSS37 | pMO2 + ybcLss | This study |
| pDHSS45 | pMO2 + livKss | This study |
| pDHSS46 | pMO2 + artJss | This study |
| pDHSS49 | pMO2 + btuFss | This study |
| pDHSS60 | pMO2 + ecoTss | This study |
| pDHSS63 | pMO2 + flgIss | This study |
| pDHSS64 | pMO2 + fepBss | This study |
| pDHSS66 | pMO2 + ansBss | This study |
| pDHSS72 | pMO2 + mepAss | This study |
| pDHSS76 | pMO2 + dsbCss | This study |
| pDHSS80 | pMO2 + ivyss | This study |
| pDH273 | pCFS119 + mature portion of dsbA | This study |
| pDH275 | pCFS122 + mature portion of dsbA | This study |
| pDH358 | pDSW204 + mature dsbA with 5′ NheI site | This study |
| pDH359 | pDH358 + sfmCss | This study |
| pDH360 | pDH358 + treAss | This study |
| pDH361 | pDH358 + pcoEss | This study |
| pDH362 | pDH358 + tolBss | This study |
| pDH378 | pDH358 + malEss | This study |
Strain and plasmid construction.
Standard genetic and molecular techniques were used in strain and plasmid construction (Table 1) (30, 36). All restriction enzymes were obtained from New England Biolabs. Plasmids pDH273 and pDH275 were made by cloning a PCR-amplified fragment of the DNA corresponding to the mature portion of DsbA cut with NcoI and XbaI into pCFS119 and pCFS122 cut with the same enzymes, respectively. A methionine residue was added to the coding sequence between the respective signal sequence and the mature portion of DsbA to accommodate an NcoI restriction site. The screening plasmid pMHO2 was constructed by ligating a PCR-amplified fragment of the trxA gene into the plasmid vector pDSW204 (43). Primers for the trxA fragment were designed so that it could be cloned into the BamHI and HindIII sites of pDSW204 and leave a unique NheI site at the 5′ end of the trxA gene. This inserted an alanine residue after the start codon so that the TrxA sequence reads MASDKII. When fusions were made to signal peptides, this alanine remained at the fusion junction (usually this was the last residue of the signal sequence). pDH358 was constructed similarly, such that a serine residue was inserted just 5′ of the coding sequence for the mature portion of DsbA (reading SAQYED). Signal sequence fusions to thioredoxin or DsbA were constructed by synthesizing overlapping primers coding for the signal sequence. The annealed primers had 5′ overhangs—CATG at the 5′ end and CTAG at the 3′ end—compatible with NcoI and NheI and were ligated into pMHO2 or pDH358 cut with NcoI and NheI.
Subcellular fractionation.
Cells were grown as described to saturation and then subcultured 1:100 and grown to an optical density at 600 nm (OD600) of 0.5. Cultures were centrifuged, and pellets were resuspended in 50 mM Tris, pH 8, 18% sucrose, 1 mM CaCl2. EDTA (0.5 mM) and lysozyme (0.5 μg/ml) were added, and samples were left on ice for 30 min before centrifugation at 3,100 × g in a benchtop centrifuge for 5 min. Supernatants and pellets were used as periplasmic and spheroplast fractions, respectively.
Fractionation of cells into soluble and membrane fractions was done by lysing whole cells and pelleting membranes by ultracentrifugation. Cultures were grown to an OD600 of 0.6 and resuspended in 50 mM Tris (pH 8.3), 1 mM EDTA, 150 mM NaCl. Cells were lysed by passage twice through a French pressure cell. Cell debris and unlysed cells were pelleted at 26,000 × g and discarded. The clarified lysate was then subjected to ultracentrifugation at >100,000 × g for 1 h. The resulting pellet fraction (membrane) was resuspended in a volume of lysis buffer equal to that of the supernatant fraction (soluble). In both fractionation protocols, fractions were precipitated with trichloroacetic acid (TCA) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Western blots.
Cell extracts were subjected to SDS-PAGE and transferred to nitrocellulose membrane using a semidry apparatus from Bio-Rad. Rabbit anti-thioredoxin-1 and anti-β-lactamase antibodies for probing membranes were obtained from Sigma and 5′-3′, respectively. All other antisera used for probing membranes were from laboratory stocks. Immunodetection was done according to the ECL protocol (Amersham) using streptavidin-horseradish peroxidase.
Pulse-chase experiments.
Pulse-chase analysis was performed as described previously (26). Cells were grown at 30°C in M63 salts supplemented with all the amino acids (except for methionine and cysteine), 0.2% glucose, and 10 μM IPTG. When the culture reached an OD600 of approximately 0.2, aliquots for the control sample were removed and 1.5 mM azide was added for 5 min. The cells were pulse-labeled with 60 μCi of [35S]methionine per ml for 15 s and chased with 10 μg/ml of cold methionine for the indicated period of time. Labeling was terminated by the addition of TCA to 5%, and the resultant precipitation was washed once with acetone to remove excess TCA. Samples were immunoprecipitated using anti-DsbA antibodies and protein A and analyzed by SDS-PAGE and autoradiography.
