Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins (original) (raw)
Abstract
Vertebrate TAP is a nuclear mRNA export factor homologous to yeast Mex67p. The middle domain of TAP binds directly to p15, a protein related to the nuclear transport factor 2 (NTF2), whereas its C-terminal domain interacts with various nucleoporins, the components of the nuclear pore complex (NPC). Here, we report that the middle domain of TAP is also similar to NTF2, as well as to regions in Ras-GAP SH3 domain binding protein (G3BP) and some plant protein kinases. Based on the known three-dimensional structure of NTF2 homodimer, a heterodimerization model of TAP and p15 could be inferred. This model was confirmed by site-directed mutagenesis of residues located at the dimer interface. Furthermore, the C-terminus of TAP was found to contain a ubiquitin-associated (UBA) domain. By site-directed mutagenesis we show that a conserved loop in this domain plays an essential role in mediating TAP–nucleoporin interaction.
INTRODUCTION
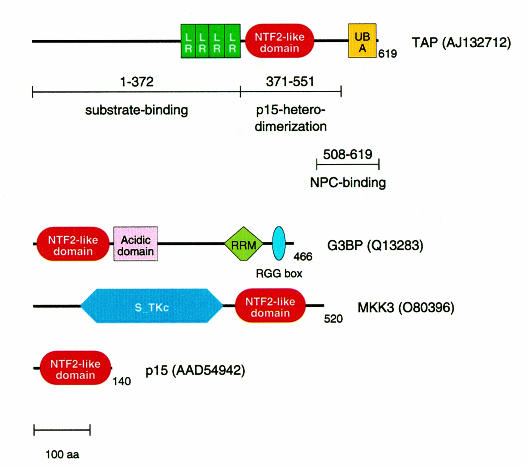
Vertebrate TAP and its yeast ortholog Mex67p are involved in the export of messenger RNAs from the nucleus (Segref et al., 1997; Grüter et al., 1998). TAP has also been implicated in the export of simian type D viral RNAs bearing the constitutive transport element (CTE) (Grüter et al., 1998; Braun et al., 1999; Kang and Cullen, 1999). Recently, several Mex67p/TAP partners have been identified. These include various nucleoporins (Katahira et al., 1999; Bachi et al., 2000); p15, a protein related to the nuclear transport factor 2 (NTF2) (Katahira et al., 1999); transportin, which mediates TAP nuclear import (Bachi et al., 2000); and several RNA-binding proteins such as E1B-AP5 (Bachi et al., 2000) and members of the Yra1p/REF family of proteins (Sträßer and Hurt, 2000; Stutz et al., 2000). Nucleoporin binding by TAP is mediated by its nuclear pore complex (NPC)-binding domain located at the very C-terminal end of the protein (residues 508–619) (Bachi et al., 2000; Figure 1). This domain directly interacts with multiple nucleoporins in vitro while in vivo it targets TAP to the NPC (Bear et al., 1999; Bachi et al., 2000). TAP binding to p15 is mediated by its middle domain (residues 371–551) (Bachi et al., 2000). The N-terminal domain of TAP (residues 1–372) represents its substrate binding domain as it exhibits RNA-binding activity, is involved in direct binding to the CTE RNA and interacts with hnRNP-like proteins (Braun et al., 1999; Kang and Cullen, 1999; Katahira et al., 1999; Bachi et al., 2000; Stutz et al., 2000).
Fig. 1. Modular architecture of proteins with NTF2-like domains. The numbers indicate the lengths of the sequences in residues; the protein names correspond to Figure 2; DDBJ/EMBL/GenBank accession Nos are given in parentheses. The binding partners of TAP and the regions involved in binding defined in previous studies (Braun et al., 1999; Bachi et al., 2000) are indicated under the TAP sequence. Abbreviations: LR, leucine-rich repeat; UBA, ubiquitin-associated domain; NPC, nuclear pore complex; RRM, RNA recognition motif; S-TKc, catalytic domain of serine/threonine protein kinase.
Here we report that the middle domain of TAP shows significant sequence similarity to p15 and NTF2, as well as to Ras-GAP SH3 domain binding protein (G3BP) and plant protein kinases. NTF2 forms homodimers (Bullock et al., 1996) while p15 monomers bind directly to TAP (Katahira et al., 1999; Bachi et al., 2000). The known three-dimensional structure of NTF2 homodimers (Bullock et al., 1996) suggests a detailed heterodimerization model of TAP and p15, which, in turn, allowed directed mutagenesis that confirmed the model. Furthermore, a similarity of the C-terminal domain of TAP to ubiquitin-associated (UBA) domains (Hofmann and Bucher, 1996) predicts a sequence motif to be involved in the binding to nucleoporins.
