Genome-wide assembly and analysis of alternative transcripts in mouse (original) (raw)
Abstract
To build a mouse gene index with the most comprehensive coverage of alternative transcription/splicing (ATS), we developed an algorithm and a fully automated computational pipeline for transcript assembly from expressed sequences aligned to the genome. We identified 191,946 genomic loci, which included 27,497 protein-coding genes and 11,906 additional gene candidates (e.g., nonprotein-coding, but multiexon). Comparison of the resulting gene index with TIGR, UniGene, DoTS, and ESTGenes databases revealed that it had a greater number of transcripts, a greater average number of exons and introns with proper splicing sites per gene, and longer ORFs. The 27,497 protein-coding genes had 77,138 transcripts, i.e., 2.8 transcripts per gene on average. Close examination of transcripts led to a combinatorial table of 23 types of ATS units, only nine of which were previously described, i.e., 14 types of alternative splicing, seven types of alternative starts, and two types of alternative termination. The 47%, 18%, and 14% of 20,323 multiexon protein-coding genes with proper splice sites had alternative splicings, alternative starts, and alternative terminations, respectively. The gene index with the comprehensive ATS will provide a useful platform for analyzing the nature and mechanism of ATS, as well as for designing the accurate exon-based DNA microarrays.
Use of genome sequences from more than two species has dramatically improved the prediction of gene structures by de novo gene-predictor programs that use only genome sequences as its input (Brent and Guigo 2004). However, the alternative use of predicted exons in each transcript requires direct biological evidence, i.e., either actual sequence reads of transcribed sequences (Kan et al. 2001; Zavolan et al. 2003) or exon-junction microarray experiments (Johnson et al. 2003; Lee and Roy 2004). Because the latter method may miss low-frequency alternative splicing patterns that fall below the sensitivity threshold and it predicted >50% false-positive alternative splicing events that were not confirmed by RT–PCR (Johnson et al. 2003), the former method, i.e., sequencing of a large number of individual full-length cDNA clones, is the only way to obtain the full sets of ATS forms of all genes. However, this task has been accomplished to a limited extent (Okazaki et al. 2002; Gerhard et al. 2004) Thus, the current assembly of alternative transcripts have to rely also on short segments of transcribed sequences, i.e., expressed sequence tags (ESTs).
Most tools for EST/transcript assembly are based on sequence homology, which was the only option before full-genome sequences became available. These include a system of clustering tools developed at The Institute for Genomic Research (TIGR) (Quackenbush et al. 2001), STACK-PACK tools developed at the South African National Bioinformatics Institute (SANBI) (Christoffels et al. 2001), and UniGene clustering at the National Center for Biotechnology Information (NCBI) (Wheeler et al. 2004). We have used these tools to assemble transcripts from ESTs obtained from the large-scale mouse cDNA project that focused on early embryos and stem cells (Sharov et al. 2003). However, homology-based assembly methods may generate chimeric transcripts, because they are based on sequence similarity in a relatively short region. Recently, several algorithms were developed for assembling transcripts from sequences aligned to the genome (Haas et al. 2003; Eyras et al. 2004; Thierry-Mieg et al. 2004; Xing et al. 2004). Their major advantage compared with homology-based methods is that the genomic location of a transcript can be identified before clustering, which reduces the probability of erroneous assemblies.
In this study, we present a fully automated computational pipeline, including a new All Alignment Assembly (AAA) algorithm that generates all potential transcripts compatible with a given set of sequences (see Methods). We also present the whole-genome analysis of ATS patterns in mouse based on a complete and nonredundant transcriptome assembly from expressed sequences (RefSeq, GenBank, dbEST, Ensembl, and NIA databases) aligned to the nearly completed mouse genome sequence. We report a combinatorial table of 23 major types of ATS units and the abundance of each type.
