Under the genomic radar: The Stealth model of Alu amplification (original) (raw)
Abstract
Alu elements are the most successful SINEs (Short INterspersed Elements) in primate genomes and have reached more than 1,000,000 copies in the human genome. The amplification of most Alu elements is thought to occur through a limited number of hyperactive “master” genes that produce a high number of copies during long evolutionary periods of time. However, the existence of long-lived, low-activity Alu lineages in the human genome suggests a more complex propagation mechanism. Using both computational and wet-bench approaches, we reconstructed the evolutionary history of the _Alu_Yb lineage, one of the most active Alu lineages in the human genome. We show that the major _Alu_Yb lineage expansion in humans is a species-specific event, as nonhuman primates possess only a handful of _Alu_Yb elements. However, the oldest existing _Alu_Yb element resided in an orthologous position in all hominoid primate genomes examined, demonstrating that the _Alu_Yb lineage originated 18–25 million years ago. Thus, the history of the _Alu_Yb lineage is characterized by ∼20 million years of retrotranspositional quiescence preceding a major expansion in the human genome within the past few million years. We suggest that the evolutionary success of the Alu family may be driven at least in part by “stealth-driver” elements that maintain low retrotranspositional activity over extended periods of time and occasionally produce short-lived hyperactive copies responsible for the formation and remarkable expansion of Alu elements within the genome.
As the most successful SINEs (Short INterspersed Elements) in primate genomes, Alu elements have enjoyed remarkable proliferation during the primate radiation and have expanded to more than 1,000,000 copies in the human genome (Lander et al. 2001; Batzer and Deininger 2002). Despite considerable progress in the understanding of their biology and distribution throughout primate taxa (Salem et al. 2003b; Singer et al. 2003; Carter et al. 2004; Hedges et al. 2004; Otieno et al. 2004; Roos et al. 2004; Ray et al. 2005), a great deal of uncertainty still remains concerning their strategy for survival. It has been generally accepted that the amplification of most Alu elements occurs through a small number of long-lived high activity “master” genes (Deininger et al. 1992; Deininger and Batzer 1993), although there is considerable debate as to the details of this amplification strategy (Matera et al. 1990; Schmid 1993; Batzer et al. 1995; Cordaux et al. 2004; Price et al. 2004). In this model, the mutations accumulated in the master genes are inherited by the copies they produced, and consequently, a series of hierarchical Alu subfamilies that share the novel diagnostic mutation(s) are generated (Slagel et al. 1987; Willard et al. 1987; Britten et al. 1988; Deininger and Slagel 1988; Jurka and Smith 1988; Deininger et al. 1992; Batzer and Deininger 2002). On the other hand, the evolutionary history of the _Alu_Ya5 lineage, one of the most active human Alu lineages, suggests that the founder gene of this Alu lineage existed long before the major expansion of the lineage within the human genome (Leeflang et al. 1993). Contrary to the prediction of the master gene model, it has maintained low retrotranspositional activity, and this founder gene itself may not be directly responsible for the propagation of the recent human _Alu_Ya5 elements (Shaikh and Deininger 1996). Thus, these studies suggest that the expansion of Alu elements may follow a more complex propagation mechanism. Unfortunately, aside from the _Alu_Ya5 lineage, little data exist concerning the evolutionary origin of other Alu subfamilies in humans, making it difficult to assess the evolutionary significance of the results reported in the original _Alu_Ya5 lineage studies.
To gain additional insight into Alu subfamily propagation, we reconstructed the evolutionary history of the _Alu_Yb lineage, one of the largest and most active Alu lineages in the human genome (Jurka 1993; Carter et al. 2004) that composes ∼40% of the human-specific Alu elements (Hedges et al. 2004). This lineage, originally termed Sb2, is characterized by a 7-nt duplication involving positions 246 through 252 of the _Alu_Y consensus sequence (Jurka 1993; Batzer et al. 1996). The _Alu_Yb lineage in the human genome is subdivided into three major subfamilies as follows: _Alu_Yb7, _Alu_Yb8, and _Alu_Yb9 (Batzer et al. 1996; Roy-Engel et al. 2001; Jurka et al. 2002; Carter et al. 2004) based on diagnostic mutations following the standardized nomenclature for Alu repeats (Batzer et al. 1996). The majority of the human _Alu_Yb elements integrated into the genome during the last 3 to 4 million years (Myr) and reached a total copy number of about 2000 elements (Carter et al. 2004). The human diseases caused by de novo _Alu_Yb8 insertions suggest that this subfamily is currently actively retrotransposing, and a comprehensive analysis of the Yb lineage indicated that about 20% of _Alu_Yb elements are polymorphic in human genome (Muratani et al. 1991; Oldridge et al. 1999; Carter et al. 2004). Previous studies suggested that the evolutionary history of the _Alu_Yb lineage is much older than its period of major expansion in the human genome, and _Alu_Yb elements have also been identified in other nonhuman primates (Zietkiewicz et al. 1994; Gibbons et al. 2004). Nevertheless, the extent to which these elements are distributed among nonhuman primate species remains undetermined.
Using both a computational approach and polymerase chain reaction (PCR) display methodology, we have determined the distribution of _Alu_Yb elements in different hominoid genomes. We find that the long-term evolutionary history of the _Alu_Yb lineage exhibits a pattern that is remarkably similar to that of the _Alu_Ya lineage. Thus, the evolution of the _Alu_Ya and _Alu_Yb lineages illustrate a common strategy for Alu element proliferation. We propose a model for the expansion and survival of Alu elements in the primate order.
Results
_Alu_Yb elements in the common chimpanzee genome
To determine the evolutionary history of the _Alu_Yb lineage, we first computationally retrieved all of the _Alu_Yb elements from the first draft of the common chimpanzee (Pan troglodytes) genomic sequence (panTro1 Nov. 2003 assembly). A total of 12 _Alu_Yb elements were identified and subjected to PCR amplification on a common chimpanzee population panel and a separate primate panel composed of human and eight additional nonhuman primates (see Methods). The number of _Alu_Yb elements recovered from the chimpanzee draft sequence is in good agreement with a previous study (Gibbons et al. 2004). Detailed information on each locus, including primer sequences, annealing temperature, PCR product sizes, and chromosomal locations are shown in Table 1. Among the 12 elements identified, 10 belong to the _Alu_Yb8 subfamily, while the other two are non-_Alu_Yb7/8/9 elements (Table 2). Of the 10 _Alu_Yb8 elements, four loci were specific to the common chimpanzee lineage, including three elements that were polymorphic within the chimpanzee population panel (Figs. 1, 2B). Five elements were shared between the pygmy chimpanzee and common chimpanzee lineage (Fig. 2C) and one was present within human, chimpanzee, and gorilla genomes. These results suggest that the initial expansion of the _Alu_Yb8 subfamily predates the divergence of gorillas and humans/chimpanzees, which is thought to have occurred ∼7 million years ago (Mya) (Goodman et al. 1998). In addition, two non_Alu_Yb7/8/9 elements displayed PCR amplicon sizes consistent with the presence of an Alu element in all of the hominoid primates we tested.
Table 1.
