Distinct role of lymphocyte function-associated antigen-1 in mediating effective cytolytic activity by cytotoxic T lymphocytes (original) (raw)
Abstract
Lymphocyte function-associated antigen-1 (LFA-1) interaction with intercellular adhesion molecules (ICAMs) facilitates T cell antigen receptor (TCR)-mediated killing. To dissect TCR and LFA-1 contributions, we evaluated cytolytic activity and granule release by cytotoxic T lymphocytes (CTL) as well as intracellular granule redistribution and morphology of CTL stimulated with natural TCR ligand in the presence or absence of LFA-1 engagement. Although other adhesion mechanisms, e.g., CD2-CD58 interaction, could substitute for LFA-1 to trigger CTL degranulation, productive LFA-1 ligation was indispensable for effective target cell lysis by the released granules. LFA-1-mediated adhesion to glass-supported bilayers containing intercellular adhesion molecule-1 was characterized by a much larger junction area, marked by LFA-1 segregation, and a more compact cell shape compared with those observed for CD2-mediated adhesion to bilayers containing CD58. A larger contact induced by intercellular adhesion molecule 1 determined a unique positioning of granules near the interface. These data provide evidence that LFA-1 delivers a distinct signal essential for directing released cytolytic granules to the surface of antigen-bearing target cells to mediate the effective destruction of these cells by CTL.
Keywords: cytolytic granules, immunological synapse, T cell receptor
Killing of virus-infected cells by cytotoxic T lymphocytes (CTL) is triggered by interactions of T cell antigen receptor (TCR) with viral peptides presented by MHC class I proteins on the surface of infected cells and can be mediated by cytotoxic granule exocytosis or FasL-Fas interaction (1, 2). This is a sensitive response often requiring less than a dozen cognate peptide-MHC [complex of antigenic peptide with MHC protein (pMHC)] complexes on the target cell (3, 4). Although productive TCR engagement is necessary and essential to induce CTL cytolytic activity, other accessory and costimulatory molecules are thought to play a role in mediating CTL degranulation and effective cytolytic activity of released granules. For example, both lymphocyte function-associated antigen-1 (LFA-1) and CD2 contribute to CTL adhesion and killing of target cells (5). Ligation of LFA-1 on CTL by high densities of intercellular adhesion molecule-1 (ICAM-1) is sufficient to initiate large-scale molecular segregation and formation of peripheral supramolecular-activating cluster (6), which typically requires antigen for helper T cells (7, 8) and CTL precursors (9). In contrast, ligation of CD2 with its natural ligand CD58 (5) mediates formation of a very small adhesion area by CTL (10). However, how specifically LFA-1, CD2, and other adhesion molecules mediate granule-induced cytotoxicity has not been defined.
Here we have investigated the role of productive LFA-1 engagement for antigen-induced granule release and target cell lysis in vitro as well as for granule polarization in CTL exposed to the glass-supported bilayer. We have found that, although blocking of LFA-1-ICAM-1 interaction abrogates specific lysis of target cells by CTL, antigen-induced release of cytolytic granules by the CTL was not inhibited. Consistent with these data, engagement of both LFA-1 and CD2 promoted polarization of granules and the Golgi complex toward the CTL-bilayer interface containing agonist pMHC. However, only engagement of LFA-1 resulted in a larger contact area with a pSMAC formation and close proximity of the granules and Golgi complex to the interface. In contrast, CD58-CD2 interaction triggered formation of a small contact area without a pSMAC and positioned the granules and Golgi complex in a stalk-like structure connecting the contact area with the rest of the cell. We conclude that productive LFA-1 engagement initiates a distinct signal required for cytoskeleton rearrangement and acquisition of characteristic CTL morphology and formation of a pSMAC. The latter is critical for directing released cytolytic granules to the surface of antigen-bearing target cells and the effective destruction of these cells by CTL.