Results and Discussion
Transcriptome assembly
Several algorithms have been developed to assemble genome-aligned expressed sequences (ESTs and mRNAs) into transcripts. An exon-based algorithm, which splits sequences into individual exons and then links them in various combinations (Xing et al. 2004), loses transcript-wide information in multiexon genes. This problem is overcome by algorithms, which directly assemble whole alignments. For example, a PASA dynamic programming algorithm maximally assembles complete transcript alignments, optimizing the total number of alignments within each assembly (Haas et al. 2003). However, this algorithm may miss some transcripts represented by several relatively rare fragments, because each rare fragment will be preferentially combined with more abundant fragments. A ClusterMerge algorithm overcomes this limitation by generating all possible extensions of each transcript (Eyras et al. 2004). These alignment-based methods are sensitive to sequence quality, and thus require a rigid filtering of sequences. For example, the ClusterMerge algorithm worked reliably only if all input sequences were correctly spliced and had genomic length greater than the median length (Eyras et al. 2004). More than 50% of ESTs has thus been excluded, resulting in potentially incomplete assemblies. These transcriptome assembly programs also utilize only the best genome alignments for each sequence. Eyras et al. (2004) allowed multiple alignments, but only if they had the same coverage. However, many genes have multiple copies (e.g., duplicated genes and pseudogenes), and therefore, sequences may have multiple, almost equally good alignments in different genomic locations. These assembly methods also do not utilize cDNA clone-linking information.
To overcome these limitations, we developed a fully automated computational pipeline, consisting of filtering of input alignments, assembly of transcripts by a new All-Alignment-Assembly (AAA) algorithm (described in Methods and Supplemental materials), and analysis of transcripts. Although the AAA algorithm is similar to one of the previous methods (Eyras et al. 2004), the advantage of this pipeline over the other methods is in mild filtering of input sequences and in the use of additional information that includes the clone-linking and the locations of promoters, CpG islands, and poly(A) signals.
We applied the pipeline/AAA algorithm to the sequences selected from the following transcript databases: RefSeq (N = 26,600; 08/24/2004) (Pruitt and Maglott 2001), Ensembl (N = 35,247; 09/14/2004) (Birney et al. 2004), GenBank (N = 129,820; 08/24/2004) (Benson et al. 2004), dbEST (N = 4,243,544; 08/24/2004) (Boguski et al. 1993), NIA (296,587 EST sequences and 55 fully sequenced clones [Sharov et al. 2003]; 19,515 old EST sequences that were not included in the earlier assembly, plus 73,873 new EST sequences generated at NIA after August 2003). Genome alignments of all sequences were generated by applying the BLAT software (Kent 2002) to the genome sequence released in May 2004 (Waterston et al. 2002). GenBank sequences duplicated in other databases were removed. To increase the speed of analysis, we also removed redundant entries from dbEST if their alignments to the genome matched entirely (±15 bp in exon fringes) to alignments of some other sequence in RefSeq, GenBank, or dbEST (except for clone-linked pairs of sequences). Only 730,886 sequences from dbEST were nonredundant and selected for analysis. Start and end sites of each intron were examined for splicing consensus (Mount 1982; Burset et al. 2000). We used canonical (GT-AG) as well as two major noncanonical (GC-AG and AT-AC) splicing consensuses, which were well validated experimentally (Burset et al. 2000).
The AAA algorithm generated 191,946 U-clusters (transcription loci) and 246,443 transcripts. U-clusters were classified as genes if they had either ORF ≥100 amino acids (aa), multiple exons, or known function. Among 39,403 genes defined in this manner, 27,497 were protein-coding genes (ORF ≥100 aa or known function), 6032 were noncoding genes or gene fragments with ORF <100 aa, 49 were genes with high repeat content (>90%), 1836 were gene models from Ensembl and RefSeq-XM with no EST or mRNA support in our assembly, and 3989 turned out to be gene copies (i.e., duplications and/or pseudogenes). The 27,497 protein-coding genes had 77,138 transcripts (average 2.8 transcripts per gene). The total number of protein-coding genes identified here (27,497) was close to the latest estimate of the total number of human protein-coding genes (20,000–25,000) (International Human Genome Sequencing Consortium 2004). It is not clear at this point whether the additional 11,906 gene candidates identified here have biological functions and can be called “genes.” However, for the sake of completeness, we included these gene candidates and used all 39,403 genes for the following analyses.