_Alu_Yb loci identified in this study
| Product size | |||||||
|---|---|---|---|---|---|---|---|
| Namea | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) | Chr. loc.bin human | Direct repeat | A. T.c | Filled | Empty |
| Pan1* | ACCAAAATGCAGGTCTCTTGTT | CCTTTTCTTTATGCGCATTTCT | 5:171753413 | AAAACATCCTC | 55 | 892 | 561 |
| Pan2* | CCTGGCCTATTGATGATTTTCT | GCCTCAGAAGGAGTTTTGTTGT | 12:42243314 | AAAACTCCCTCTGAG | 58.6 | 481 | 129 |
| Pan3* | CCGATATCATGCATTTTCCATA | TGGCAAGAAAGACATGATTGAA | 3:155790154 | AAAAATAAATACCA | 60 | 728 | 412 |
| Pan4* | AGGGCAATTACTATGTTTCAGGA | TTTTTCACGTTCTTACAATAGAACA | 18:66254955 | ACAATAGAACATTCCT | 55 | 403 | 52 |
| Pan5* | GCTTCATTTCTGCCTGCTTATT | TCTGCAAATTTAACTCCAAACC | 10:97871078 | AAAAACAGCAAGT | 55 | 504 | 181 |
| Pan6* | GGCATTTTAAGCTCTTTGATAGC | CATGCTAGACATGAGAACAAACA | 2:183429337 | GAAATAGTTCCTGCT | 60 | 749 | 257 |
| Pan7* | GCAGCTGCTTTCTGTCTCTGTA | TCAGCAACAATAAGGAACGAAG | 3:49401325 | AAAAGAAACCAGTCAC | 55 | 502 | 175 |
| Pan8* | CCCAGATTGATTCTTCCCTTTA | ATGCCAGTTCCATTATTTCCAC | 8:101149947 | AAGAGAGAAGAAA | 55 | 834 | 506 |
| Pan9* | CCAGGACCCAGAGCTTAGTAGT | TCCAAAGGAATATGATGTCACAG | 14:52808462 | AAAGAAATTGATT | 55 | 434 | 111 |
| Pan10* | TGTGGCAGATTTTATTGTAGACTT | TCCATGCTCTTGGAGTAAATGA | 4:72958275 | AAAAAAATTCATCTG | 55 | 500 | 174 |
| Pan11* | AGGAGCTCTTCTTAGAAAATCCA | AAAGATCCACGACTAGGCACAT | 2:214354697 | AAAGAAAAAGGAAAAAAC | 60 | 521 | 193 |
| Pan12* | CGTTAGCTCTGGATTTTTCATGT | TATTCTCCACCATGACCAAGTG | X:93270745 | AAAAATTAGCCGGGCA | 60 | 871 | 548 |
| Gorilla7# | TGAAGAATTGGGAGGAAAAGAA | ACTGCAAGATTGCTGTTCACAT | 13:32666822 | AAAAAATGACTAACAG | 55 | 583 | 261 |
| Gorilla8# | GCACTCATCCGTACCTGACTTA | TCAGAGCATCTCTTTCTGTCCA | 2:229645741 | AAAAAAGGAGGCAA | 60 | 540 | 219 |
| Gorilla16# | AATTCTTTGGGGTAGGTGGAAT | CACAACGTACACCCTAAAATGG | X:24004038 | AAGAGTTGCAGTTGCTCA | 55 | 531 | 202 |
| Gorilla19# | CAAGTTGTGTTATGTGAGGTTTTGA | GGAGCCCTAATGTATAGCATGG | 7:10262937 | AAGAGAGTAGATCTT | 59.5 | 597 | 279 |
| Gorilla21# | AACAAGAGATGCTAGAAAGCCAAT | CGGAGTTGGACACATTTCTTTT | 1:51666132 | AAAGAAAGGAGGA | 55 | 749 | 434 |
| Pygmy4# | GCAGTTTTTCCCATTTGCTCTA | TTGAGTCTTTTTCTGGGCTTTC | 2:130058820 | AAGACTTACTATA | 55 | 600 | 281 |
Table 2.
Primate phylogenetic PCR panel results for chimpanzee _Alu_Yb elements
| Name | Chimpanzee panel | Human (HeLa) | Pygmy chimpanzee | Gorilla | Orangutan | Gibbon | Siamang | Green monkey | Owl monkey |
|---|---|---|---|---|---|---|---|---|---|
| Pan1 | Fixed present (_Alu_Yb8) | - | Polymorphic (_Alu_Yb8) | - | - | - | - | × | × |
| Pan2 | Fixed present (_Alu_Yb6) | + (_Alu_Yb6) | + (_Alu_Yb6) | + (_Alu_Yb6) | + (_Alu_Yb6) | + (_Alu_Yb6) | + (_Alu_Yb6) | + (_Alu_Sg) | × |
| Pan3 | Fixed present (_Alu_Yb) | + (_Alu_Yb) | + (_Alu_Yb) | + (_Alu_Yb) | + (_Alu_Y) | + (_Alu_Y) | + (_Alu_Y) | - | × |
| Pan4 | Fixed present (_Alu_Yb8) | + (_Alu_Yb8) | + (_Alu_Yb8) | + (_Alu_Yb8) | - | - | - | - | - |
| Pan5 | Polymorphic (_Alu_Yb8) | - | - | - | - | - | - | - | × |
| Pan6 | Fixed present (_Alu_Yb8) | - | + (_Alu_Yb8) | - | - | - | - | - | - |
| Pan7 | Fixed present (_Alu_Yb8) | - | + (_Alu_Yb8) | - | - | - | - | - | - |
| Pan8 | Fixed present (_Alu_Yb8) | - | - | - | - | - | - | - | - |
| Pan9 | Fixed present (_Alu_Yb8) | - | + (_Alu_Yb8) | - | - | - | - | - | - |
| Pan10 | Fixed present (_Alu_Yb8) | - | + (_Alu_Yb8) | - | - | - | - | - | - |
| Pan11 | Polymorphic (_Alu_Yb8) | - | - | - | - | - | - | - | × |
| Pan12 | Polymorphic (_Alu_Yb8) | - | - | - | - | - | - | - | - |
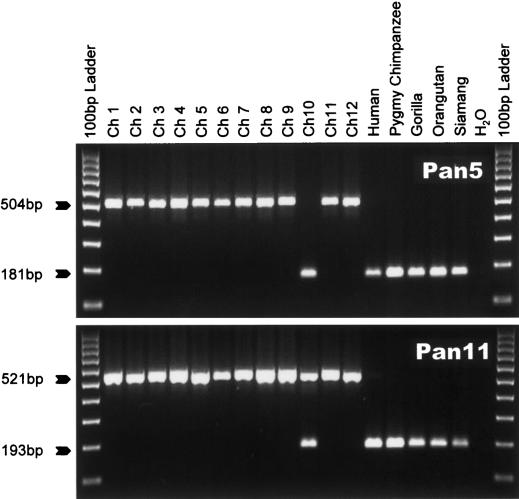
Figure 1.
Polymorphic _Alu_Yb8 elements in the common chimpanzee genome. Gel chromatographs of two loci are shown. The locus name is shown at right. The product sizes for filled and empty alleles (pre-integration size) are indicated at left. The DNA panel is composed of 12 unrelated common chimpanzee individuals and other primate species. The template used in each reaction is listed at top.