Materials and Methods
Cells. The human CTL clone 68A62 specific for the HIV reverse transcriptase-derived peptide ILKEPVHGV (IV9) (11, 12) was kindly provided by Bruce D. Walker (Massachusetts General Hospital, Boston), and CTL CER43 that recognizes GILGFVFTL (GL9) peptide from the matrix protein of influenza virus (13, 14) was a gift from Antonio Lanzavecchia (Institute of Immunology, Bellinzona, Italy).
The human HLA-A2+ lymphoblastoid cell line JY was grown in RPMI medium 1640 containing 10% FCS, 10 mM Hepes, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol (referred to as R10).
Proteins and Peptides. HLA-A2 molecules containing a 6-histidine tag at the C terminus were expressed and purified as described (15). Empty HLA-A2 (67 μM) was loaded with peptide of interest at a peptide concentration of 100 μM overnight at 4°C. Unbound peptide was removed by using Micro Bio-Spin 6 column (Bio-Rad) immediately before the experiment.
Hybridomas producing anti-LFA-1 mAb TS1/22 and TS2/4 (16) and the hybridoma secreting ICAM-1-specific mAb HB9580 (17) were purchased from American Type Culture Collection. mAb 24 recognizing activated LFA-1 (18) was a kind gift from Nancy Hogg (Imperial Cancer Research Fund, London). Anti-DNP antibody 2A1 was a kind gift of Ronald Taylor (University of Virginia, Charlottesville). Fab fragments of TS1/22 antibody were prepared by using the Immuno Pure IgG1 Fab kit (Pierce) followed by gel filtration on Superdex 200HR.
Peptide ILKEPVHGV, which serves as an epitope for 68A62 CTL, was kindly provided by Herman Eisen (Massachusetts Institute of Technology, Cambridge). Peptide GILGFVFTL, recognized by CER43 CTL, was synthesized by Research Genetics (Huntsville, AL). The peptide purity was confirmed by HPLC and mass spectrometry.
Cytolytic Assay. Various amounts of peptide in 50 μl of Dulbecco's PBS were combined with 51Cr-labeled target cells (JY) and 2.5 × 104 CTL in 150 μl of complete medium in round-bottomed 96-well plates at an effector-to-target cell ratio of 5:1 or 1:1. The plates were incubated in a CO2 incubator for 4 h at 37°C. Percent specific lysis was determined as described (4).
Granule Release Assay. To quantify granule release, 1 × 105 target cells (JY) were pulsed with various concentrations of cognate peptide for 1 h at 37°C and combined with CTL at effector-to-target cell ratio 1:1 in a round-bottomed 96-well plate in a total volume of 200 μl. After 4 h at 37°C in a CO2 incubator, 100 μl of the culture supernatant was collected, and the activity of serine esterase, an essential component of secreted granules (19), was measured as described (20).
Planar Bilayers and Imaging of Cytolytic Granules and Golgi Complex. Planar bilayers were formed in parallel plate flow cell as described (6). The density of ICAM-1 in the bilayers was adjusted to 200-300 molecules per μm2, the physiological density on antigen-presenting cells (21). The density of HLA-A2 molecules in the bilayers was varied from 500 to 5,000 molecules per μm2 as established by quantitative flow cytometry of beads covered with lipid bilayer containing MHC molecules. Bilayers were blocked with 5% nonfat dry milk containing 100 μM NiCl2. In some experiments, GPI-linked CD58 was incorporated into the lipid bilayers at a density of 400 molecules per μm2. The flow cell was warmed to 37°C, and cells were injected in 500 μl of assay media (10 mM Hepes, pH 7.0/0.14 M NaCl/1 mM Ca2+/2 mM Mg2+/1% human serum albumin) and imaged on a confocal fluorescence microscope (Zeiss LSM510) (22).
Immunofluorescence Staining and Image Analysis. For granule and Golgi apparatus staining, CTLs were labeled with LysoTracker Red DND-99 (Molecular Probes) at 60 nM for 30 min at 37°C and with Bodipy NBD C6 ceramide at 5 μM for 30 min at 4°C, respectively, and injected in 500 μl into a flow cell. In other experiments, unstained CTLs in flow cells were fixed with warm 2% paraformaldehyde in assay media for 12 min and washed thoroughly with assay media. Then the cells were stained with fluorescently labeled mAb for 30 min at 4°C.