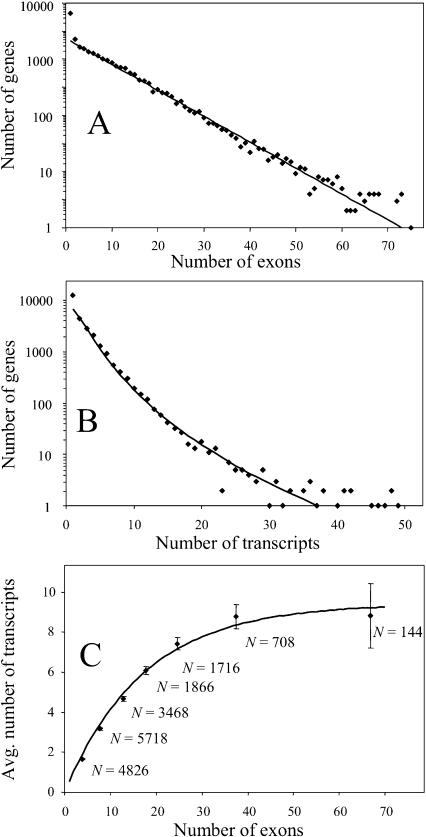
The frequency distribution of protein-coding genes versus the number of exons was exponential (linear in the log-scale) (Fig. 1A). The largest number of exons was found in Neb (160 exons) and the second largest, in lgh-4 (128 exons). The frequency distribution of protein-coding genes versus the number of transcripts turned out to be curvilinear in the log-scale (Fig. 1B). The largest number of transcripts was found in Rtel (130 transcripts). We observed that the average number of transcripts increased with the increasing number of exons, and then leveled off (Fig. 1C). The number of transcripts in large genes with >50 exons was possibly underestimated, because many of these genes had a limited number of supporting mRNA/EST alignments.
Figure 1.
Frequency distribution of protein-coding genes by the number of exons (A) and transcripts (B), and the relation between the number of exons and average number of transcripts per gene (C). Regression lines are as follows: (A) log(N) = 3.44–0.0485_x_; (B) log(N) = 3.72–1.32[log(x)]2; (C) N = 9.44[1–exp(–0.0574_x_)].
Comparison with other gene indexes
Genes and transcripts generated here were compared with four other whole-genome mouse gene indexes (downloaded on 11/09/2004) as follows: TIGR (Quackenbush et al. 2001), UniGene (Pontius et al. 2003), DoTS (The Computational Biology and Informatics Laboratory 2004), and ESTGenes (Eyras et al. 2004). Because UniGene did not have assembled sequences, we selected the best representative from each cluster. Transcripts of each gene index were aligned to the genome using BLAT, and then the overlap with the NIA transcripts was determined by combining genome alignments. Transcripts were considered matching if at least 30% of their length matched within genome boundaries and at least 5% length matched to exons.
The NIA Mouse Gene Index and ESTGenes were compiled from genome alignments; thus, all transcripts had a genome match (except for a few ESTGenes that were not aligned properly due to repeats) and very few sequences had a wrong orientation. Other gene indexes had more entries with no genome match (from 9.8% to 21.3%) and with wrong orientation (from 8.7 to 31.6) (Table 1). The orientation of a transcript was considered wrong if the transcript was overlapped by >50% length with a better-supported transcript in the opposite strand, although some of them may be real antisence transcripts. TIGR and DoTS gene indexes had >300 transcripts with hybrid orientation (with more than one intron in a positive orientation and more than one intron in a negative orientation). NIA Mouse Gene Index and ESTGenes had no transcripts with hybrid orientation.
Table 1.
Number of transcripts in the NIA Mouse Gene Index and other gene indexes
| Gene index | Number of transcripts | No genome match (%) | Wrong orientation (%) | Hybrid orientation (%) |
|---|---|---|---|---|
| NIA | 246,443 | 0.0 | 1.2 | 0.00 |
| UniGene | 46,543 | 9.8 | 8.7 | 0.00 |
| TIGR | 718,567 | 21.3 | 9.0 | 0.05 |
| DoTS | 786,526 | 15.0 | 31.6 | 0.04 |
| ESTGenes | 51,792 | 0.6 | 0.7 | 0.00 |
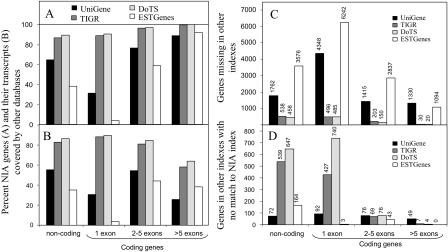
TIGR and DoTS gene indexes had a more complete coverage of genes and transcripts than UniGene and ESTGenes. Especially, ESTGenes had the largest number of missing genes and transcripts (Fig. 2A–C). The observed deficiency of UniGene compared with TIGR and DoTS may have resulted from our selection of only the best representative from each UniGene cluster. Protein-coding genes with more than five exons were best represented in all gene indexes (almost 100% by TIGR and DoTS). Noncoding and single-exon coding genes had the largest number of missing entries in all gene indexes. Among UniGene, TIGR, DoTS, and ESTGenes transcripts that did not match to the NIA Mouse Gene Index, only 5%–8% were considered genes (ORF ≥100 aa, multiple exons, or known function). The majority of these genes were noncoding or single-exon coding genes (Fig. 2D). A few genes with more than five exons that were missing in the NIA Mouse Gene Index, were mostly gene models without EST/mRNA support.