Figure 2.
Lineage-specific _Alu_Yb8 insertions. Four examples of gel chromatographs are shown. The locus designation is shown at right, while the product sizes for filled and empty alleles (pre-integration size) are indicated at left. The DNA template used in each reaction is listed at top. (A) Pygmy chimpanzee-specific _Alu_Yb8 insertion. (B) Common chimpanzee-specific _Alu_Yb8 insertion. (C) Chimpanzee lineage-specific _Alu_Yb8 insertion. (D) Gorilla-specific _Alu_Yb8 insertion.
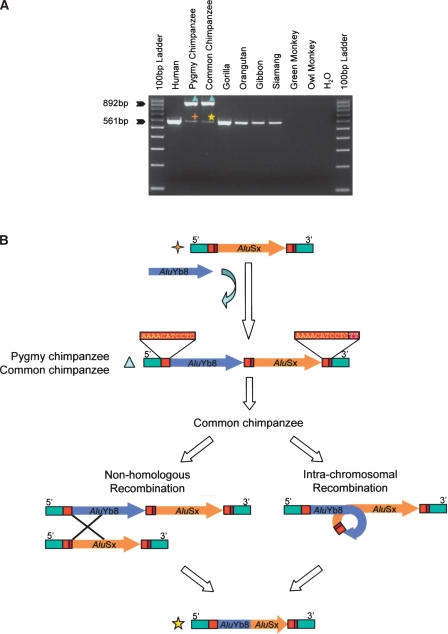
To confirm that the PCR products were derived from authentic _Alu_Yb elements, all of the filled amplicons were cloned and sequenced. This sequencing effort revealed additional insight into the evolutionary history of the following four loci: Pan1, Pan2, Pan3, and Pan4. At the Pan1 locus, an _Alu_Yb8 element was inserted into the chimpanzee lineage (pygmy and common) after the divergence of humans and chimps. However, the pygmy chimpanzee and common chimpanzee showed both filled and empty size amplicons in their PCR amplifications, suggesting the presence of an Alu insertion polymorphism in both species (Fig. 3A). Sequence analysis of the locus showed that the _Alu_Yb8 element inserted immediately upstream of an _Alu_Sx element in the same orientation, and that the integration site is partially shared (three copies of direct repeats). More recently, a nonhomologous recombination or intrachromosomal recombination event in the common chimpanzee genome generated a hybrid Alu element that is composed of the first half of the older _Alu_Sx element and the second half of the newly inserted _Alu_Yb8 element. In contrast, the smaller allele amplified in other primates appeared to be the preintegration site of the _Alu_Yb8 element (Fig. 3B).
Figure 3.
Sequence analysis of the Pan1 locus. (A) The gel chromatographs of PCR amplification results are shown. The template used in each lane is listed at the top of the gel. The product sizes for filled and empty sites (pre-integration size) are indicated at left. (B) Schematic diagrams for the possible evolutionary scenarios. Light-blue triangles denote the amplicons with an _Alu_Yb8 insertion; orange crosses denote the pre-integration products, and the yellow star denotes the recombination product in the common chimpanzee genome. Flanking sequences are shown as green boxes; target site direct repeats are shown in red and pink boxes. Alu elements are shown as arrows and the direction of arrow indicates the orientation (5′→3′), with the head of the arrow denoting the end of the Alu elements.
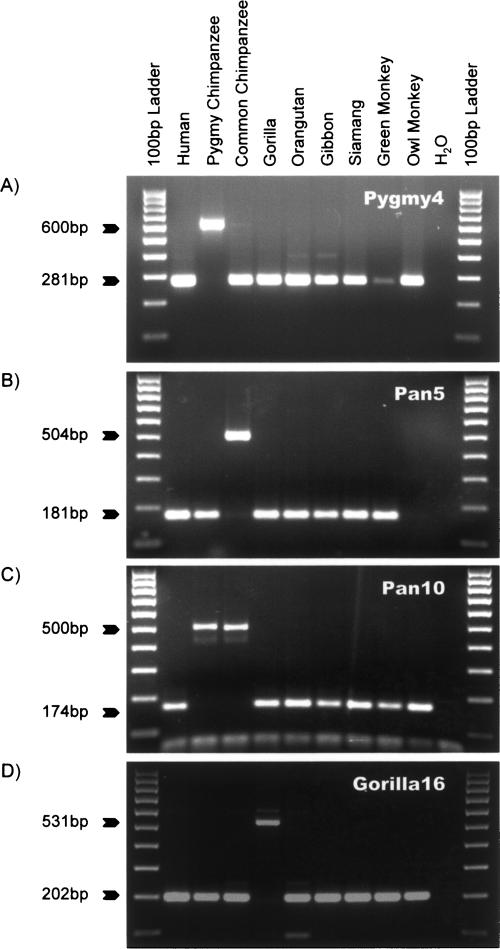
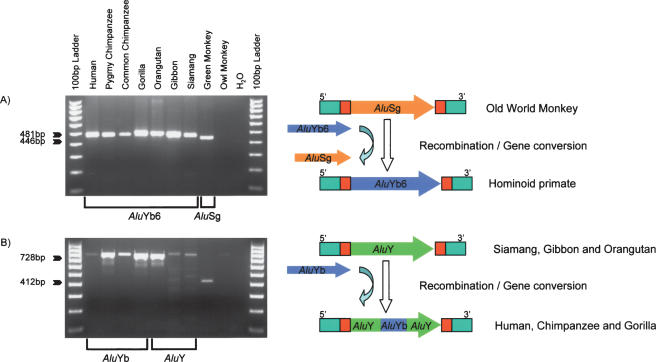
DNA sequence analysis of the Pan2 and Pan3 loci resulted in the recovery of two gene conversion events. The Pan2 locus appears to be the oldest Alu element that we recovered. PCR analysis of the locus showed the presence of an Alu element or filled allele in all of the hominoid primates we examined. The green monkey also showed a filled size amplicon, but the amplicon was slightly smaller than the predicted size (Fig. 4A). DNA sequence analysis showed that all hominoid primates possessed an _Alu_Yb element at the Pan2 locus with the _Alu_Yb lineage diagnostic duplication and five of the eight additional mutations that characterize the _Alu_Yb8 consensus sequence. However, an _Alu_Sg element was found in the orthologous locus in green monkey. This result suggests that the _Alu_Yb6 element (missing two diagnostic mutations compared with the _Alu_Yb8 consensus) at the Pan2 locus was introduced into the genome via a gene conversion event after the divergence of Old World monkeys and hominoids, but before the radiation of the hominoid primates. Thus, the _Alu_Yb6 element at the Pan2 locus is 18–25 Myr old (Goodman et al. 1998). Given the sequence identity between the _Alu_Yb and the _Alu_Sg consensus sequences, we estimate that the starting point of the gene conversion event was located within the first 75 bp of the Alu element, and that the 3′ terminus was located between positions #267 and #310.
Figure 4.