RESULTS AND DISCUSSION
The NTF2-like domain: a novel mobile module mediating TAP–p15 heterodimerization
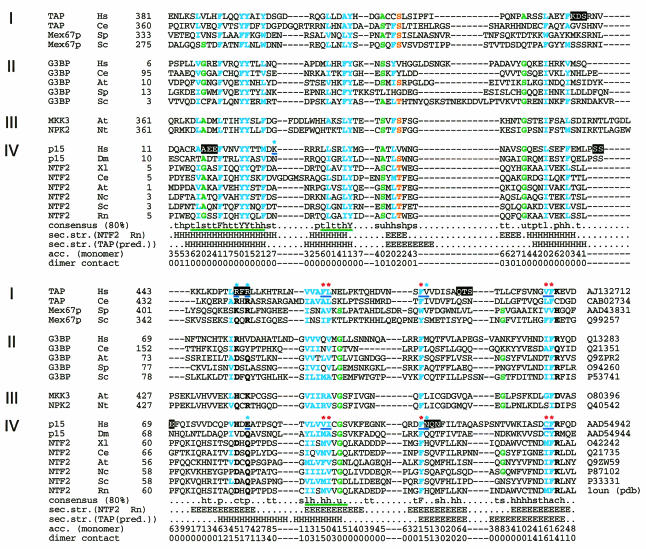
The recently published sequence of TAP-interacting partner p15 (Katahira et al., 1999) enlarged the family of NTF2-like sequences and related Ras-GAP SH3 domain binding protein (G3BP). Indeed, database searches with p15 using PSI-BLAST (Altschul et al., 1997; http://www.ncbi.nlm.nih.gov/blast/psiblast.cgi), which searches against sequence databases iteratively with a position-specific scoring matrix, converge after four iterations. However, TAP was the highest ranking hit (expected ratio of false positive, E = 3.0) among the sequences without significant similarity. The respective sequence region (residues 381–503) did correspond to the experimentally defined TAP–p15 interaction domain (residues 371–551; Figure 1). This prompted a more detailed analysis using the MACAW alignment program (Schuler et al., 1991), which confirmed the statistical significance of the sequence similarity (probability of chance match, P <3 × 10–14 for several alignment blocks; Figure 2, green underlines). Exhaustive database searches with other known NTF2-like family members also identified MAP kinase kinase 3 (MKK3) of Arabidopsis thaliana and a protein kinase of Nicotiana tabacum as distant homologs (for example, using G3BP as a query, PSI-BLAST E values of 0.22 were obtained and verified using MACAW analysis). When including the plant kinases into the iterations, TAP sequences also ranked highest (E values in the order of 1.0) among the sequences without significant similarity. A multiple sequence alignment of all identified family members indicates the presence of many conserved positions (Figure 2). The modular architecture of the proteins with NTF2-like domains is shown in Figure 1.
Fig. 2. Multiple alignment of selected NTF2-like domains. The sequences are grouped into four subfamilies according to the domain organization of entire proteins (Figure 1): (I) TAP; (II) G3BP; (III) plant protein kinases; and (IV) NTF2. First column, protein names; second column, species names: At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Nc, Neurospora crassa; Nt, Nicotiana tabacum; Rn, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Xl, Xenopus laevis; third column, positions of the first aligned residues in each of the sequences; last column, DDBJ/EMBL/GenBank database accession Nos. The positions conserved in 80% of the sequences are indicated in the consensus line: c (DEHKR), charged; h (ACFGHIKLMRTVWY), hydrophobic; l (ILV), aliphatic; p (CDEHKNQRST), polar; s (ACDGNPSTV), small; t (ACDEGHKNQRST), turn like; u (AGS), tiny. The statistically significant alignment blocks verified by MACAW are indicated by the green underlines in the consensus line. The positions conserved among the four subfamilies are indicated in bold characters with colors: cyan, hydrophobic; green, tiny; orange, hydroxyl; black, polar. The random mutations of tripeptides to alanines are highlighted. The designed point mutations that are mentioned in the text are indicated by blue underlines; the colors of the corresponding asterisks indicate the phenotypic effect: red, impaired binding; cyan, no obvious effect. The known secondary structure of rat NTF2 is indicated below the consensus line (H, α-helix; E, β-strand) and agrees well with the predicted secondary structure (Rost and Sander, 1994) of TAP sequences. The surface accessibility is shown below the predicted secondary structure. The dimer contact is shown at the bottom.