Figure 2.
Comparison of the NIA Mouse Gene Index with other indexes (UniGene, TIGR, DoTS, and ESTGenes). (A) Gene coverage; (B) transcript coverage; (C) genes missing in other gene indexes; (D) genes missing in the NIA Mouse Gene Index.
In contrast to the good coverage of the gene set, the coverage of individual transcripts in all public databases was substantially incomplete (Fig. 2B). The proportion of matching transcripts was estimated as the ratio of NIA transcripts that matched best to at least one sequence in another database to the total number of transcripts. Protein-coding genes with more than five exons had the largest number of transcripts that were missing in the existing databases. The most complete was the DoTS database that covered 71% transcripts of genes with experimental supports by mRNA or ESTs; TIGR covered 66%, UniGene covered 33%, and ESTGenes covered 35% transcripts. The NIA Mouse Gene Index had 19,186 additional transcripts of protein-coding genes that consisted of combinations of exons or their parts (>30 bp) not found in UniGene, TIGR, DoTS, or ESTGenes databases (Supplemental Table 3).
If multiple sequences in other databases were mapped to the same NIA transcript, they were considered redundant. Redundancy was very limited in the UniGene and ESTGenes databases, but very high in TIGR and DoTS (282,488 and 368,557 redundant transcripts, respectively). For example, a transcript of Hbb1-b1 had 2445 entries in TIGR and 333 entries in DoTS. Elimination of redundancy in the NIA Mouse Gene Index was achieved mainly by assembling transcripts from genome alignments.
In general, transcript assemblies in the existing gene indexes had smaller numbers of exons and introns with correct splice sites, and shorter ORFs than the NIA Mouse Gene Index (Supplemental Fig. S1). ESTGenes appeared to be the most deficient in the number of exons and ORF length in comparison with other gene indexes.
Classification of alternative transcription/splicing (ATS)
Previous classification of ATS patterns was based on limited data sets, and thus, was incomplete. The most comprehensive study has used only human UniGene clusters, which do not include all EST sequences from the dbEST database and do not represent all genes and transcripts that can be found in TIGR or DoTS gene indexes (Modrek et al. 2001). Other studies have analyzed even smaller number of genes and transcripts (Kan et al. 2002; Gupta et al. 2004; Hui et al. 2004). The ATS types described thus far include exon skipping, alternative donor site, alternative acceptor site, intron retention, mutually exclusive exons, multiple exon skipping, alternative first exon, alternative last exon, alternative termination in intron, and alternative polyadenylation (Mironov et al. 1999; Beaudoing and Gautheret 2001; Modrek et al. 2001; Kan et al. 2002; Kondrashov and Koonin 2003; Landry et al. 2003; Nurtdinov et al. 2003; Zhou et al. 2003; Galante et al. 2004; Zheng 2004). Recent development of a binary code system for exon–intron structures has identified several new patterns of ATS, although these patterns have not been shown (Nagasaki et al. 2003).