Gene conversion of _Alu_Yb elements. Gel chromatographs of PCR products derived from a phylogenetic analysis of the Pan2 locus (A) and Pan3 locus (B) are shown at left. The DNA template used in each lane is shown at top. The product sizes for filled and empty alleles (pre-integration size) are indicated at left. The schematic diagrams depict the potential evolutionary scenarios on the right. Flanking sequences are shown as green boxes; target site direct repeats are shown in red boxes. Alu elements are shown as arrows, and the direction of the arrow indicates the orientation (5′→3′), with the head of the arrow denoting the end of the Alu elements.
The Pan3 locus also contains a gene conversion event (Fig. 4B). Siamang, gibbon, and orangutan possess an Alu element that does not contain the _Alu_Yb lineage diagnostic duplication and has the highest sequence identity to the _Alu_Y consensus. However, the Alu elements in the gorilla, chimpanzee, and human genomes belong to the _Alu_Yb lineage, as indicated by the presence of the _Alu_Yb diagnostic duplication in these elements. This indicates that a gene conversion event occurred after the divergence of gorillas and orangutans, but prior to the divergence of gorillas, humans, and chimpanzees. During this process, a small proportion of the ancestral _Alu_Y element has been converted to an _Alu_Yb-like sequence, including the _Alu_Yb diagnostic duplication and another adjacent tightly linked diagnostic mutation. The gene-converted region could be as small as 8 bp (251–259) or as long as 30 bp (237–267) in this case. Thus, the overall gene conversion rate (2/12) we observed here seems to be much higher than in previous studies (Maeda et al. 1988; Kass et al. 1995; Roy-Engel et al. 2002; Salem et al. 2003a). However, this is not surprising, since the difference may be due to the small sample size in the current study or reflect the longer evolutionary time period that each element may have been subjected to gene conversion.
The Pan4 locus contains the oldest _Alu_Yb8 element we identified. Interestingly, the Pan4 _Alu_Yb8 element in the human genome only has one CpG mutation (G→A) at position #5, compared with the _Alu_Yb8 consensus sequence, while the chimpanzee and gorilla _Alu_Yb8 elements at the orthologous Pan4 locus each have accumulated five species-specific mutations compared with the _Alu_Yb8 consensus sequence. Since no mutations are shared by the human, chimpanzee, and gorilla _Alu_Yb8 elements at the Pan4 locus, we believe that the ancestral _Alu_Yb8 element at the Pan4 locus was a canonical _Alu_Yb8 element. Using the BLAT program (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start), we identified three additional _Alu_Yb8 elements in the human genome with 100% sequence identity to the Pan4 element. This result suggests that the Pan4 element may still be retrotranspositionally active in the human genome, although we cannot completely rule out the possibility that these four elements independently mutated at position #5. Multiple alignments of the Pan1, Pan2, Pan3, and Pan4 loci are available on our Web site under publications (http://batzerlab.lsu.edu).
_Alu_Yb insertions in other primate genomes
To further investigate the propagation of the _Alu_Yb lineage, we next searched for the presence of _Alu_Yb elements in additional nonhuman primate genomes. Since no complete draft genomic sequences are available other than human and common chimpanzee, we used a modified PCR display method to identify _Alu_Yb elements from other primate genomes (see Methods). The display procedure was performed for four hominoid primates: pygmy chimpanzee, gorilla, orangutan, and gibbon; three Old World monkeys: green monkey, rhesus monkey, and pig-tailed macaque. In addition, this approach was also applied to the common chimpanzee to identify additional _Alu_Yb elements absent from the common chimpanzee genomic sequence. Two restriction enzymes were used for every template, and a minimum of 72 colonies were sequenced for each species. For the common chimpanzee, a total of seven _Alu_Yb elements were retrieved, all of which had previously been identified from the draft sequence. This suggested that our method involving two restriction enzymes yielded a ∼60% coverage (7/12) of the _Alu_Yb elements in the genome. One pygmy chimpanzee-specific and five gorilla-specific _Alu_Yb8 insertions were identified and confirmed by a PCR analysis of nonhuman primates (Fig. 2A,D) and DNA sequencing. No _Alu_Yb element was recovered from the orangutan genome using the PCR display approach. The only locus identified within the gibbon genome was Pan2, which had been previously identified in the chimpanzee genome using the computational approach. Despite at least two trials for each species, no _Alu_Yb elements were identified in the three Old World monkeys (green monkey, rhesus monkey, and pig-tailed macaque) examined. All of the new _Alu_Yb loci identified by PCR display are listed in Table 2.
Age estimates for the _Alu_Yb8 insertions in chimpanzee and gorilla
To estimate the average age of _Alu_Yb8 elements in the chimpanzee genome, CpG and non-CpG mutation densities were calculated for 10 chimpanzee-specific _Alu_Yb8 elements as reported previously (Xing et al. 2004) using the chimpanzee _Alu_Yb8 consensus. The 10 elements contained a total of nine non-CpG mutations of 2420 nucleotides and 10 CpG mutations of 460 CpG nucleotides. The mutation densities were 0.37% ± 0.30% (average ± standard deviation) and 2.17% ± 2.29% for the non-CpG nucleotides and CpG nucleotides, respectively. Using a neutral mutation rate of 0.0015/site/Myr for non-CpG sites and a mutation rate of 0.0090/site/Myr for CpG sites (Xing et al. 2004), the average non-CpG and CpG mutation densities yield age estimates of 2.48 ± 2.03 and 2.42 ± 2.55 Myr, respectively. For the gorilla-specific _Alu_Yb8 elements, a total of five elements were analyzed. The mutation densities were 0.17% ± 0.23% and 3.48% ± 3.30% for the non-CpG and CpG sites, yielding age estimates of 1.10 ± 1.51 and 3.86 ± 3.66 Myr, respectively.
Phylogenetic analysis of _Alu_Yb elements in the human genome
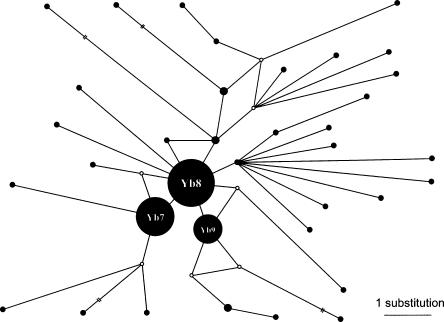
The presence of _Alu_Yb8 elements in different primate genomes suggests that the origin of the _Alu_Yb8 subfamily may be much older than its major expansion in humans. In addition, the presence of _Alu_Yb8, but not _Alu_Yb7 or _Alu_Yb9 elements in the chimpanzee genome suggests that the _Alu_Yb8 subfamily may be the ancestral component of the human _Alu_Yb lineage and would, therefore, predate the other two major human _Alu_Yb subfamilies (i.e., _Alu_Yb7 and _Alu_Yb9). To test this hypothesis, we examined the phylogenetic relationships of the different components of the _Alu_Yb lineage in the human genome, using a median-joining network approach (Cordaux et al. 2004) as implemented in the software NETWORK 4.1 (Bandelt et al. 1999) available at http://www.fluxus-engineering.com/sharenet.htm. From the _Alu_Yb elements identified by Carter et al. (2004) in the human genome, we removed truncated elements and members of the _Alu_Yb7, _Alu_Yb8, and _Alu_Yb9 subfamilies, leaving 36 previously unclassified _Alu_Yb elements. After deleting the middle A-rich region and poly-A tail of the elements, a network of the _Alu_Yb lineage was reconstructed using the 36 non-_Alu_Yb7/8/9 elements and the consensus sequences of the _Alu_Yb7, _Alu_Yb8, and _Alu_Yb9 subfamilies. Collectively, these three subfamilies comprise ∼2000 copies. A preliminary analysis suggested that several nucleotide positions may have mutated more than once. Thus, the _Alu_Yb network was calculated with these putative hypervariable positions down-weighted to 1 (positions #64, #98, and #144) or 5 (position #174 and position #211), while other positions were given a weight of 10. The resulting network (Fig. 5) shows that the _Alu_Yb8 subfamily occupies a central position in the network. In addition, the _Alu_Yb8 node is associated with the highest number of direct branches (nine), as compared with the _Alu_Yb7 and _Alu_Yb9 nodes (four branches each). Finally, 72% of the non-_Alu_Yb7/8/9 elements are more closely related to _Alu_Yb8 than to _Alu_Yb7 or _Alu_Yb9. Taken together, these results are strongly suggestive that the _Alu_Yb8 elements are ancestral to the other _Alu_Yb subfamilies in the human genome (Posada and Crandall 2001; Cordaux et al. 2004).