For determination of granule and Golgi apparatus location, serial axial (z) sections were acquired at 0.5- to 1-μm intervals through the volume of the cells. Image processing and bilayer calibration were performed by using Zeiss software.
To quantify distribution of the Golgi complex, the z sections, in which Golgi complex staining was centered, were identified for each adherent cell. Every cell was divided equally into three parts along the z axis, i.e., bottom, middle, and top, each containing an equal number of sections. The percentage of cells with staining in each third of the cell was then calculated. Cytolytic granules in each z section were quantified based on groups of bright pixels to exclude single pixel noise. Every cell was divided into three thirds as above, and the average percentage of the granules in each third was calculated for all adherent cells.
The shape index was determined from the ratio of the long axis of the cell to the diameter of the adhesion area. The distance between the bilayer and the top of the cell was taken as the long axis. The diameter of the contact area was approximated by the diameter of a circle with best fit to the contact area, as established by interference reflection microscopy (IRM).
Results
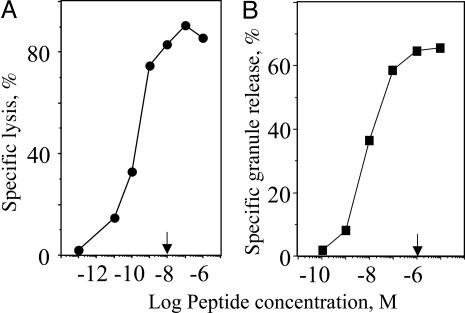
Specific Lysis of Target Cells Is More Sensitive than Measurable Release of Cytotoxic Granules by Human CTL. Specific lysis of target cells was quantified by measuring chromium release from target cells, whereas CTL ability to release cytolytic granules was determined by measurement of serine esterases in the extracellular medium (19). Target cells were treated with antigenic peptide at various concentrations to determine the relative sensitivity of killing and granule release. Although 10-8 M antigenic peptide was sufficient to reach maximal specific lysis (≈85-90%) of target cells, the amount of released granules was further increased at higher peptide concentration and approached its maximum (≈60%) at 10-6 M (Fig. 1). At a low peptide concentration, i.e., 10-10 M, specific lysis was clearly detectable, whereas the amount of released esterases was too small to detect. The destruction of target cells under these conditions was calcium-dependent, as established by blocking with EGTA (data not shown), which is consistent with perforin- but not Fas-mediated killing (1, 2). The difference in the sensitivity of the two responses is determined by the requirement of multiple rounds of secretion by individual CTL to achieve maximal granule release, whereas only a single round is sufficient to induce specific lysis. These data indicate that the release of a small amount of available granules (23) suffices for target cell lysis.
Fig. 1.
Maximal granule release is not necessary for maximal target cell lysis. The concentration of GL9 peptide (arrow) required to achieve maximal specific lysis of HLA-A2+ target cells (JY) is ≈2 orders of magnitude lower than the peptide concentration necessary to induce maximal granule release by human Flu-specific CTL CER43. A representative experiment of five independent experiments is shown.