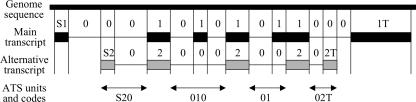
To characterize ATS patterns, we compared each alternative transcript with the main transcript that was selected for having the longest ORF with a significant number of supporting alignments (for details, see Supplemental Methods). We defined an ATS unit as a genomic interval where the main and/or alternative transcripts have one or several nonmatching exons (or a portion of exon) within intron(s) of another transcript. An ATS unit is flanked by common exons or by the end of a transcript (Fig. 3). We improved the coding system that indicated the sequence of exons and introns of the main and alternative transcripts within the ATS unit, which was originally developed by Nagasaki et al (2003). Exons of the main sequence were denoted by “1”, exons of the alternative transcript were denoted by “2”, and introns were denoted by “0”. Additional symbols “S” and “T” were used to denote the start and termination of a transcript, respectively. For example, a skipped exon was coded as “010”, whereas an alternative start was coded “S20” (Fig. 3). ATS patterns were defined as complementary if one code can be obtained by replacing all symbols “1” in the other code by “2”, and vice versa. For example, a skipped exon and an inserted exon are complementary, because the only difference between them is whether the exon belongs to the main or alternative transcript. Compared with the previous coding system (Nagasaki et al. 2003), our coding system combined two codes for the main and alternative transcripts into one, and redefined the end of ATS units for alternative starts and terminations by the shorter transcript. For example, alternative starts (or terminations) that follow the first or second exon of the main transcript have the same codes in our system, but different codes in the system by Nagasaki et al. (2003).
Figure 3.
Coding system for alternative transcription/splicing (ATS).
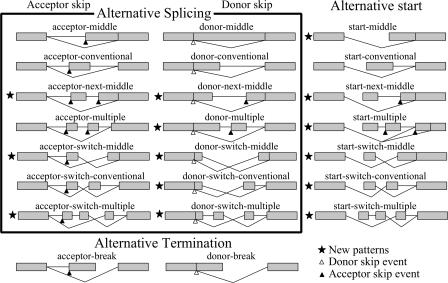
Based on the coding system, we developed a new combinatorial classification of 23 major patterns of ATS units (Fig. 4, Table 2), which include 14 types of alternative splicing, seven types of alternative start, and two types of alternative termination. This classification resulted from the combination of three kinds of alternative start-patterns and eight kinds of alternative end-patterns. ATS units started either from a splicing acceptor failure that led to exon skipping, a splicing donor failure that led to a partial or complete intron retention, or alternative transcription start. The ends of ATS units were more variable; the most common patterns were the endings at the start of the next exon (conventional ending), at an additional acceptor site within the closest exon (middle ending) or in more distant exons (multiple ending). Some ATS units had mutually exclusive exons (denoted as “switch” in Fig. 4). If a transcript terminated in one of the additional exons, then the ATS unit had a break ending. This classification does not include sequential alternative poly(A) signals, which may cause transcript truncation, because our method of transcript assembly would combine a truncated transcript with a longer transcript.
Figure 4.
Combinatorial classification of ATS units.
Table 2.
Types of ATS units
| Types of ATS units | Codesa | Total | In ORF | Complementary % |
|---|---|---|---|---|
| Acceptor-middle | 01 | 3604 | 3239 | 42 |
| Accepter-conventional | 010 | 8114 | 7264 | 35 |
| Acceptor-next-middleb | 0101 | 212 | 183 | 32 |
| Acceptor-multiple | 01010, 010101,... | 1871 | 1735 | 34 |
| Acceptor-switch-middleb | 0102 | 198 | 182 | 52 |
| Acceptor-switch-conventional | 01020 | 295 | 247 | 48 |
| Acceptor-switch-multipleb | 010102, 0102020,... | 208 | 188 | 54 |
| Acceptor-break | 01T, 0101T, 010201T,... | 2878 | 2447 | 79 |
| Donor-middle | 1 | 1357 | 1085 | 91 |
| Donor-conventional | 10 | 3339 | 2715 | 48 |
| Donor-next-middleb | 101 | 293 | 270 | 28 |
| Donor-multipleb | 1010, 10101, 101010,... | 419 | 361 | 30 |
| Donor-switch-middleb | 102 | 181 | 168 | 45 |
| Donor-switch-conventionalb | 1020 | 196 | 148 | 57 |
| Donor-switch-multipleb | 10102, 102020,... | 155 | 139 | 45 |
| Donor-break | 1T, 101T, 10201T,... | 715 | 551 | 78 |
| Start-middleb | S1 | 358 | 224 | 77 |
| Start-conventional | S10 | 4117 | 2961 | 72 |
| Start-next-middleb | S101 | 85 | 52 | 66 |
| Start-multipleb | S1010, 10101, 101010,... | 450 | 327 | 65 |
| Start-switch-middleb | S102 | 86 | 59 | 86 |
| Start-switch-conventionalb | S1020 | 167 | 124 | 72 |
| Start-switch-multipleb | S10102, S102020,... | 94 | 70 | 63 |
| Total | 29,392 | 24,739 |
We analyzed ATS units in 20,323 multiexon protein-coding genes with splicing sites, except ATS units that lacked splicing consensus or evidence from real expressed sequences. Additional filtering was needed to exclude erroneous transcription starts and terminations, which often resulted from cDNA-cloning artifacts. Thus, we analyzed only those alternative starts that had a promoter or a CpG island within a 2-kb distance, and those alternative terminations that had a canonical poly(A) signal (AATAAA or ATTAAA) within the span of the last exon. Promoters were identified with the FirstEF software, P ≥ 0.75 (Davuluri et al. 2001), and CpG islands, with the CpG-proD software (Ponger and Mouchiroud 2002).