Figure 5.
Median-joining network of the human-specific _Alu_Yb elements. The network of the _Alu_Yb lineage was reconstructed using the 36 non-Yb7/8/9 elements and the consensus sequences of the Yb7, Yb8, and Yb9 subfamilies as representatives of these three subfamilies. Black circles denote sequence types. Reconstructed nodes are identified as empty circles. The size of circles indicates the number of Alu loci with this sequence type, while arbitrary sizes were chosen for the Yb7/8/9 nodes to represent the relative sizes of the three subfamilies. Lines denote substitution steps, with a one-step distance being indicated in the bottom–right corner. Broken lines indicate that the length of the branch is not drawn to scale.
Discussion
The origin of the _Alu_Yb lineage
The _Alu_Yb lineage is one of the most active Alu lineages in the human genome, with an estimated copy number of ∼2000 (Carter et al. 2004). To obtain further insight into the origin of the _Alu_Yb lineage in the primate order, we analyzed the draft sequence of the common chimpanzee genome and identified 12 _Alu_Yb insertions, 10 of which are members of the _Alu_Yb8 subfamily, while the other two are non-Yb7/8/9 elements. The presence of an _Alu_Yb element at the Pan2 locus within siamang and gibbon genomes suggested the _Alu_Yb lineage originated before the divergence of all hominoid primates. However, no _Alu_Yb elements have been identified in multiple old-world monkey genomes; thus, the origin of the _Alu_Yb lineage was after the divergence of Old World monkeys and apes. These results place the origin of the _Alu_Yb lineage at the early stage of the hominoid evolution, about 18–25 Mya (Goodman et al. 1998) It is worth noting that although the _Alu_Yb6 insertion at the Pan2 locus is the oldest _Alu_Yb element we identified, it was most likely generated via a gene conversion event. Therefore, it may not be the founder gene of the _Alu_Yb lineage, but rather an early offspring of the _Alu_Yb founder gene, which was subsequently lost in extant primates.
The Pan4 _Alu_Yb element appears to have been fixed in the human, chimpanzee, and gorilla genomes, and contains all eight diagnostic mutations that characterize the _Alu_Yb8 consensus sequence. This suggests that within the 10 Myr or so after the _Alu_Yb lineage initially arose, it was not very active in terms of retrotransposition, if at all. However, this lineage retained its retrotranspositional potential during this extended period of time. Furthermore, our results suggest that in gorilla and chimpanzee genomes, the copy number of _Alu_Yb elements is two orders of magnitude lower than that in human. Therefore, over several million years following the insertion of the _Alu_Yb8 at the Pan4 locus, the _Alu_Yb lineage still retained a very low retrotransposition activity (∼1.5 fixed copies/Myr in the chimpanzee genome) until the major expansion of the _Alu_Yb lineage in the human genome within the past 3–4 Myr (Carter et al. 2004; Xing et al. 2004).
The “stealth driver” model of Alu evolution
The long evolutionary history of _Alu_Yb lineage leads to the conclusion that the _Alu_Yb lineage has remained in the genome with little or no retrotransposition activity for an extended period of time, while retaining the ability to generate an appreciable number of new copies later in a species-specific manner (Fig. 6). This scenario is different from the classic “master” gene model, in which a master gene is defined as an element that is highly active over long periods of time (Deininger et al. 1992). In general, the amplification dynamics of the _Alu_Yb lineage show a striking similarity to that of the _Alu_Ya5 lineage (Leeflang et al. 1993; Shaikh and Deininger 1996). Although the existence of low-activity Alu source genes has previously been suggested for the _Alu_Ya5 subfamily (Shaikh and Deininger 1996), here we provide evidence that low retrotransposition activity Alu source genes should be recognized as a major factor driving Alu expansion and evolution.
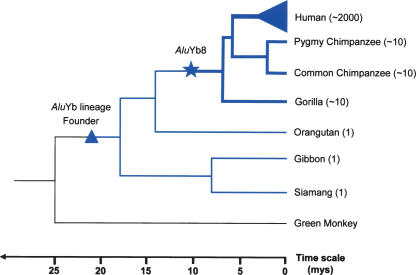
Figure 6.
Putative evolutionary scenario for the _Alu_Yb lineage. A schematic diagram of the hominid primates is shown with the approximate time scale shown on the bottom in million years. The blue lines indicate the expansion of the _Alu_Yb lineage, and the thickness of the lines represents its relative retrotransposition activity. The estimated copy number of _Alu_Yb elements in various primates is shown after their names. The blue triangle represents the estimated integration time period of _Alu_Yb founder gene, and the star represents the estimated integration time period of the oldest known _Alu_Yb8 element (Pan4).
We propose a model of Alu evolution and retrotransposition, in which the low-activity Alu elements are termed “stealth drivers.” In contrast to master genes, stealth drivers are not responsible for generating the majority of new Alu copies, but rather for maintaining genomic retrotransposition capacity over extended periods of time. By generating new Alu copies at a slow rate, a stealth driver may generate some daughter elements that are capable of much higher retrotransposition rates. These hyperactive daughter elements may act as master genes for the amplification of Alu subfamilies and are responsible for producing the majority of the subfamily members. In addition, the new master genes may also give birth to additional retrotranspositionally active Alu copies that serve as secondary master genes or sprouts and also contribute to the expansion of the subfamily (Deininger and Batzer 1993). This view is supported by the fact that recently integrated human Alu subfamilies typically contain ∼15% of such secondary master genes, in addition to the primary subfamily master gene (Cordaux et al. 2004). While the highly active master genes that are produced from the stealth driver would be deleterious and generally be subject to negative selection, the low-activity stealth driver itself will not be subject to such selection and would allow the lineage to persist for extended periods of time. Furthermore, both stealth driver and its daughter elements will also generate new elements that exhibit low levels of retrotransposition activity, effectively becoming new stealth-driver elements. In fact, it is possible that the continuation of a lineage over extended periods of time may depend on the production of new driver elements.