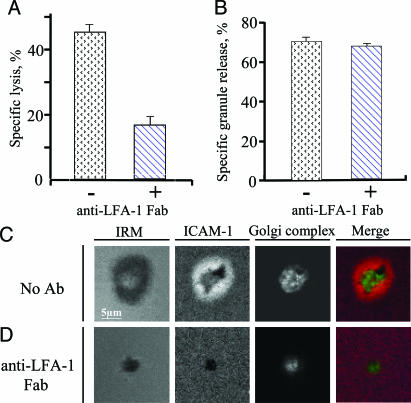
The inhibition of LFA-1-ICAM-1 Interaction Has a Differential Effect on Antigen-Mediated Target Cells Lysis and Release of Cytolytic Granules by CTL. To probe the significance of LFA-1 engagement in target cell lysis and granule release, we compared the effect of the TS1/22 antibody (16), which blocks LFA-1-mediated adhesion, on specific lysis of target cells and release of cytolytic granules by human CTL clone CER43. To prevent crosslinking of cell-surface LFA-1 and induction of unwanted LFA-1-mediated signaling, we used monovalent Fab fragments of the TS1/22 antibody. Fig. 2 A and B shows that the anti-LFA-1 Fab fragments added to the extracellular medium at saturating concentration inhibited CTL cytolytic activity without inhibiting granule release. In control experiments, TS2/4, a nonblocking LFA-1 antibody, had no inhibitory effect on either specific target cell lysis or granule release by CTL (not shown). Similar data have been produced with another human CTL clone 68A62 (data not shown). Thus, at the conditions used, LFA-1 ligation is essential for efficient killing but not for TCR-mediated granule release.
Fig. 2.
Inhibitory effect of anti-LFA-1 Fab on antigen-mediated cytolytic activity and the ability to form pSMAC by CTL. Inhibition of specific lysis of target cells (A) and release of cytolytic granules (B) by CTL at various peptide concentrations by Fab fragments of blocking TS1/22 anti-LFA-1 antibody are shown. Both assays were performed at effector-to-target cell ratio 1:1 and saturating peptide concentration (10-6 M) during 4 h. A representative result of five independent experiments is shown. Standard deviations are indicated. (C and D) TS1/22 anti-LFA-1 Fab blocks the formation of pSMAC (ICAM-1 ring) by CTL exposed to a glass-supported bilayer containing ICAM-1 and cognate pMHC. Images are taken at the level of the bilayer. The Golgi complex is recruited to the bilayer regardless of LFA-1-ICAM-1 interactions but to a different proximity. The Golgi complex was located 1.2 ± 0.4 and 1.9 ± 0.5 μm above the bilayer surface containing ICAM-1 + pMHC in the absence or presence of anti-LFA-1 Fab, respectively (P < 0.005). Merge image: ICAM-1 is red, and the Golgi complex is green.
Anti-LFA-1 Fab Fragments Effectively Block the Formation of Outer Ring Junction. Although LFA-1 and ICAM-1 are the commonly used markers for the development of pSMAC by CTL (9, 24), it has not been demonstrated whether a pSMAC, which is recognized by the large area of contact around the cSMAC, may be formed when LFA-1-ICAM-1 interactions are inhibited. Fig. 2_C_ shows that CTL exposed to the bilayer containing ICAM-1 and cognate pMHC form a cSMAC and pSMAC within a large contact area, as expected from our previous work (6). In contrast, in the presence of TS1/22 Fab, the cells formed a small contact area about the size of the cSMAC, which was apparently mediated by the TCR-pMHC interactions likely in collaboration with CD8 but completely failed to form a pSMAC, as indicated by the absence of any contact area with ICAM-1 accumulation outside the cSMAC-like contact (Fig. 2_D_). When LFA-1-ICAM-1 interactions were allowed in the absence of the antibody, the Golgi complex was tightly juxtaposed to the bilayer in the central opening of the pSMAC, as described (Fig. 2_C_). In the presence of the anti-LFA-1 Fab, the Golgi was still polarized but was not closely apposed to the small contact region (Fig. 2_D_). Thus, close juxtaposition of the Golgi complex to the pMHC-bearing surface requires LFA-1 engagement. These data indicate that laterally mobile TCR-pMHC interactions are adequate for adhesion at high pMHC density and polarization of the Golgi complex, but the cells formed a small contact area without a pSMAC.