Alternative splicing was detected in 9470 multiexon coding genes (47%), alternative transcription start was detected in 3689 genes (18%), and alternative transcription termination was detected in 2893 genes (14%). We detected and analyzed 29,392 ATS units, including 20,442 alternative splicings, 5357 alternative transcription starts, and 3593 alternative transcription terminations (Fig. 4, Table 2). The most frequent start of ATS was acceptor failure (59%), followed by the donor failure (22%) and alternative start (18%). The most common ending of ATS units was “conventional-ending” (N = 15,570; 53%), followed by “middle-ending” (N = 5319; 18%), “break-ending” (N = 3593; 12%), and “multiple-ending” (N = 2740; 9%). Other endings were infrequent (∼2% cases each). The close examination of the ATS units revealed 14 new patterns of ATS that have not been reported (Fig. 4, Table 2).
Perspectives
This study presents an assembly of a gene index with a comprehensive coverage of ATS units from the alignments of expressed sequences to the genome sequence. Compared with previous versions (Sharov et al. 2003), the current version thus includes more genes and gene candidates (39,403 compared with 29,810 in version 1 and 32,114 in version 2) and has a larger set of transcripts for each gene. Comparison with other whole-genome mouse gene indexes (UniGene, TIGR, DoTS, and ESTGenes) supported the notion that this gene index is the most comprehensive one with least redundancy. The software package, which is made freely available to the research community, should also be useful to build gene indexes of other species.
The current NIA Mouse Gene Index will be a valuable resource for future studies directed on validation of structure and function of genes and transcripts. In particular, it will be useful for designing microarrays targeted at genes, transcripts, or specific ATS patterns. For example, it is now possible to validate each ATS unit by developing a DNA microarray representing all possible exon junctions based on the current gene index. Alternative strategy will be high-throughput sequencing of more full-length cDNA clones, but the diminution of yield will make it difficult to exhaust all possible combination of ATS (Modrek et al. 2001).
The classification of possible ATS patterns, including new patterns identified in this report, will also provide valuable tools to understand the mechanisms of alternative splicing and its evolution. It will be interesting to examine which ATS patterns are more often utilized for modification and/or disruption of specific protein domains in the main ORF. Finally, with the comprehensive database of ATS patterns, it may be possible to identify consensus sequences at the exon/intron boundaries that are specific to a particular ATS pattern.
Methods
Full description of methods can be found in the Supplemental materials.
Genome alignments were selected if ≥30% length matched to the genome, and the ratio of the total alignment length to the best alignment was at least 0.9. Low-quality alignments (percent identity, PID <70%) were removed. Alignment artifacts (e.g., additional small exons; Volfovsky et al. 2003) generated by BLAT were removed if they had no splice sites. Sequence orientation was validated using intron splice sites and overlap with genes with already established orientation. We did not intend to assemble unspliced antisense transcripts, because they could not be effectively distinguished from the genomic contamination, and their biological function was unclear. Also, we removed flanking exons if they (or adjacent introns) overlapped with other genes.
The proposed All Alignment Assembly (AAA) algorithm assembled the set of all longest transcripts from EST/mRNA sequences aligned to the genome. Two alignments were considered compatible if each sequence had no elements mapped to an intron of another sequence. Alignments in each chromosome and each strand were grouped into nonoverlapping clusters, and then each cluster was processed sequentially by the AAA algorithm. The proposed AAA algorithm consisted of four steps as follows: (1) find all nonredundant left extensions for each alignment; (2) identify all right-end alignments that cannot be extended to the right; (3) assemble transcripts starting from right to left by branching the extension of each alignment to the left; (4) remove redundant and low-quality transcripts. The pseudo-code for the algorithm is available in the Supplemental information. Transcripts were assembled starting from the rightmost alignments, which were then combined with all possible nonredundant left extensions. Compatible transcripts that contained pairs of clone-linked sequences were grouped together. Partially overlapping transcripts (>5% length) in the same strand were grouped into a U-cluster.