But the question remains as to why, after persisting relatively quietly for millions of years, a lineage can show a sudden increase in numbers, as appears to be the case for _Alu_Ya and _Alu_Yb lineages. Under the stealth-driver model, the master or stealth locus does not need to be “turned on,” as one or more such drivers has been active during the entire history of the lineage. There are multiple scenarios that may account for the sudden expansion of the Alu lineage. In the scenario that we favor, periods of rapid expansion may be related to the ability of highly active daughter elements to escape selection at the population level and, consequently, produce more progeny. The change in the efficiency of natural selection in weeding out overactive elements may be related to population bottlenecks or other demographic factors (Hedges et al. 2004). Alternatively, periodic increases in element numbers may simply be due to the stochastic nature by which active daughter elements are produced. Yet another possibility is that, contingent on the relative abundance of available L1 retrotransposition machinery, there may be fluctuation in Alu expansion rates (Dewannieux et al. 2003). Less likely is the possibility that molecular host defense mechanisms, which were previously suppressing the activity of Alu elements, failed for some reason during these periods.
In the _Alu_Yb lineage, multiple lines of evidence suggest that the _Alu_Yb8 element at the Pan4 locus may be a recent stealth-driver gene in human-specific Alu retrotransposition. First, the Pan4 element is the oldest _Alu_Yb8 element we identified; second, it only accumulated one point mutation over the last 7 Myr; third, the presence of three identical human _Alu_Yb8 elements is consistent with recent low levels of retrotransposition of this element. On the other hand, since the Pan4 _Alu_Yb8 element has accumulated five mutations in the chimpanzee and gorilla genome, and there are no other _Alu_Yb elements in these genomes with the same mutations, the _Alu_Yb8 element at Pan4 locus is unlikely to be the current driver gene in the chimpanzee and gorilla genomes. The reasons why the Pan4 _Alu_Yb8 element may be a stealth driver in human, but not chimpanzee and gorilla, are unclear. However, there is a striking correlation between sequence similarity to the _Alu_Yb8 consensus sequence and total number of _Alu_Yb copies in these species, raising the possibility of mutational inactivation of the Pan4 _Alu_Yb8 element in chimpanzee and gorilla, but not in the human lineage.
Previous studies have shown that the amplification of the Alu family reached its peak about 30 Mya and subsequently underwent retrotranspositional quiescence (Shen et al. 1991; Britten 1994). Although we still do not know the underlying mechanisms for the retrotranspositional quiescence, the stealth-driver model may explain why the Alu lineage has been subjected to periods of retrotranspositional quiescence interspersed with episodic bursts of amplification, as suggested by the accumulation of at least 5000 human-specific Alu elements since the human–chimpanzee divergence (Carroll et al. 2001; Batzer and Deininger 2002; Xing et al. 2003; Carter et al. 2004; Otieno et al. 2004). The _Alu_Ya and _Alu_Yb lineages that comprise >60% of the human-specific Alu elements may be just two successful examples of this strategy. Similar patterns of amplification have also been observed in the retrotransposition of rodent SINE family, ID, (Kim et al. 1994) and the rodent LINE (Long INterspersed Element) family, Lx (Pascale et al. 1993). In addition to these evolutionary observations, there is also experimental evidence that indicates that some varieties of mobile elements evolve strategies to attenuate their own activity. In Alu, the acquisition of a second monomer to form its dimeric structure has been linked to decreased retrotranspositional activity (Li and Schmid 2004). In vitro modifications to currently active L1 elements can produce orders of magnitude increases in the L1 amplification rate (Han and Boeke 2004). In addition, cryptic polyadenylation sites throughout the L1 sequence may serve to quell the number of full-length, retrotransposition-competent L1 copies generated (Perepelitsa-Belancio and Deininger 2003). Taken together, this evidence suggests that the stealth-driver model may not be a unique feature of the Alu family itself, but rather may be one variant of a common survival strategy for SINE and LINE elements. More generally, the ability of mobile elements to maintain low to moderate levels of amplification activity, rather than more rapid duplication rates, may be a common feature of long-lived, successful families of transposons.
Methods
Computational identification of _Alu_Yb elements
A 31-bp (TGCGCCACTGCAGTCCGCAGTCCGGCCTGGG) oligonucleotide that included the _Alu_Yb lineage-specific duplication was used to screen the common chimpanzee genome draft sequences (panTro1 Nov. 2003 assembly, http://www.ncbi.nlm.nih.gov; R. Waterston, pers. comm.) using the Basic Local Alignment Search Tool (BLAST) program available at http://www.ensembl.org/multi/blastview (Altschul et al. 1990). All Alu elements that have the diagnostic 7-bp duplication were selected and extracted along with 1000 bp of unique DNA sequence adjacent to both ends of the elements. The program RepeatMasker (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker) was then used to annotate all known repeat elements within the DNA sequence. Flanking oligonucleotide primers for the PCR amplification of each Alu element were then designed using Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The primers were subsequently screened against the GenBank NR database using BLAST queries to determine whether they resided in unique DNA sequences and would only amplify the Alu elements of interest.
Identification of _Alu_Yb elements with PCR display
The Alu element PCR display methodology has been reported previously (Ray et al. 2005). Using this approach, 500 ng of genomic DNA was partially digested using restriction endonucleases NdeI or MseI as recommended by the manufacturer (New England Biolabs) in 120 μL reactions. Digestion products were then ligated with double-stranded linkers and amplified and “displayed” using the primer LNP (5′-GAATTCGTCAACATAG CATTTCT-3′) and an _Alu_Yb-specific primer (5′-GGCCGGA CTGCGGACT-3′) to acquire partial Alu sequences and the accompanying flanking unique sequences from each template. Since the Alu sequence in the amplicon is about 300 bp long, the PCR products were then purified by BD CHROMAS SPIN-400 columns (BD Biosciences) to select the fragments larger than 400 bp, so that enough unique flanking sequence can be obtained to locate the orthologous sequences in the draft sequence of the human genome. A second-round amplification was performed using the LNP oligonucleotide and a second nested _Alu_Yb-specific primer (5′-AATCTCGGCTCACTGCAAGCTCCGCT-3′) to increase the specificity of the amplicons. The second-round PCR products were separated on a 2% agarose gel, and fragments larger than 400 bp were excised and extracted from the gel using the Wizard gel-purification kit (Promega). The purified products were then cloned into the TOPO-TA cloning vector (Invitrogen). At least 72 clones were randomly isolated from each template, and DNA sequences were determined from both strands using chain-termination sequencing (Sanger et al. 1977) on an ABI 3100 automated DNA sequencer.
After obtaining the sequences, the BLAST-Like Alignment Tool (BLAT) program (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start) (Kent 2002) was used to compare the resulting partial _Alu_Yb8 element and its adjacent flanking sequence with human draft sequences to identify orthologous sequences. Using a combination of the unique flanking sequence and the orthologous human DNA sequences, oligonucleotide primers were designed around each newly identified _Alu_Yb element as outlined above.