Computational analysis.
For our data set, we chose a set of proteins consisting of all of the E. coli K-12 proteins in the SwissProt databank with the word signal appearing in either the keyword or feature lines of the annotations. This list was further narrowed down to 171 representative signal sequences by eliminating all proteins with the words “potential,” “by similarity,” “probable,” or “TAT” in the annotation.
The hydrophobicity of the signal sequences was assessed using an algorithm modified from Boyd et al. (5). The software scans a given sequence using varying window lengths (in amino acids) and records the average hydrophobicity of the most hydrophobic window. We optimized our algorithm by varying the window size and scale used. Scales used were those derived by Chothia, Eisenberg et al., Engleman et al., Hopp and Woods, Janin, Kyte and Doolittle, and Wimley and White (7, 11, 12, 15, 17, 24, 45). Additional scales used were those contained in the AAindex database (http://www.genome.ad.jp/dbget/aaindex.html) (23) and the 88 scales in the SPLIT web server (http://garlic.mefos.hr/split/) (20). Where not noted, the scale used was the JTT2 scale (5).
RESULTS
Signal sequence screen.
We set out to determine which E. coli cleavable signal sequences are capable of exporting thioredoxin to the periplasm and which are not. Our previous results suggested that thioredoxin could be used to distinguish those signal sequences that are SRP dependent from those that promote posttranslational export (37). By comparing large numbers of signal sequences that directed proteins to the SRP pathway to those that did not, we hoped to determine the key features of the SRP-dependent signal sequences. We have previously concluded that the DsbA signal sequence is significantly more hydrophobic than either the PhoA or MalE signal sequence. We have proposed that the difference in the behavior of cleavable signal sequences was based on the varying hydrophobicity of their hydrophobic cores (4, 37).
Therefore, we selected signal sequences to test based on a hydrophobicity analysis, choosing a collection of signal sequences that included those with the highest hydrophobicities and those with lower hydrophobicities. Our approach was to fuse a small subset of native E. coli signal sequences to thioredoxin and assay for export by subcellular fractionation. With these results, we then assessed the hydrophobicity of the signal sequences, using a number of different hydrophobicity scales and varying the number of residues chosen (i.e., window length) over which the hydropathy calculation was made. The maximum hydropathy value was then taken to represent the hydrophobicity of the signal sequence. We chose the scale and window length that best distinguished between the two classes of signal sequences. Our analysis was done in an iterative fashion. That is, after we analyzed the thioredoxin export results from the initial subset of signal sequences, we examined the export properties of an additional collection of signal sequences, choosing them based on the hydropathy scale and window that best separated the two classes of signal sequences already tested. With results from the new subset, we repeated the hydropathy analysis to find the optimum parameters. By this means, we hoped to arrive at a method for calculating hydrophobicity that would cleanly distinguish between SRP-dependent and non-SRP-dependent signal sequences.
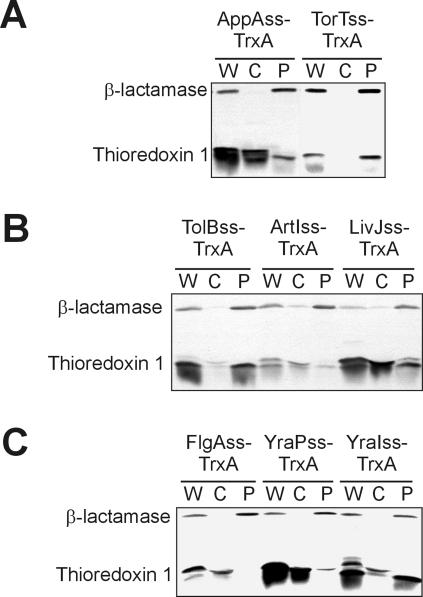
Our initial studies, with only the three signal sequences mentioned above, suggested that we might be able to use the JTT2 hydropathy scale and a window length of 11 amino acids to distinguish SRP-dependent from non-SRP-dependent signal sequences (37). Using these parameters, the DsbA signal sequence has a higher hydrophobicity than both PhoA and MalE, and the differences in hydrophobicities between the former and the latter is at a maximum. When we used these conditions to analyze a representative set of 171 signal sequences from E. coli, we noticed that several signal sequences were more hydrophobic than the DsbA signal sequence. We fused two of these, AppA and TorT, to thioredoxin. However, only the TorT signal sequence promoted translocation of thioredoxin (Fig. 1A).
FIG. 1.