Ligation of Either LFA-1 or CD2 Makes Different Contributions to CTL Polarity and Contact Formation. To investigate mechanisms of cytolytic granule polarization, CTL were exposed to different bilayers containing various combinations of cognate pMHC, ICAM-1, and CD58 and intracellular distribution of cytolytic granules, and the Golgi complex was determined by using confocal microscopy. The Golgi complex is a compact structure that is colocalized with the microtubule organizing center (MTOC) (25, 26). This approach provides a unique opportunity to single out the effects of the productive engagement of LFA-1, CD2, and TCR on recruitment of cytolytic granules and Golgi complex polarization without influence of other molecular interactions that are impossible to exclude using live target cells.
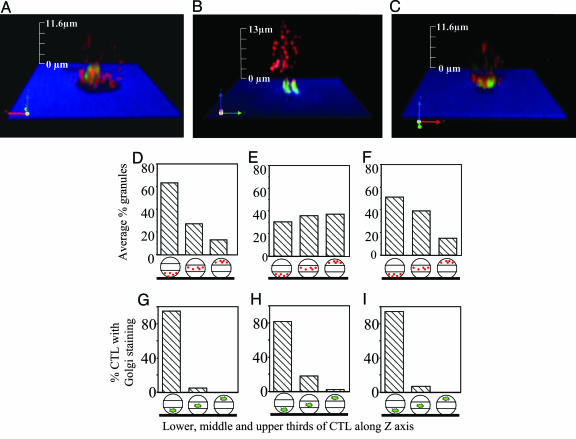
Fig. 3 shows an example of CTL imaged on various bilayers as well as results of the quantitation of the granules and Golgi complex distribution (see also Table 1 and Fig. 5, which is published as supporting information on the PNAS web site). Consistent with our prior report (6), both the Golgi complex and the granules in CTL on ICAM-1 + pMHC bilayer are concentrated in the third closest to the bilayer (Fig. 3 D and G and Table 1). CTL exposed to bilayers containing cognate pMHC at high density (≈2,000 molecules per μm2) did adhere to the bilayer and polarized the Golgi apparatus, which was an average of 2.9 ± 1.6 μm from the contact area (Fig. 3_B_), but granules were not localized to any section of the cells more frequently than another (Fig. 3 E and H and Table 1). At lower density of the pMHC on bilayer (≈500 molecules per μm2), which was still sufficient to mediate adhesion, albeit at a lower percentage of the cells, polarization of the Golgi complex was no longer observed with a mean distance of 4.8 ± 2.4 μm between the Golgi complex and the contact plane (not shown). Thus, TCR engagement alone with its natural ligand may induce Golgi complex polarization but is not sufficient to induce granule polarization.
Fig. 3.
Distribution of cytolytic granules and the Golgi complex in CTL exposed to various glass-supported bilayers. The distribution of the Golgi complex (green) and cytolytic granules (red) is recorded in various z sections (see Fig. 5) of CTL exposed to the bilayer containing either cognate pMHC and ICAM-1 (A), pMHC alone (B), or pMHC and CD58 (C); interference reflection microscopy indicates the level of bilayer (blue) and the CTL contact area (black). z sections were used to generate 3D images of Golgi complex (green) and cytolytic granule (red) intracellular distribution by using volocity software. Average percent of the granules (D-F) or Golgi complex (G-I) in the lower, middle, and upper thirds of all analyzed CTL exposed to bilayers containing ICAM-1 + pMHC (D and G), pMHC (E and H), or CD58 + pMHC (F and I) is shown.
Table 1. Quantitative comparison of morphology and polarization of CTL exposed to various glass-supported bilayers.