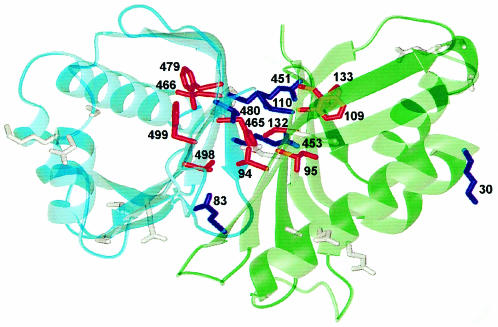
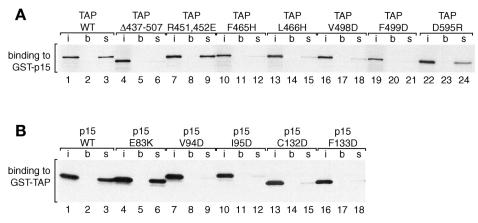
As the three-dimensional (3D) structure of NTF2 has already been determined as a homodimer (Bullock et al., 1996), a detailed model for heterodimerization between TAP and p15 could be inferred (Figure 3). Subtraction of accessible surface area of the NTF2 dimer from that of the monomer allowed the identification of residues that are located in the dimer interface (high numbers in the dimer contact line in Figure 2). To verify that these residues were involved in the TAP–p15 interaction, we performed two types of mutagenesis experiments. (i) Mutagenesis of six tripeptides to alanines in random positions was carried out (i.e. three times three mutations in each, TAP and p15). None of these 18 mutations (Figure 2, black high- lights) disrupted binding to the respective wild-type partner. (ii) Directed mutagenesis was performed for the residues that were predicted to contribute to the interaction and some neighboring residues in both TAP (double mutation R451,453E, and single mutations F465H, L466H, F479W, V480W, V498D and F499D) and p15 (E83K, V94D, I95D, F109W, N110W, C132D and F133D), respectively, as well as one control mutation (K30E) in a region of p15 that should not interfere with TAP binding (Figure 2, asterisks and blue underlines). Whereas the random mutations did not show any effect, 10 of the 15 designed mutations impaired TAP–p15 interaction (in TAP: F465H, L466H, F479W, V498D and F499D and equivalents in p15: V94D, I95D, F109W, C132D and F133D; Figure 4 and data not shown). All the mutations in TAP and the corresponding ones in p15 show exactly the same phenotypic effects, which strongly points to a homology and similar binding features (Figure 4). Furthermore, all mutations with a phenotypic effect are part of the predicted hydrophobic interface to which the β-sheets of the One mutation of a hydrophobic residue in an interface position only slightly reduced binding to the wild-type protein (in TAP: V480W and the equivalent but hydrophilic mutant in p15: N110W) (data not shown). This further supports our model, as the hydrophobicity in this particular position is not conserved, indicating that the effects will be milder. The two remaining positions of the interface (TAP R451,453E and p15 E83K corresponding to the latter) whose mutation was without phenotypic effect are hydrophilic. They only seem to be needed to shield the hydrophobic core and are consequently also not conserved within the family (Figure 2).
Fig. 3. Proposed model of the interaction between the middle domain of TAP and p15. The model structure was calculated (Sali and Blundell, 1993) based on the 3D structure of the homodimer of NTF2 (Bullock et al., 1996). Cyan, TAP; green, p15. The side chains are shown for positions where mutations were introduced: red, affected binding; dark blue, binding efficiency similar to the wild-type proteins. Residue numbers are indicated for mutation set (ii) (see the text). Numbers below and above 400 denote residues in p15 and TAP, respectively. The triple alanine mutations are shown in gray. The figure was prepared using MOLMOL (Koradi et al., 1996).
Fig. 4. GST pull-down assays with TAP and p15 mutants. (A) Binding assays were performed with [35S]methionine-labeled TAP or various TAP mutants and immobilized GST–p15 on glutathione agarose beads. One tenth of the input (i) and one quarter of the bound fractions (s) were analyzed on SDS–PAGE followed by fluorography. Lanes (b) show the background obtained with glutathione agarose beads coated with GST. (B) Binding assays were performed with [35S]methionine-labeled p15 or various p15 mutants and immobilized GST–TAP on glutathione agarose beads. Symbols are as in (A).