A subset of signal sequences can export thioredoxin-1 to the periplasm. Western analysis of cells (DRH223) expressing signal sequence-thioredoxin fusions fractionated into cytoplasmic and periplasmic fractions by spheroplasting. (A) Cells expressing thioredoxin (TrxA) fused to the AppA (pMO16) and TorT (pMO15) signal sequences (AppAss and TorTss, respectively) from the first iteration of the signal sequence screen. (B) Representative samples from the second iteration of signal sequence screen: TrxA fused to the TolB (pDHSS14), ArtI (pDHSS19), and LivJ (pDHSS29) signal sequences (TolBss, ArtIss, and LivJss, respectively). (C) Representative samples from the third iteration of signal sequence screen: TrxA fused to the FlgA (pDHSS17), YraP (pDHSS18), and YraI (pDHSS20) signal sequences (FlgAss, YraPss, and YraIss, respectively). β-Lactamase is included in all cases as a periplasmic control. W, whole-cell extract; C, cytoplasm plus cytoplasmic membrane fraction; P, periplasm fraction.
Using two scales (JTT2 and Kyte-Doolittle) and a range of window lengths from 7 to 19 amino acids (excluding even-numbered window lengths for theoretical reasons), we found the best ranking of the signal sequences was obtained using the JTT2 scale with a window length of 17. Under these conditions, the DsbA and TorT signal sequences were among the most hydrophobic and the PhoA, MalE, and AppA signal sequences were among the least hydrophobic. Using this refined method, we selected 17 signal sequences for fusion to thioredoxin which had hydrophobicities ranging from the most hydrophobic to the lower hydrophobicity of the MalE signal sequence. (The MalE signal sequence had the highest hydrophobicity of those signal sequences that did not utilize the SRP pathway.) Of these, only two, the SfmC and TolB signal sequences, support export of thioredoxin (examples of results are shown in Fig. 1B). In addition, we found that, with these newly added sequences, a window length of 15 with the JTT2 scale gave a better fit to the data. The only signal sequences to this point that would export thioredoxin were among the top 20 in terms of their hydrophobicity. This continued to point to hydrophobicity as the key distinguishing factor.
To more precisely establish a cutoff in hydrophobicity required for thioredoxin export, we screened as many as possible of the remaining signal sequences that scored between the most hydrophobic and the 31st most hydrophobic. Of these, two were not expressed well (Trf5, AggD), five could not be cloned by the method used (YcbF, HdeA, FimC, YehC, PrsJ), one was predicted to be exported by an alternative pathway (the TAT pathway—NrfA), and one was a misannotated transmembrane protein (DegS). Of the 13 most hydrophobic signal sequences that were testable, five (FocC, NikA, CcmH, YraI, and FlgI) promoted export of thioredoxin (examples of results are shown in Fig. 1C).
Our fractionation method does not distinguish between cytoplasmic proteins and proteins bound to the cytoplasmic membrane. We worried that the apparent inability of some signal sequences to export thioredoxin as deduced from our fractionation studies could be due to an inability of the signal sequence to be processed from thioredoxin after export. This would result in significant amounts of thioredoxin tethered to the cytoplasmic membrane and appearing in what we termed the cytoplasmic fraction. To test this possibility, we fractionated lysed cells expressing several highly hydrophobic signal sequence fusions that do not export thioredoxin into soluble and membrane-bound fractions by ultracentrifugation. All fusion proteins tested were found to be entirely in the soluble fraction (Fig. 2), whereas a membrane protein control, FtsQ, was entirely in the pellet fraction (data not shown). We conclude from these studies that those proteins classified as nonexported by our first fractionation procedure were, in fact, located in the cytoplasm.
FIG. 2.
Thioredoxin fusion proteins are not tethered to the cytoplasmic membrane by their signal sequence. Cells (DRH223) expressing thioredoxin (TrxA) fused to the DsbA (pCFS123), TreA (pDHSS9), FlgI (pDHSS17), and PcoE (pDHSS13) signal sequences (DsbAss, TreAss, FlgIss, and PcoEss, respectively) were fractionated into membrane-bound and soluble fractions by ultracentrifugation and analyzed by Western blotting against thioredoxin. W, whole-cell extract; M, 100,000 × g centrifugation membrane pellet fraction; S, soluble supernatant fraction.
In summary, we screened a total of 36 signal sequences for their ability to promote export of thioredoxin. These signal sequences were among those with hydrophobicities between the most hydrophobic and the 85th most hydrophobic according to our analysis. Of these, only nine were able to export thioredoxin. All nine were among the 26 most hydrophobic signal sequences according to the method we used to analyze them. Essentially, no export of thioredoxin was observed with any signal sequence that ranked lower than 26th (of 171 analyzed for hydrophobicity).
SRP dependence.