PCR analysis of _Alu_Yb elements
All of the _Alu_Yb loci were screened on a panel composed of human genomic DNA (HeLa cell line ATCC CCL-2) and DNA samples from the following nonhuman primate species: Pan troglodytes (common chimpanzee), Pan paniscus (bonobo or pygmy chimpanzee), Gorilla gorilla (lowland gorilla), Pongo pygmaeus (orangutan), Hylobates syndactylus (siamang), Hylobates lar (white handed gibbon), Chlorocebus aethiops sabaeus (green monkey), and Aotus trivirgatus (owl monkey). The nonhuman primate DNA is available as a primate phylogenetic panel (PRP00001) from the Coriell Institute for Medical Research. The chimpanzee-specific _Alu_Yb loci were also screened for insertion presence/absence using a common chimpanzee population panel composed of 12 unrelated individuals of unknown geographic origin, which was provided by the Southwest Foundation for Biomedical Research.
PCR amplification of each locus was performed in 25-μL reactions using 10–50 ng of target DNA, 200 nM of each oligonucleotide primer, 200 μM dNTP's in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.4), and 2.5 units Taq DNA polymerase. Each sample was subjected to an initial denaturation step of 94°C for 150 sec, followed by 32 cycles of 1 min of denaturation at 94°C, 1 min of annealing at optimal annealing temperature, 1 min of extension at 72°C, followed by a final extension step at 72°C for 10 min. Resulting PCR products were fractionated on a 2% agarose gel with 0.25 μg of ethidium bromide and visualized using UV fluorescence.
DNA sequence analysis
To confirm the presence of _Alu_Yb elements, all PCR products suggesting the presence of an Alu element were gel purified using the Wizard gel-purification kit (Promega). Purified PCR products were then cloned into vectors using the TOPO TA cloning kit (Invitrogen) and sequenced using chain-termination sequencing (Sanger et al. 1977) on an Applied Biosystems 3100 automated DNA sequencer. All clones were sequenced in both directions to confirm the sequence. The DNA sequences generated in this study are available in the GenBank under accession numbers AY791249–AY791290.
Acknowledgments
We thank Dr. David Ray for critical reading and suggestions during the preparation of this manuscript. We also thank Anthony Carter for providing the human _Alu_Yb sequences. This research was supported by the Louisiana Board of Regents Millennium Trust Health Excellence Fund HEF (2000-05)-01 (M.A.B.), Na-tional Science Foundation BCS-0218338 (M.A.B.) and EPS-0346411 (M.A.B.), National Institutes of Health RO1 GM59290 (M.A.B.), and the State of Louisiana Board of Regents Support Fund (M.A.B.).
[The sequence data from this study have been submitted to GenBank under accession nos. AY791249–AY791290.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3492605.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215**:** 403–410. [DOI] [PubMed] [Google Scholar]
- Bandelt, H.J., Forster, P., and Rohl, A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16**:** 37–48. [DOI] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3**:** 370–379. [DOI] [PubMed] [Google Scholar]
- Batzer, M.A., Rubin, C.M., Hellmann-Blumberg, U., Alegria-Hartman, M., Leeflang, E.P., Stern, J.D., Bazan, H.A., Shaikh, T.H., Deininger, P.L., and Schmid, C.W. 1995. Dispersion and insertion polymorphism in two small subfamilies of recently amplified human Alu repeats. J. Mol. Biol. 247**:** 418–427. [DOI] [PubMed] [Google Scholar]
- Batzer, M.A., Deininger, P.L., Hellmann-Blumberg, U., Jurka, J., Labuda, D., Rubin, C.M., Schmid, C.W., Zietkiewicz, E., and Zuckerkandl, E. 1996. Standardized nomenclature for Alu repeats. J. Mol. Evol. 42**:** 3–6. [DOI] [PubMed] [Google Scholar]
- Britten, R.J. 1994. Evidence that most human Alu sequences were inserted in a process that ceased about 30 million years ago. Proc. Natl. Acad. Sci. 91**:** 6148–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten, R.J., Baron, W.F., Stout, D.B., and Davidson, E.H. 1988. Sources and evolution of human Alu repeated sequences. Proc. Natl. Acad. Sci. 85**:** 4770–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, M.L., Roy-Engel, A.M., Nguyen, S.V., Salem, A.H., Vogel, E., Vincent, B., Myers, J., Ahmad, Z., Nguyen, L., Sammarco, M., et al. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311**:** 17–40. [DOI] [PubMed] [Google Scholar]
- Carter, A.B., Salem, A.H., Hedges, D.J., Keegan, C.N., Kimball, B., Walker, J.A., Watkins, W.S., Jorde, L.B., and Batzer, M.A. 2004. Genome-wide analysis of the human Alu Yb-lineage. Hum. Genomics 1**:** 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux, R., Hedges, D.J., and Batzer, M.A. 2004. Retrotransposition of Alu elements: How many sources? Trends Genet. 20**:** 464–467. [DOI] [PubMed] [Google Scholar]
- Deininger, P.L. and Batzer, M.A. 1993. Evolution of retroposons. In Evolutionary biology (eds. M.K. Hecht, et al.), pp. 157–196. Plenum Press, New York.