Taken together, these results strongly support a detailed model for TAP–p15 heterodimerization with a defined binding interface (Figure 3). A similar dimer formation can also be predicted for NTF2-like domains in the other newly identified family members (Figure 1); their different modular architecture indicates that NTF2 is the prototype of a novel mobile module (Bork, 1992).
A UBA-like domain is implicated in the interaction of TAP with nucleoporins
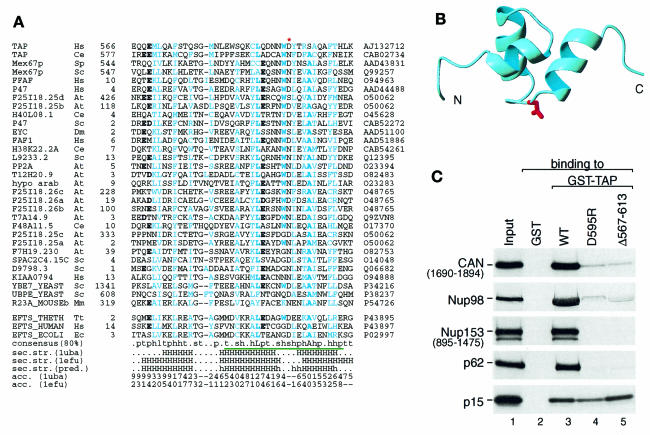
The nucleoporin-binding domain of TAP comprises residues 508–619. In HeLa cells, this domain is necessary and sufficient to target green fluorescent protein (GFP)–TAP fusions to the nuclear rim (Bear et al., 1999; Bachi et al., 2000). Moreover, the isolated domain strongly competes multiple export pathways in vivo, probably by blocking binding sites on the NPC that are shared with other transport receptors (Bachi et al., 2000). A significant sequence similarity of the C-terminal domain of TAP (residues 566–608) to UBA domains was identified using PSI-BLAST (Altschul et al., 1997) (Figure 5A). The UBA domain is a sequence motif present in multiple enzymes of the ubiquitylation pathway, but also in proteins involved in DNA repair and certain protein kinases (Hofmann and Bucher, 1996). Several UBA-like domains were highest ranking in the twilight zone hits with E values of 0.76 and above. The similarity was confirmed by MACAW analysis for about a half length of the alignment (P values <1 × 10–50; Figure 5A, green underline). The known structure of UBA domains (Dieckmann et al., 1998) suggests a conserved loop (NWD at position 593 in human TAP) to be involved in the interaction with nucleoporins (Figure 5B). Substitution of these three residues by alanines was previously shown to block the ability of TAP to promote CTE-dependent export of a precursor mRNA in quail cells (Kang and Cullen, 1999). Moreover, a single amino acid change in this loop, D595R, was sufficient to disrupt binding of TAP to the nucleoporins CAN, Nup98, Nup153 and p62 (Figure 5C). Strikingly, this single point mutation had the same effect on nucleoporin binding as the deletion of the entire domain (TAPΔ567–613). Together, these results indicate that the conserved loop of the UBA-like domain has a critical role in the interaction of TAP with nucleoporins.
Fig. 5. The NPC-binding domain of TAP contains a UBA-like motif. (A) Multiple alignment of UBA domains. Description of the columns is as in Figure 2. Species names (other than those in Figure 2): Mm, Mus musculus; Tt, Thermus thermophilus; Ec, Escherichia coli. Conserved hydrophobic positions are colored in cyan with highly conserved residues in bold characters. Highly conserved positions occupied with negatively charged residues are indicated by bold characters. The mutation, D595R, which interrupts binding to nucleoporins, is indicated by a red asterisk. The alignment consists of two groups: top, UBA domains found in TAP and the sequences with significant similarity; bottom, structurally related elongation factor Ts (EF-Ts). See the legend to Figure 2 about the consensus line. The known secondary structures of 1uba and 1efu are indicated below the consensus line (H, α-helix). The predicted secondary structure, which follows the known secondary structures, was calculated by the PHD program (h and H, α-helix predicted with expected average accuracy below and above 82%, respectively; Rost and Sander, 1994). The surface accessibility below (0 low, 9 high) was calculated using the DSSP program (Kabsch and Sander, 1983). (B) Proposed model for the UBA-like domain of TAP based on the structure of the UBA domain (PDB: 1uba) (Dieckmann et al., 1998. The red side chain indicates the position corresponding to TAP D595. The direction of the chain is indicated at both termini. The figure was prepared using MOLMOL (Koradi et al., 1996). (C) Mutations in the conserved loop of the UBA-like domain disrupt TAP–nucleoporin interaction. The assays were performed using the [35S]methionine-labeled nucleoporins indicated on the left, and recombinant GST, GST–TAP, GST–TAP D595R or GST–TAPΔ567–613 as indicated above the lanes. [35S]methionine-labeled p15 served as an internal control, as its interaction with TAP is not affected by mutations or deletions that prevent nucleoporin binding Bachi et al., 2000). One tenth of the input and one quarter of the bound fractions were analyzed on SDS–PAGE followed by fluorography.