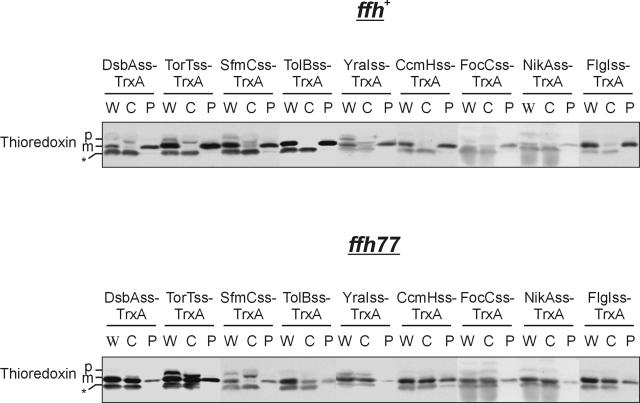
Previously, we reported that export of the DsbA signal sequence/thioredoxin fusion protein is SRP dependent. We wished to determine whether this was true of the newly characterized signal sequences which efficiently exported thioredoxin. We found that all eight of these signal sequences (DsbA was previously tested) were defective for export in a strain bearing the SRP mutation ffh77, supporting our hypothesis (Fig. 3) (41).
FIG. 3.
Signal sequences that export thioredoxin are dependent on the SRP. Western analysis of ffh + cells (CFS456) or ffh77 mutant cells (CFS459) expressing thioredoxin (TrxA) fused to the DsbA (pCFS123), TorT (pMO15), SfmC (pDHSS1), TolB (pDHSS14), YraI (pDHSS20), CcmH (pDHSS21), FocC (pDHSS22), NikA (pDHSS24), and FlgI (pDHSS63) signal sequences (DsbAss, TorTss, SfmCss, TolBss, YraIss, CcmHss, FocCss, NikAss, and FlgIss, respectively) fractionated by spheroplasting. We have previously observed the presence of mature thioredoxin in the cytoplasm of cells expressing signal sequence fusions. We attribute this to the degradation of the signal sequence in the cytoplasm due to its unstructured nature. Bands: *, cross-reacting cytoplasmic protein; p, precursor-containing signal sequence; m, mature protein. Lanes: W, whole-cell extract; C, cytoplasm plus cytoplasmic membrane fraction; P, periplasm fraction.
Further computational analysis.
Approximately half of the signal sequences above the cutoff in hydrophobicity required for thioredoxin export nonetheless do not support export of thioredoxin. We considered the possibility that the imprecision in fully separating SRP-dependent signal sequences from non-SRP-dependent signal sequences was due to limitations in the method we were using. We therefore sought to systematically optimize our prediction method.
To test whether we could identify SRP-dependent signal sequences either by other hydrophobicity scales or by using properties other than hydrophobicity, we altered our computer program to use scales measuring a variety of properties. These included the 494 scales contained in the AAindex databank (http://www.genome.ad.jp/aaindex/), 88 scales contained in the SPLIT web server (http://garlic.mefos.hr/split/), and 8 commonly used hydrophobicity scales. In addition, we systematically varied the window size between 7 and 19, including even numbered windows.
All scales that performed well in this reanalysis were hydrophobicity scales, confirming the assumption that the major factor distinguishing SRP-dependent signal sequences from the others was this property. In addition, we found the optimal window length to be 12 for most of the hydrophobicity scales tested. The scale that performed the best was that derived by Wertz and Scheraga (44) and measures the probability of a given amino acid being buried in the interior of a folded protein (Table 2). Other scales that performed well were our original JTT2 scale, the Goldman-Engleman-Steitz scale, and the Wimley-White scale (5, 12, 45). However, no scale or method tested is perfectly compatible with the experimental data. That is, no scale or method could completely separate the signal sequences into the two distinct classes (SRP versus non-SRP). Because with each iteration we progressively chose to analyze the most hydrophobic signal sequences, the differences between the hydrophobicities of the two groups using any scale are small. This makes assessing the statistical significance of the differences between scales difficult in terms of which scale is best for predicting SRP dependence. It is possible with testing of additional signal sequences that one of these other hydrophobicity scales will prove to be more accurate.
TABLE 2.
Optimized hydrophobicity calculation using the Wertz-Scheraga scalea
| Rank | Signal sequence | WS score |
|---|---|---|
| 1 | TorT | 0.758 |
| 2 | SfmC | 0.75 |
| 4 | TraU | 0.737 |
| 7 | FocC | 0.731 |
| 8 | TreA | 0.722 |
| 10 | CcmH | 0.72 |
| 11 | FecB | 0.718 |
| 12 | YraI | 0.718 |
| 13 | TolB | 0.712 |
| 16 | AsmA | 0.708 |
| 19 | NikA | 0.703 |
| 24 | FlgI | 0.698 |
| 26 | DsbA | 0.691 |
| 27 | AppA | 0.69 |
| 33 | PcoE | 0.683 |
| 34 | BtuF | 0.682 |
| 44 | PapJ | 0.673 |
| 46 | YbcL | 0.672 |
| 53 | DsbC | 0.667 |
| 58 | ArtJ | 0.664 |
| 62 | ArtI | 0.662 |
| 65 | YraP | 0.659 |
| 71 | YcfS | 0.658 |
| 72 | FlgA | 0.656 |
| 76 | LivK | 0.654 |
| 83 | Agp | 0.652 |
| 92 | ModA | 0.649 |
| 93 | MalE | 0.648 |
| 94 | PhoA | 0.648 |
| 104 | LivJ | 0.643 |
| 119 | FepB | 0.635 |
| 122 | EcoT | 0.633 |
| 142 | MepA | 0.621 |
| 155 | AnsB | 0.61 |
| 158 | Ivy | 0.608 |
Export of DsbA.