Analysis of transcripts included identification of (1) ORF, (2) repeat regions, (3) main transcript for each U-cluster, (4) duplicated U-clusters, (5) U-clusters with suspicious orientation, and (6) generating annotations for transcripts and U-clusters. ORF was detected using the ORF Finder software (Wheeler et al. 2004) with both standard and alternative genetic code options. Because generated transcripts might have contained ORF shifts resulting from single nucleotide insertions/deletions, we analyzed not just individual ORFs, but also composite ORFs consisting of a pair of overlapping ORFs if each portion was longer than 100 aa. Main transcripts for each U-cluster were identified based on the score S = L_·(1 + 0.25·_N/_N_max), if N ≥ 10, or S = L, if N < 10, where L is ORF length, N is the average number of supporting mRNA/EST sequences for each intron (RefSeq sequences were weighted as 10), and _N_max is the maximum value for N among all transcripts of the gene. A U-cluster was considered a copy of another U-cluster if <30% of its members were best matches. Annotations for transcripts were generated from annotations of member sequences. The preference was given to member sequences from Refseq, GenBank, and to sequences with a valid symbol.
The NIA Mouse Gene Index can be accessed at http://lgsun.grc.nia.nih.gov/geneindex4/. All the data and software tools are available for download at http://lgsun.grc.nia.nih.gov/geneindex4/download.html. In addition to Gene Index Assembly Software, a Perl script (psl2gff.pl) is available to convert the PSL format (BLAT) to GFF format (http://www.sanger.ac.uk/Software/formats/GFF/).
Acknowledgments
We thank David Schlessinger, Ramaiah Nagaraja, and Vincent Vanburen for discussion and critical reading of the manuscript, Ramana V. Davuluri (Department of Molecular Virology, Immunology and Medical Genetics, Ohio State University) for providing promoter locations of mouse genes generated using their First Exon Annotator program, and Yulan Piao, Yong Qian, Patrick Martin, and Uwem Bassey for helping the generation of new mouse ESTs reported in this manuscript. This study utilized the high-performance computational capabilities of the Biowulf/LoBoS3 cluster at the National Institutes of Health.
[Supplemental material is available online at www.genome.org and http://lgsun.grc.nia.nih.gov/geneindex4/. The sequence data from this study have been submitted to GenBank under accession nos. CK329321–CK334090; CF891695–CF906652; CF906741–CF916750; CK334091–CK347104; CK387035–CK393993; CN660032–CN690720; CN690721–CN725493.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3269805\. Freely available online through the Genome Research Immediate Open Access option.
References
- Beaudoing, E. and Gautheret, D. 2001. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 11**:** 1520–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L. 2004. GenBank: Update. Nucleic Acids Res. 32**:** D23–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Andrews, D., Bevan, P., Caccamo, M., Cameron, G., Chen, Y., Clarke, L., Coates, G., Cox, T., Cuff, J., et al. 2004. Ensembl 2004. Nucleic Acids Res. 32**:** D468–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. 1993. dbEST—Database for “expressed sequence tags.” Nat. Genet. 4**:** 332–333. [DOI] [PubMed] [Google Scholar]
- Burset, M., Seledtsov, I.A., and Solovyev, V.V. 2000. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28**:** 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels, A., van Gelder, A., Greyling, G., Miller, R., Hide, T., and Hide, W. 2001. STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucleic Acids Res. 29**:** 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Computational Biology and Informatics Laboratory. 2004. DoTS: A database of transcribed sequences for human and mouse genes. Center for Bioinformatics, University of Pennsylvania, Philadelphia, PA.