Although we cannot currently rule out that some of the mutations described above affect binding by causing the proteins to be misfolded, we consider this possibility unlikely for the following reasons. First, the mutants described above could be expressed in Escherichia coli as efficiently as the wild-type protein (data not shown). This is in contrast with the deletion mutants TAPΔ437–505 and TAPΔ567–613 (deletions of the NTF2-like and UBA-like domains) and some of the triple alanine-substitution mutants that can hardly be expressed in E. coli as full-length proteins. Secondly, mutations in the NTF2-like domain of TAP only impaired binding to p15 but not to nucleoporins or to the CTE RNA (data not shown). Similarly, mutation D595R interferes with nucleoporin binding but not with p15 interaction (Figure 5C). Finally, mutations in the NTF2-like domain of TAP reduced but did not abolish its ability to export a reporter mRNA in human 293 cells (I.C. Braun and E. Izaurralde, unpublished), indicating that these proteins are properly folded.
The NTF2-like domain and the UBA-like domain occur in the Caenorhabditis elegans TAP homolog and in Mex67p, the Schizosaccharomyces pombe and Saccharomyces cerevisiae homologs (Figures 2 and 5), indicating that the overall structural domain organization of the protein has been evolutionarily conserved. However, the functional role of these domains in Mex67p is unknown. First, there is no obvious p15 homolog encoded by the yeast genome. Secondly, although a fraction of Mex67p localizes to the NPC, a direct interaction of Mex67p with nucleoporins has not yet been demonstrated. On the contrary, in S. cerevisiae, the association of Mex67p with the NPC appears to be mediated by a protein known as Mtr2p (Santos-Rosa et al., 1998). Mtr2p does not exhibit obvious sequence similarities with p15 or NTF2, and the domain of Mex67p implicated on its binding has not been defined. Prediction of Mtr2p secondary structure (data not shown) suggests that it could be a p15 analog and therefore may interact with the NTF2-like domain of Mex67p. Consistent with this is the observation that co-expression of human TAP and p15 in yeast partially restores growth of the otherwise lethal mex67/mtr2 double knockout strain. Thus, Mtr2p and p15 may have a similar function (Katahira et al., 1999).
In summary, based on sequence similarity we have predicted that the C-terminal half of TAP and TAP homologs contain NTF2- and UBA-like domains. In higher eukaryotes these domains are implicated in p15 and nucleoporin binding, respectively. This prediction successfully guided the rational design of mutations with particular phenotypic effects, i.e. impairment of TAP–p15 or TAP–nucleoporin interaction, which is often hard to achieve by random mutagenesis. These mutants are useful tools for the analysis of TAP and p15 function in vivo. Moreover, the prediction of these structural domains allows the re-investigation of their function in Mex67p. More generally, our data indicate that TAP/Mex67p belong to a family of mRNA export factors having a conserved modular architecture.
METHODS
Sequence and structure analysis. Sequence similarity searches were performed using PSI-BLAST (Altschul et al., 1997) against the non-redundant protein sequence database with the BLOSUM62 scoring matrix. Multiple sequence alignment of the sequences detected by PSI-BLAST was constructed by CLUSTAL W (Thompson et al., 1994) and manually refined on the SEAVIEW alignment editor (Galtier et al., 1996). The statistical significance of the alignments was verified by the MACAW alignment program (Schuler et al., 1991), which can evaluate the significance of aligned sequence blocks of similarity using a mathematical theory (Karlin and Altschul, 1990). Secondary structure prediction based on the multiple alignments was carried out using the PHD program (Rost and Sander, 1994) at the PredictProtein server (http://dodo.cpmc.columbia.edu/pp/predictprotein.html). The surface accessibility (0 low, 9 high) was calculated using the DSSP program (Kabsch and Sander, 1983). The dimer contact was derived by subtracting accessible surface area of the dimer from that of the monomer (i.e. the higher the numbers, the more involvement in the dimer contact). The 3D structure model was calculated using the method described by Sali and Blundell (1993).