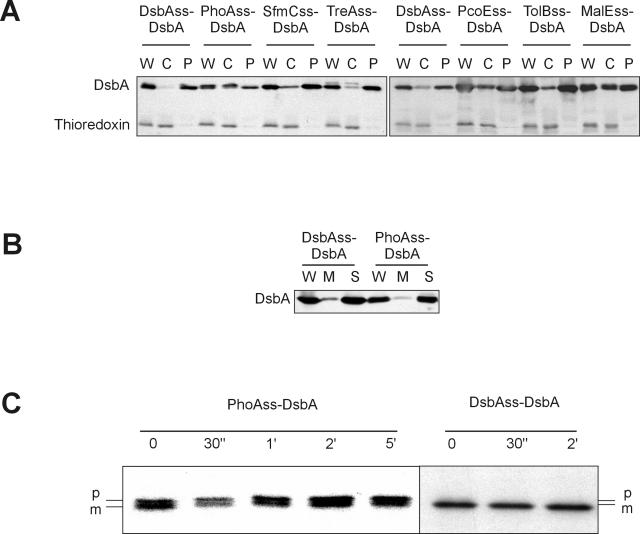
We are interested in the question of why some (a small fraction) E. coli proteins with cleavable signal sequences utilize the SRP pathway. We wished to test whether directing such proteins to the cotranslational pathway was necessary for export of such proteins or whether these proteins could be equally well exported by the posttranslational mechanism. To this end, we compared the export of DsbA when it contained its native signal sequence and when the DsbA signal sequence was replaced by the PhoA signal sequence. Export of the PhoAss-DsbA fusion was severely defective, as revealed by high steady-state levels of DsbA in the cytoplasmic fraction (Fig. 4A). Further, all of the precursor protein in the soluble fraction of cells separated into membrane and soluble fractions, indicating that the fusion is not membrane anchored (Fig. 4B). In addition, the kinetics of export of the PhoA signal sequence fusion is severely retarded compared with wild-type DsbA, as assayed by pulse-chase analysis. The half-life of proteolytic processing of the DsbA signal sequence from DsbA is less than our pulse point (<30 s), while that of the PhoA signal sequence fusion is ∼5 min (Fig. 4C).
FIG. 4.
DsbA requires an SRP-dependent signal sequence for efficient export to the periplasm. (A) Cells (MB68) expressing DsbA fused to the DsbA (pCFS123), PhoA (pCFS126), SfmC (pDH359), TreA (pDH360), PcoE (pDH362), TolB (pDH361), and MalE (pDH378) signal sequences (DsbAss, PhoAss, SfmCss, TreAss, PcoEss, TolBss, and MalEss, respectively) were fractionated by spheroplasting and analyzed by Western blotting. W, whole-cell extract; C, cytoplasm plus cytoplasmic membrane fraction; P, periplasm fraction. Thioredoxin is included as a cytoplasmic control. (B) Cells (MB68) expressing DsbA fused to the DsbA (pCFS123) or PhoA (pCFS126) signal sequence were fractionated by ultracentrifugation and analyzed by Western blotting. W, whole-cell extract; M, 100,000 × g centrifugation membrane pellet fraction; S, soluble supernatant fraction. (C) Pulse-chase immunoprecipitation of MB68 (Δ_dsbA_) cells expressing DsbA fused to either the PhoA (pDH275) or DsbA (pDH273) signal sequence (PhoAss or DsbAss, respectively) using anti-DsbA antibodies. Chase times are indicated: 0 or 30 min or 1, 2, or 5 h. p, precursor-containing signal sequence; m, mature protein.
To establish that these results were not specific to either the PhoA or DsbA signal sequences, we fused the DsbA mature protein to two signal sequences that can export thioredoxin (SfmC, TolB) and three that cannot (PcoE, TreA, MalE). The two SRP-dependent signal sequences promoted efficient export of DsbA, and two of the non-SRP-dependent signal sequences were clearly defective in their ability to export DsbA (Fig. 4A) (see Discussion for comments on the one non-SRP-dependent signal sequence that supports efficient translocation of DsbA). These results suggest that export of DsbA is generally much more efficient when it undergoes cotranslational targeting to the secretion machinery by SRP.