| Shape and adhesion area | Position of Golgi complex | Granule distribution | Cell polarization | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bilayer | n* | Adhesion area, μm2 | P | Shape index | P | n* | Golgi complex, μm | P | Percent polarized‡ | n* | Granule amount in the lower one-third of CTL, % | P | n* | Percent polarized§ |
| ICAM-1 + pMHC | 46 (3) | 54.9 ± 14.8 | NS† | 1.4 ± 0.4 | NS† | 40 (2) | 1.3 ± 0.5 | <0.0001† | 95 | 59 (3) | 58 ± 23.1 | <0.0005† | 40 (2) | 73 |
| ICAM-1 | 55 (2) | 58.6 ± 21.2 | 1.2 ± 0.3 | 51 (2) | 5.9 ± 2.5 | 24 | 62 (2) | 29.8 ± 20.6 | 51 (2) | 12 | ||||
| CD58 + pMHC | 83 (3) | 17.5 ± 8.9 | <0.05† | 3.0 ± 0.9 | NS† | 42 (2) | 2.5 ± 0.5 | <0.0001† | 93 | 55 (3) | 50.5 ± 23.6 | <0.0005† | 42 (2) | 63 |
| CD58 | 24 (2) | 13.5 ± 6.5 | 3.3 ± 0 | 53 (2) | 4.9 ± 2.0 | 34 | 55 (2) | 32.4 ± 22 | 48 (2) | 15 | ||||
| pMHC | 81 (3) | 14.3 ± 10.6 | 3.8 ± 1.4 | 63 (2) | 2.9 ± 1.6 | 81 | 62 (2) | 33.7 ± 24 | 61 (2) | 26 |
Similar to ICAM-1, CD58 also promoted recruitment of the granules (Fig. 3 C and F and Table 1) and the Golgi complex (Fig. 3_I_ and Table 1) to the third of the CTL closest to the contact area. The Golgi complex, however, was less closely apposed on average (at 2.5 ± 0.5 μm from the contact area) than in cells on the ICAM-1 + pMHC bilayers (Fig. 3_C_ and Table 1). Although CD58 alone caused CTL to adhere, granule or Golgi complex polarization was not observed, i.e., the granules and the Golgi were localized in the middle of the cell (Table 1). Thus, LFA-1 is not the only adhesion molecule facilitating recruitment of cytolytic granules to the area of CTL contact with target cells in the presence of pMHC, but it is more efficient than CD2 at generating close apposition of the Golgi complex and granules to the contact area.
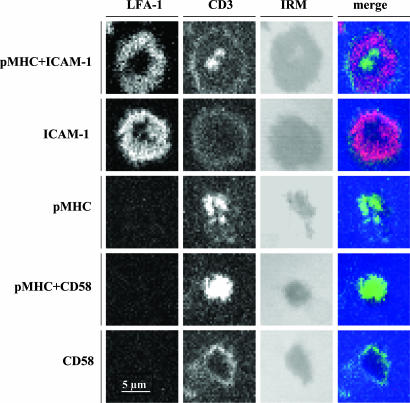
Productive Engagement of LFA-1 Is Indispensable for pSMAC Formation. Formation of pSMAC requires migration of LFA-1 toward the interface. To evaluate whether TCR ligation alone would induce LFA-1 recruitment to the interface and pSMAC formation, we examined TCR and LFA-1 distribution on the surface of CTL exposed to bilayers containing cognate pMHC with the presence or absence of LFA-1 ligand. We used anti-CD3 antibody to observe distribution of TCR and TS2/4 to follow LFA-1. CTL exposed to bilayers containing pMHC complexes alone polarized TCR but not LFA-1 (Fig. 4; see also Fig. 6, which is published as supporting information on the PNAS web site). Although the addition of bivalent TS1/22 IgG did induce migration of LFA-1 toward the interface, the pSMAC, i.e., accumulation and segregation of LFA-1 at the bilayer level, was not formed (Fig. 6). Addition of irrelevant isotype-matched antibodies did not have any affect (Fig. 6). These data show that TCR engagement alone with cognate pMHC on the bilayer is not sufficient for LFA-1 migration to the interface and of the formation pSMAC, and that the latter requires TCR and LFA-1 ligands to be confined to the same surface.
Fig. 4.
Productive engagement of LFA-1 is required for its recruitment to the CTL contact surface and formation of the outer ring junction. CTL were exposed to various bilayers (indicated on the left), fixed, and stained with anti-LFA-1 mAb 24 (red), recognizing activated LFA-1 and anti-CD3 mAb (green). IRM shows the CTL contact area. LFA-1 ring staining is evident only on bilayers containing ICAM-1, regardless of the presence of cognate pMHC.