- Davuluri, R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29**:** 412–417. [DOI] [PubMed] [Google Scholar]
- Eyras, E., Caccamo, M., Curwen, V., and Clamp, M. 2004. ESTGenes: Alternative splicing from ESTs in Ensembl. Genome Res. 14**:** 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante, P.A., Sakabe, N.J., Kirschbaum-Slager, N., and de Souza, S.J. 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 10**:** 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard, D.S., Wagner, L., Feingold, E.A., Shenmen, C.M., Grouse, L.H., Schuler, G., Klein, S.L., Old, S., Rasooly, R., Good, P., et al. 2004. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC). Genome Res. 14**:** 2121–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Zink, D., Korn, B., Vingron, M., and Haas, S.A. 2004. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics 20**:** 2579–2585. [DOI] [PubMed] [Google Scholar]
- Haas, B.J., Delcher, A.L., Mount, S.M., Wortman, J.R., Smith Jr., R.K., Hannick, L.I., Maiti, R., Ronning, C.M., Rusch, D.B., Town, C.D., et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31**:** 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, L., Zhang, X., Wu, X., Lin, Z., Wang, Q., Li, Y., and Hu, G. 2004. Identification of alternatively sliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene 23**:** 3013–3023. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431**:** 931–945. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302**:** 2141–2144. [DOI] [PubMed] [Google Scholar]
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11**:** 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, Z., States, D., and Gish, W. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12**:** 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12**:** 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A. and Koonin, E.V. 2003. Evolution of alternative splicing: Deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 19**:** 115–119. [DOI] [PubMed] [Google Scholar]
- Landry, J.R., Mager, D.L., and Wilhelm, B.T. 2003. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 19**:** 640–648. [DOI] [PubMed] [Google Scholar]
- Lee, C. and Roy, M. 2004. Analysis of alternative splicing with microarrays: Successes and challenges. Genome Biol. 5**:** 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9**:** 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29**:** 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount, S.M. 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10**:** 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki, H., Suwa, M., and Gotoh, O. 2003. An algorithm for classification of alternative splicing and transcriptional initiation and its genome-wide application. Genome Inform. 14**:** 424–425. [Google Scholar]
- Nurtdinov, R.N., Artamonova, I.I., Mironov, A.A., and Gelfand, M.S. 2003. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 12**:** 1313–1320. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420**:** 563–573. [DOI] [PubMed] [Google Scholar]
- Ponger, L. and Mouchiroud, D. 2002. CpGProD: Identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics 18**:** 631–633. [DOI] [PubMed] [Google Scholar]
- Pontius, J.U., Wagner, L., and Schuler, G.D. 2003. UniGene: A unified view of the transcriptome. In The NCBI handbook. National Center for Biotechnology Information, Bethesda, MD.
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29**:** 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29**:** 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov, A.A., Piao, Y., Matoba, R., Dudekula, D.B., Qian, Y., VanBuren, V., Falco, G., Martin, P.R., Stagg, C.A., Bassey, U.C., et al. 2003. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 1**:** E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg, D., Thierry-Mieg, J., Potdevin, M., and Sienkiewicz, M. 2004. Identification and functional annotation of cDNA-supported genes in higher organisms using AceView. http://www.aceview.org/.
- Volfovsky, N., Haas, B.J., and Salzberg, S.L. 2003. Computational discovery of internal micro-exons. Genome Res. 13**:** 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520–562. [DOI] [PubMed] [Google Scholar]
- Wheeler, D.L., Church, D.M., Edgar, R., Federhen, S., Helmberg, W., Madden, T.L., Pontius, J.U., Schuler, G.D., Schriml, L.M., Sequeira, E., et al. 2004. Database resources of the National Center for Biotechnology Information: Update. Nucleic Acids Res. 32**:** D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., Resch, A., and Lee, C. 2004. The multiassembly problem: Reconstructing multiple transcript isoforms from EST fragment mixtures. Genome Res. 14**:** 426–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D.A., Hayashizaki, Y., and Gaasterland, T. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13**:** 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11**:** 278–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Zhou, C., Ye, L., Dong, J., Xu, H., Cai, L., Zhang, L., and Wei, L. 2003. Database and analyses of known alternatively spliced genes in plants. Genomics 82**:** 584–595. [DOI] [PubMed] [Google Scholar]
Web site references
- http://lgsun.grc.nia.nih.gov/geneindex4/; NIA Mouser Gene Index.
- http://www.sanger.ac.uk/Software/formats/GFF/; GFF format.
- http://lgsun.grc.nia.nih.gov/geneindex4/download.html; All data and software.