Plasmids, mutagenesis, recombinant protein expression and purification. TAP and p15 constructs used in this study were previously described (Braun et al., 1999; Bachi et al., 2000). All mutations were introduced using an oligonucleotide-directed in vitro mutagenesis system from Stratagene (Quick-change site-directed mutagenesis) following the instructions of the manufacturer. TAP mutants were generated on pBSSKHA-TAP 1–619 and subsequently subcloned into pGEXCS-TAP 1–619. p15 mutants were generated on pRN3zz-p15 and subcloned in pGEX-5X-1. Mutations were confirmed by restriction mapping and by sequencing. For generation of 35S-labeled in vitro translated proteins plasmids pBSSKHA-TAP and pRN3zz-p15 bearing the mutations described in this manuscript were used as templates in the combined in vitro transcription/translation (TnT) kit from Promega. Reactions were carried out at 30°C for 2 h. Translation was checked by SDS–PAGE and subsequent fluorography using intensify solution from Amersham (Amplify). TAP and p15 glutathione _S-_transferase (GST) fusions were expressed in E. coli using pGEXCS-TAP and pGEX-5X-1-p15 constructs. Recombinant proteins were prepared as described by Grüter et al. (1998).
GST pull-down assays. About 5 µg of GST-tagged recombinant protein immobilized on 20 µl of packed glutathione agarose beads were used per binding reaction. Following binding of GST-tagged recombinant proteins, beads were washed three times with 0.5 ml of IPP buffer (10 mM Tris–HCl pH 8.0, 50 mM NaCl, 10% glycerol, 0.1% Triton X-100). Between 2 and 5 µl of in vitro synthesized proteins were used per binding reaction in a final volume of 200 µl of IPP buffer supplemented with bovine serum albumin (BSA) at 0.05 mg/ml. Binding was for 1 h at 4°C. Beads were washed three times with 0.5 ml of IPP buffer, and once with the same buffer without Triton X-100. Bound proteins were eluted with SDS-sample buffer and analyzed by SDS–PAGE followed by fluorography.
Acknowledgments
ACKNOWLEDGEMENTS
The technical assistance of Michaela Rode is gratefully acknowledged. We wish to thank Maria Macias for help in modeling. We are grateful to Iain W. Mattaj for critical reading of the manuscript. This study was supported by the European Molecular Biology Organization (EMBO), the Human Frontier Science Program Organisation (HFSPO) and the Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J., Tan, W., Zolotukhin, A.S., Tabernero, C., Hudson, E.A. and Felber, B.K. (1999) Identification of novel import and export signals of human TAP, the protein that binds to the CTE element of the type D retrovirus mRNAs. Mol. Cell. Biol., 19, 6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. (1992) Mobile modules and motifs. Curr. Opin. Struct. Biol., 2, 413–421. [Google Scholar]
- Braun I.C., Rohrbach, E., Schmitt, C. and Izaurralde, E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock T.L., Clarkson, W.D., Kent, H.M. and Stewart, M. (1996) The 1.6 Å resolution crystal structure of nuclear transport factor 2 (NTF2). J. Mol. Biol., 260, 422–431. [DOI] [PubMed] [Google Scholar]
- Dieckmann T., Withers-Ward, E.S., Jarosinski, M.A., Liu, C.F., Chen, I.S. and Feigon, J. (1998) Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nature Struct. Biol., 5, 1042–1047. [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy, M. and Gautier, C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- Grüter P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K. and Izaurralde, E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Bucher, P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. [PubMed] [Google Scholar]
- Kabsch W. and Sander, C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kang Y. and Cullen, B.R. (1999) The human TAP protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev., 13, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. and Altschul, S.F. (1990) Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA, 87, 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Sträßer, K., Podtelejnikov, A., Mann, M., Jung, J.U. and Hurt E. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter, M. and Wüthrich, K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Rost B. and Sander, C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- Sali A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Panté, N. and Hurt, E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G.D., Altschul, S.F. and Lipman, D.J. (1991) A workbench for multiple alignment construction and analysis. Proteins, 9, 180–190. [DOI] [PubMed] [Google Scholar]
- Segref A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R. and Hurt, E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. and Hurt, E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi, A., Doerks, T., Braun, I.C., Séraphin, B., Wilm, M., Bork, P. and Izaurralde, E. (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]