- Deininger, P.L. and Slagel, V.K. 1988. Recently amplified Alu family members share a common parental Alu sequence. Mol. Cell. Biol. 8**:** 4566–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger, P.L., Batzer, M.A., Hutchison III, C.A., and Edgell, M.H. 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8**:** 307–311. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., Esnault, C., and Heidmann, T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35**:** 41–48. [DOI] [PubMed] [Google Scholar]
- Gibbons, R., Dugaiczyk, L.J., Girke, T., Duistermars, B., Zielinski, R., and Dugaiczyk, A. 2004. Distinguishing humans from great apes with _Alu_Yb8 repeats. J. Mol. Biol. 339**:** 721–729. [DOI] [PubMed] [Google Scholar]
- Goodman, M., Porter, C.A., Czelusniak, J., Page, S.L., Schneider, H., Shoshani, J., Gunnell, G., and Groves, C.P. 1998. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9**:** 585–598. [DOI] [PubMed] [Google Scholar]
- Han, J.S. and Boeke, J.D. 2004. A highly active synthetic mammalian retrotransposon. Nature 429**:** 314–318. [DOI] [PubMed] [Google Scholar]
- Hedges, D.J., Callinan, P.A., Cordaux, R., Xing, J., Barnes, E., and Batzer, M.A. 2004. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 14**:** 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. 1993. A new subfamily of recently retroposed human Alu repeats. Nucleic Acids Res. 21**:** 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. and Smith, T. 1988. A fundamental division in the Alu family of repeated sequences. Proc. Natl. Acad. Sci. 85**:** 4775–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J., Krnjajic, M., Kapitonov, V.V., Stenger, J.E., and Kokhanyy, O. 2002. Active Alu elements are passed primarily through paternal germlines. Theor. Popul. Biol. 61**:** 519–530. [DOI] [PubMed] [Google Scholar]
- Kass, D.H., Batzer, M.A., and Deininger, P.L. 1995. Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol. Cell. Biol. 15**:** 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12**:** 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Martignetti, J.A., Shen, M.R., Brosius, J., and Deininger, P. 1994. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc. Natl. Acad. Sci. 91**:** 3607–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M. Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409**:** 860–921. [DOI] [PubMed] [Google Scholar]
- Leeflang, E.P., Liu, W.M., Chesnokov, I.N., and Schmid, C.W. 1993. Phylogenetic isolation of a human Alu founder gene: Drift to new subfamily identity [corrected]. J. Mol. Evol. 37**:** 559–565. [DOI] [PubMed] [Google Scholar]
- Li, T.H. and Schmid, C.W. 2004. Alu's dimeric consensus sequence destabilizes its transcripts. Gene 324**:** 191–200. [DOI] [PubMed] [Google Scholar]
- Maeda, N., Wu, C.I., Bliska, J., and Reneke, J. 1988. Molecular evolution of intergenic DNA in higher primates: Pattern of DNA changes, molecular clock, and evolution of repetitive sequences. Mol. Biol. Evol. 5**:** 1–20. [DOI] [PubMed] [Google Scholar]
- Matera, A.G., Hellmann, U., Hintz, M.F., and Schmid, C.W. 1990. Recently transposed Alu repeats result from multiple source genes. Nucleic Acids Res. 18**:** 6019–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani, K., Hada, T., Yamamoto, Y., Kaneko, T., Shigeto, Y., Ohue, T., Furuyama, J., and Higashino, K. 1991. Inactivation of the cholinesterase gene by Alu insertion: Possible mechanism for human gene transposition. Proc. Natl. Acad. Sci. 88**:** 11315–11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldridge, M., Zackai, E.H., McDonald-McGinn, D.M., Iseki, S., Morriss-Kay, G.M., Twigg, S.R., Johnson, D., Wall, S.A., Jiang, W., Theda, C., et al. 1999. De novo _Alu_-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am. J. Hum. Genet. 64**:** 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otieno, A.C., Carter, A.B., Hedges, D.J., Walker, J.A., Ray, D.A., Garber, R.K., Anders, B.A., Stoilova, N., Laborde, M.E., Fowlkes, J.D., et al. 2004. Analysis of the human Alu Ya-lineage. J. Mol. Biol. 342**:** 109–118. [DOI] [PubMed] [Google Scholar]
- Pascale, E., Liu, C., Valle, E., Usdin, K., and Furano, A.V. 1993. The evolution of long interspersed repeated DNA (L1, LINE 1) as revealed by the analysis of an ancient rodent L1 DNA family. J. Mol. Evol. 36**:** 9–20. [DOI] [PubMed] [Google Scholar]
- Perepelitsa-Belancio, V. and Deininger, P. 2003. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 35**:** 363–366. [DOI] [PubMed] [Google Scholar]
- Posada, D. and Crandall, K.A. 2001. Intraspecific gene genealogies: Trees grafting into networks. Trends Ecol. Evol. 16**:** 37–45. [DOI] [PubMed] [Google Scholar]
- Price, A.L., Eskin, E., and Pevzner, P.A. 2004. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 14**:** 2245–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, D.A., Xing, J.C., Hedges, D.J., Hall, M.A., Laborde, M.E., Anders, B.A., White, B.R., Stoilova, N., Fowlkes, J.D., Landry, K.E., et al. 2005. Alu insertion loci and platyrrhine primate phylogeny. Mol. Phylogenet. Evol. (in press). [DOI] [PubMed]
- Roos, C., Schmitz, J., and Zischler, H. 2004. Primate jumping genes elucidate strepsirrhine phylogeny. Proc. Natl. Acad. Sci. 101**:** 10650–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Carroll, M.L., Vogel, E., Garber, R.K., Nguyen, S.V., Salem, A.H., Batzer, M.A., and Deininger, P.L. 2001. Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159**:** 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Carroll, M.L., El-Sawy, M., Salem, A.H., Garber, R.K., Nguyen, S.V., Deininger, P.L., and Batzer, M.A. 2002. Non-traditional Alu evolution and primate genomic diversity. J. Mol. Biol. 316**:** 1033–1040. [DOI] [PubMed] [Google Scholar]
- Salem, A.H., Kilroy, G.E., Watkins, W.S., Jorde, L.B., and Batzer, M.A. 2003a. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 20**:** 1349–1361. [DOI] [PubMed] [Google Scholar]
- Salem, A.H., Ray, D.A., Xing, J.C., Callinan, P.A., Myers, J.S., Hedges, D.J., Garber, R.K., Witherspoon, D.J., Jorde, L.B., and Batzer, M.A. 2003b. Alu elements and hominid phylogenetics. Proc. Natl. Acad. Sci. 100**:** 12787–12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74**:** 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, C.W. 1993. How many source Alus? Trends Genet. 9**:** 39. [DOI] [PubMed] [Google Scholar]
- Shaikh, T.H. and Deininger, P.L. 1996. The role and amplification of the HS Alu subfamily founder gene. J. Mol. Evol. 42**:** 15–21. [DOI] [PubMed] [Google Scholar]
- Shen, M.R., Batzer, M.A., and Deininger, P.L. 1991. Evolution of the master Alu gene(s). J. Mol. Evol. 33**:** 311–320. [DOI] [PubMed] [Google Scholar]
- Singer, S.S., Schmitz, J., Schwiegk, C., and Zischler, H. 2003. Molecular cladistic markers in New World monkey phylogeny (Platyrrhini, Primates). Mol. Phylogenet. Evol. 26**:** 490–501. [DOI] [PubMed] [Google Scholar]
- Slagel, V., Flemington, E., Traina-Dorge, V., Bradshaw, H., and Deininger, P. 1987. Clustering and subfamily relationships of the Alu family in the human genome. Mol. Biol. Evol. 4**:** 19–29. [DOI] [PubMed] [Google Scholar]
- Willard, C., Nguyen, H.T., and Schmid, C.W. 1987. Existence of at least three distinct Alu subfamilies. J. Mol. Evol. 26**:** 180–186. [DOI] [PubMed] [Google Scholar]
- Xing, J.C., Salem, A.H., Hedges, D.J., Kilroy, G.E., Watkins, W.S., Schienman, J.E., Stewart, C.B., Jurka, J., Jorde, L.B., and Batzer, M.A. 2003. Comprehensive analysis of two Alu Yd subfamilies. J. Mol. Evol. 57**:** S76–S89. [DOI] [PubMed] [Google Scholar]
- Xing, J.C., Hedges, D.J., Han, K., Wang, H., Cordaux, R., and Batzer, M.A. 2004. Alu element mutation spectra: Molecular clocks and the effect of DNA methylation. J. Mol. Biol. 344**:** 675–682. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz, E., Richer, C., Makalowski, W., Jurka, J., and Labuda, D. 1994. A young Alu subfamily amplified independently in human and African great apes lineages. Nucleic Acids Res. 22**:** 5608–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://batzerlab.lsu.edu; Batzer laboratory.
- http://www.ensembl.org/multi/blastview; Basic Local Alignment Search Tool (BLAST). [DOI] [PubMed]
- http://www.fluxus-engineering.com/sharenet.htm; NETWORK 4.1.
- http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.
- http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi; Primer3.
- http://genome.ucsc.edu/cgi-bin/hgBlat?command=start; BLAT.