We also examined the presence of an activation epitope of LFA-1 at the interface using mAb 24 (27). CTL exposure to bilayers containing only cognate pMHC in the absence of LFA-1 ligands led to TCR accumulation at the interface but showed no evidence of LFA-1 activation (Fig. 4). TCR recruitment was enhanced on the bilayer containing pMHC and CD58 (Fig. 4 and Table 2), consistent with the notion that CD58 binding to costimulatory receptor CD2 enhances the TCR-mediated signal (28, 29). Bilayers containing ICAM-1 with or without pMHC generated similar levels of mAb 24 LFA-1 staining in a pSMAC pattern (Fig. 4). Thus, ICAM-1 with or without TCR signaling induces a similar LFA-1 activation in the pSMAC.
Table 2. TCR recruitment to the interface between CTL and various bilayers.
| Bilayer | CD3 accumulation, % cells | Area of CD3 accumulation, μm2 | Number of analyzed cells |
|---|---|---|---|
| pMHC | 50 | 4.4 ± 2.4 | 74 |
| pMHC + CD58 | 75 | 6.3 ± 2.8 | 72 |
| CD58 | 9 | 3.8 ± 2 | 41 |
LFA-1, but Not CD2, Engagement Promotes Morphological Change of CTL. CTL-mediated killing is characterized by morphological changes in the transition from an elongated migratory cell with a uropod to a more compactly spread cell shape without the evident uropod (30). Although CTL adhered to all tested bilayers, the morphology of the attached CTL was very different and depended on the nature of the molecules on the bilayer. Adhesion mediated by TCR-pMHC and/or CD2-CD58 interactions did not lead to significant changes of the CTL shape (Table 1; see also Figs. 7 and 8 and Movie 1, which are published as supporting information on the PNAS web site) that appeared to be elongated (31), typical for T cells with migratory morphology. CTL adhesion on ICAM-1-contaning bilayers resulted in a dramatic change of the shape and acquisition by CTL morphology of activated cells. The latter is characterized by a much larger area of adhesion and the absence of a uropod-like structure (Fig. 8 and Table 1). Thus, productive engagement of LFA-1 but not CD2 or TCR initiates drastic cytoskeleton redistribution and acquisition of characteristic morphology of activated CTL.
Discussion
Granule-mediated killing constitutes several steps that involve recruitment of the granules to the CTL contact area and their release and delivery into the target cell. Our data presented here indicate that the release of the granules does not guarantee their effective cytolytic activity, and that the latter is strongly influenced by LFA-1 engagement. This is implied by experiments in which the blocking of LFA-1-ICAM-1 interactions at the condition preserving CTL target cell adhesion precludes pSMAC formation and leads to inhibition of specific lysis without detectable decrease of the granule release. Thus, LFA-1-ICAM-1 interactions may be dispensable for the release of cytolytic granules but are essential for effective target cell lysis.
Although LFA-1-ICAM-1 interactions can induce formation of a ring junction similar to a pSMAC even in the absence of antigen, this does not cause granule redistribution and recruitment of Golgi/MTOC toward the surface of CTL contact without TCR signaling (6). To dissect TCR- and LFA-1-mediated signals, we used bilayers containing only cognate pMHC and found that CTL could adhere to the bilayer, but granules did not polarize. Thus, productive engagement of both LFA-1 and TCR is necessary for granule recruitment to the CTL contact area. These data suggest that TCR and LFA-1 initiate different intracellular molecular events. In support of this conclusion, experiments using plastic beads covered with anti-CD3 antibody and antigen-presenting cells expressing ICAM-1 show that TCR and LFA-1 ligation could lead to T cell polarization in two different directions: cytoplasmic protein talin is accumulated at the site of LFA-1 engagement, whereas the MTOC is recruited toward ligated TCR on the opposite side of the T cell (32). Note that such dual T cell polarization is likely possible only in response to strong TCR stimulation. Consistent with this, we have observed Golgi/MTOC polarization in CTL exposed to pMHC-containing bilayers only at high density of the pMHC. In addition, we have shown that TCR and LFA-1 can be independently recruited to the CTL-bilayer interface. These data indicate that the signals induced by TCR and LFA-1 are distinct and complement each other to mediate effective destruction of target cells by CTL. It is likely that the TCR and the LFA-1 are localized in different membrane domains, and that each receptor may use a distinct signaling platform. To stimulate effective granule-mediated cytotoxicity, engaged TCR and LFA-1 should be colocalized at the CTL contact area to bring about overlapping of the two signaling platforms, resulting in a qualitatively different signaling.