DISCUSSION
Our results show that the export of thioredoxin provides a sensitive method for detecting SRP-dependent signal sequences. Signal sequences fall into two distinct classes when fused to thioredoxin—those that can export thioredoxin and those that cannot. Those signal sequences that can export thioredoxin are all SRP dependent; the export of thioredoxin in these cases is strongly inhibited by a mutation in the gene ffh, which encodes a component of the SRP. We presume that those signal sequences that are incapable of supporting export of thioredoxin are not SRP dependent.
This method of detecting SRP-dependent signal sequences has allowed us to define criteria that largely separate the two classes of signal sequences. Through an iterative process, we found that hydropathy analysis over a defined length of amino acid sequence helped us define a cutoff point, below which no signal sequence promoted export of thioredoxin. Twenty-six of the 171 signal sequences subjected to hydropathy analysis were above this cutoff (that is, ∼15% were above this cutoff). Of these 26, 13 were tested by fusion to thioredoxin and nine supported SRP-dependent thioredoxin export (∼69% of those tested). If this result were to hold for the remainder of the 26 signal sequences, it would indicate that roughly 10% were SRP-dependent (15% of 69%). That is, if our assumptions are correct, about 10% of all periplasmic proteins are likely exported by the SRP pathway. There are predicted to be approximately 742 E. coli signal sequences (22). Therefore, we estimate that there may be 74 SRP-dependent cleavable signal sequences in the E. coli proteome. All calculations of hydrophobicity in this case were made using the conditions we found to be optimal (window size = 12, Wertz-Scheraga scale). This scale is based on the tendency of residues to be buried or solvent exposed in a number of known X-ray crystal structures.
Does DsbA require an SRP-dependent signal sequence for export? Our findings have led us to ask why some proteins with cleavable signal sequences have evolved to utilize the SRP pathway. To examine this question, we have studied the export of DsbA, whose native signal sequence we have classified as SRP dependent. DsbA is efficiently exported by other SRP-dependent signal sequences and is inefficiently exported by most of the signals we have defined as not SRP dependent (based on the export of thioredoxin). This is the first example, to our knowledge, in which signal sequences from native exported proteins cannot be interchanged to give efficient export. We propose that the defect in export is due to the inherent rapid folding of DsbA to its native conformation in the cytoplasm, thus requiring a signal sequence that functions cotranslationally for efficient export.
The one case of a non-SRP-dependent signal sequence that exported DsbA efficiently was that of the TreA signal sequence. While this result may be significant, we point out that this is the one non-SRP-dependent signal sequence that has acted aberrantly in certain phenotypic assays when fused to thioredoxin (data not included). However, this apparent discrepancy may also be explained by the observation that varying amounts of DsbA can be exported depending on the non-SRP-dependent signal sequence used. For example, even with the worst signal sequence, a considerable amount of DsbA can be observed in the periplasmic fraction. This is in contrast to thioredoxin, which is always close to fully cytoplasmic when fused to a non-SRP-dependent signal sequence. The difference in exportability between thioredoxin and DsbA may reflect a difference in the folding properties of the proteins. Larger proteins, such as DsbA, may have to sample more conformations in their folding pathways and thus have substantially slower folding kinetics than smaller proteins of similar fold, such as thioredoxin. In addition, the TreA signal sequence consistently ranked as more hydrophobic than many SRP-dependent signal sequences. Thus, the TreA signal sequence may represent a very good non-SRP-dependent signal sequence that promotes posttranslational translocation at a much earlier stage of polypeptide elongation than most other non-SRP-dependent signal sequences.
Hydrophobicity may not be the sole factor for SRP recognition.
No permutation of our hydrophobicity calculation yielded a perfect fit with the experimental data, i.e., providing a way of separating SRP-dependent and non-SRP-dependent signal sequences in the high hydrophobicity range. We can conceive of three explanations for this imprecision of prediction. (i) None of the existing hydrophobicity scales predict with sufficient accuracy for our purposes what they are designed to predict. While this explanation is possible, we point out that we have used a large number of hydrophobicity scales, most of which are based on different approaches but which yield the same imprecision.
(ii) Another feature of signal sequences, in addition to hydrophobicity, plays a role in SRP recognition. Other groups have suggested that the position of the basic residues or the presence of helix-breaking residues in the hydrophobic core may play roles in SRP discrimination (1, 33). We are investigating a possible role of the N-terminal positive charge in orienting the signal sequence for SRP recognition. We have found no evidence supporting the involvement of helix-breaking residues such as glycine and proline in SRP recognition. Helix-breaking residues were found in both SRP-dependent and non-SRP-dependent signal sequences. However, recent evidence suggests some involvement of such residues for eukaryotic signal sequences recognized by the SRP (29). In addition, some loose sequence conservation or the context of the hydrophobic core in the overall signal sequence may contribute to making a signal sequence SRP dependent.
(iii) Our classification of some of the highly hydrophobic signal sequences as not SRP dependent is too simplistic. It is possible that some of the signal sequences in the high-hydrophobicity range that do not allow export of thioredoxin are capable of promoting SRP-dependent export, but the particular combination of those signal sequences with thioredoxin is not productive. For instance, there may be features of the thioredoxin mature protein sequence to which a signal sequence is fused that interfere with the functioning of that particular signal sequence. This might explain the ability of the TreA signal sequence to promote export of DsbA but not thioredoxin. While we have not excluded this possibility, we feel it is unlikely for the following reason. In a cotranslational export pathway, where ribosomes are thought to be transferred to the membrane-bound SecYEG translocase as soon as the signal sequence appears from the ribosome, there is no opportunity for interactions between the signal and the mature sequences. However, other more complex mechanisms may be involved.
The SRP-dependent proteins we identified have a wide variety of structures even for such a limited data set. The only class of proteins for which we found more than one SRP-dependent signal sequence was the pilin-fimbrial chaperone proteins, which share an immunoglobulin fold. However, PapJ, a well characterized fimbrial chaperone, does not have an SRP-dependent signal sequence. Also, while we propose that DsbA is a substrate for SRP-dependent export, the signal sequence of DsbC, another disulfide oxidoreductase with a thioredoxin fold, does not promote SRP-dependent export. This may be because DsbC, unlike DsbA, contains a structural disulfide bond required for its stability (27). Thus, DsbC may fold slowly in the cytoplasmic compartment where cysteines are kept as sulfhydryls, allowing it to be exported posttranslationally. We hypothesize that the only common property of proteins requiring cotranslational export is their rapid folding kinetics in the cytoplasm.
A mechanism for the evolution of a periplasmic protein.
How might the oxidative periplasmic DsbA, which is a member of the thioredoxin family, have evolved from the reductive cytoplasmic thioredoxin? We speculate that the first step in the evolutionary pathway could have been the fusion of a cytoplasmic thioredoxin to an export signal in the form of a membrane protein's transmembrane segment. In this way, the thioredoxin would be exported efficiently to the periplasm by the SRP pathway, a process necessitated by the rapid folding of thioredoxin. The thioredoxin would now be facing the periplasm and anchored to the membrane by the transmembrane segment. Such fusion proteins are already known; CcmG is a periplasmic thioredoxin protein anchored to the cytoplasmic membrane by a transmembrane segment (10).
When thioredoxin is exported to the periplasm, it is capable of acting as an oxidant and introducing disulfide bonds into substrate proteins, albeit weakly and even though its redox potential is quite low (8, 9). Thus, the fusion may have provided a selective advantage to the cell carrying it. The thioredoxin moiety of this membrane protein may then have evolved to exhibit a more oxidizing redox potential by mutations like those already observed in the laboratory (32). This protein could have further evolved via mutation(s) that allowed cleavage of the signal sequence (transmembrane segment) from the now DsbA-like protein, thus releasing the oxidative protein into the periplasm. Such a mutation has been observed in studies on the cytoplasmic membrane protein SecY (2).
Finally, how might this evolved DsbA have acted as an oxidant when the DsbB protein, the enzyme that reoxidizes DsbA, was not present? This is not a problem, as small molecules such as oxidized glutathione or cystine can restore some DsbA activity to a dsbB mutant strain apparently by directly oxidizing DsbA (3). DsbB may have evolved at a later stage to promote more efficient oxidation of DsbA.
Acknowledgments
We thank members of the Beckwith lab for many helpful conversations.
J.B. is an American Cancer Society Professor. This work was supported by grant no. GM41883 from the NIH to J.B. and grant GM054160-02 to M.G. Y.X. is supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
REFERENCES
- 1.Adams, H., P. A. Scotti, H. De Cock, J. Luirink, and J. Tommassen. 2002. The presence of a helix breaker in the hydrophobic core of signal sequences of secretory proteins prevents recognition by the signal-recognition particle in Escherichia coli. Eur. J. Biochem. 269**:**5564-5571. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, Y., T. Inada, Y. Nakamura, and K. Ito. 1990. SecY, a multispanning integral membrane protein, contains a potential leader peptidase cleavage site. J. Bacteriol. 172**:**2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90**:**1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers, C. W., F. Lau, and T. J. Silhavy. 2003. Secretion of LamB-LacZ by the signal recognition particle pathway of Escherichia coli. J. Bacteriol. 185**:**5697-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., C. Schierle, and J. Beckwith. 1998. How many membrane proteins are there? Protein Sci. 7**:**201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138**:**179-207. [DOI] [PubMed] [Google Scholar]
- 7.Chothia, C. 1976. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 105**:**1-12. [DOI] [PubMed] [Google Scholar]
- 8.Debarbieux, L., and J. Beckwith. 1998. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc. Natl. Acad. Sci. USA 95**:**10751-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debarbieux, L., and J. Beckwith. 2000. On the functional interchangeability, oxidant versus reductant, of members of the thioredoxin superfamily. J. Bacteriol. 182**:**723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edeling, M. A., L. W. Guddat, R. A. Fabianek, L. Thony-Meyer, and J. L. Martin. 2002. Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment. Structure (Cambridge) 10**:**973-979. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299**:**371-374. [DOI] [PubMed] [Google Scholar]
- 12.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15**:**321-353. [DOI] [PubMed] [Google Scholar]
- 13.Fekkes, P., and A. J. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63**:**161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy, S. J., and L. L. Randall. 1991. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science 251**:**439-443. [DOI] [PubMed] [Google Scholar]
- 15.Hopp, T. P., and K. R. Woods. 1983. A computer program for predicting protein antigenic determinants. Mol. Immunol. 20**:**483-489. [DOI] [PubMed] [Google Scholar]
- 16.Jander, G., J. E. Cronan, Jr., and J. Beckwith. 1996. Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J. Bacteriol. 178**:**3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janin, J. 1979. Surface and inside volumes in globular proteins. Nature 277**:**491-492. [DOI] [PubMed] [Google Scholar]
- 18.Jonda, S., M. Huber-Wunderlich, R. Glockshuber, and E. Mossner. 1999. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 18**:**3271-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josefsson, L. G., and L. L. Randall. 1983. Analysis of cotranslational proteolytic processing of nascent chains using two-dimensional gel electrophoresis. Methods Enzymol. 97**:**77-85. [DOI] [PubMed] [Google Scholar]
- 20.Juretic, D., and A. Lucin. 1998. The preference functions method for predicting helical turns with membrane propensity. J. Chem. Inf. Comput. Sci. 38**:**575-585. [Google Scholar]
- 21.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72**:**111-135. [DOI] [PubMed] [Google Scholar]
- 22.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338**:**1027-1036. [DOI] [PubMed] [Google Scholar]
- 23.Kawashima, S., H. Ogata, and M. Kanehisa. 1999. AAindex: amino acid index database. Nucleic Acids Res. 27**:**368-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157**:**105-132. [DOI] [PubMed] [Google Scholar]
- 25.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98**:**3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, G., T. B. Topping, and L. L. Randall. 1989. Physiological role during export for the retardation of folding by the leader peptide of maltose-binding protein. Proc. Natl. Acad. Sci. USA 86**:**9213-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, X., and C. C. Wang. 2001. Disulfide-dependent folding and export of Escherichia coli DsbC. J. Biol. Chem. 276**:**1146-1151. [DOI] [PubMed] [Google Scholar]
- 28.Luirink, J., and B. Dobberstein. 1994. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 11**:**9-13. [DOI] [PubMed] [Google Scholar]
- 29.Matoba, S., and D. M. Ogrydziak. 1998. Another factor besides hydrophobicity can affect signal peptide interaction with signal recognition particle. J. Biol. Chem. 273**:**18841-18847. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 31.Missiakas, D., F. Schwager, and S. Raina. 1995. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 14**:**3415-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mossner, E., M. Huber-Wunderlich, and R. Glockshuber. 1998. Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol/disulfide oxidoreductases. Protein Sci. 7**:**1233-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson, J. H., C. A. Woolhead, and H. D. Bernstein. 2003. Basic amino acids in a distinct subset of signal peptides promote interaction with the signal recognition particle. J. Biol. Chem. 278**:**46155-46162. [DOI] [PubMed] [Google Scholar]
- 34.Randall, L. L. 1983. Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell 33**:**231-240. [DOI] [PubMed] [Google Scholar]
- 35.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93**:**13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 37.Schierle, C. F., M. Berkmen, D. Huber, C. Kumamoto, D. Boyd, and J. Beckwith. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185**:**5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sijbrandi, R., M. L. Urbanus, C. M. ten Hagen-Jongman, H. D. Bernstein, B. Oudega, B. R. Otto, and J. Luirink. 2003. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J. Biol. Chem. 278**:**4654-4659. [DOI] [PubMed] [Google Scholar]
- 39.Teschke, C. M., J. Kim, T. Song, S. Park, C. Park, and L. L. Randall. 1991. Mutations that affect the folding of ribose-binding protein selected as suppressors of a defect in export in Escherichia coli. J. Biol. Chem. 266**:**11789-11796. [PubMed] [Google Scholar]
- 40.Thom, J. R., and L. L. Randall. 1988. Role of the leader peptide of maltose-binding protein in two steps of the export process. J. Bacteriol. 170**:**5654-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian, H., and J. Beckwith. 2002. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 184**:**111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian, H., D. Boyd, and J. Beckwith. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. USA 97**:**4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181**:**508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wertz, D. H., and H. A. Scheraga. 1978. Influence of water on protein structure. An analysis of the preferences of amino acid residues for the inside or outside and for specific conformations in a protein molecule. Macromolecules 11**:**9-15. [DOI] [PubMed] [Google Scholar]
- 45.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3**:**842-848. [DOI] [PubMed] [Google Scholar]