That the release of cytolytic granules may occur without LFA-1 engagement suggested that other adhesion molecules could substitute for LFA-1. Indeed, we found that CD2 ligation in the absence of LFA-1-ICAM-1 interactions leads to polarization of cytolytic granules and Golgi/MTOC, suggesting an LFA-1-independent mechanism of the recruitment of granules. Although CD2 is known to mediate cellular adhesion, it does not induce a distinct signaling but rather enhances TCR-mediated signals through recruitment of the Src family kinase Fyn (33, 34). The difference in LFA-1- and CD2-mediated signaling was manifested in a profoundly different morphology of CTL adhered to ICAM-1- or CD58-containing bilayers. The CTL on CD58 bilayers had a morphology very similar to that of a cell attached through the uropod (35), the trailing structure of migrating cells based on the small adhesion area and a large shape index (36). One signal that LFA-1 may deliver to the cytoskeleton facilitating large contact area and pSMAC formation includes activation of ezrin-radixin-moesin (ERM) protein dephosphorylation to induce cytoskeletal relaxation leading to the changes of CTL morphology (37).
CTL interaction with the bilayer containing a high density of ICAM-1 is sufficient to form an adhesion ring that anchors LFA-1-associated talin and actin cytoskeleton (6), whereas addition of the TCR signal is necessary for microtubule coupling to the ring near the junction between cSMAC and pSMAC (38, 39). It is believed this is important for activation of the dynein motor that pulls microtubules and consequently MTOC toward the CTL contact surface (39, 40). This mechanism likely reflects a unique function of LFA-1/TCR synergy, explaining a stronger polarization of Golgi/MTOC on ICAM-1- and pMHC-containing bilayers compared with pMHC alone or CD58 and pMHC, an essential prerequisite for granule polarization (see Fig. 3). In contrast, TCR and CD2 ligation in the absence of ICAM-1-LFA-1 interactions may allow Golgi/MTOC positioning in the uropod by a default mechanism. The augmented TCR signaling induced by CD2-CD58 interaction and other adhesion molecules may facilitate granule polarization and release by human CTL missing LFA-1, accounting for residual granule-mediated cytotoxic activity in humans with leukocyte adhesion deficiency (41). It remains to be determined whether FasL-mediated killing by CTL shares this special reliance on LFA-1 with granule-mediated killing or a more general dependence that could be served equally well by other adhesion mechanisms. It is possible that recent reports describing pSMAC-independent killing by CTL may be based on FasL-Fas mechanisms (42, 43). These data show that both LFA-1 and CD2 are indispensable for granule polarization, but they likely use different mechanisms for directional granule release.
We propose that the unique role of LFA-1 in the effective lysis of target cells by CTL is attributed to mediating very tight contact between membranes of target cells and CTL. This membrane junction accounts for strong stable adhesion between membranes of two cells and functions as a gasket to concentrate released granules in a limited space between the two membranes. This notion is consistent with findings by Griffiths and colleagues (24), who analyzed in detail the ultrastructure of the CTL-target cell interface by using electron microscopy and found contact between the two membranes so tight that it forms areas of membrane fusion (24), surrounding clefts that contain cytolytic granules (24, 44). Although the location of these clefts within cell-cell contact cannot be defined from electron micrographs, they are most likely formed within cSMAC or at the edge between cSMAC and pSMAC. The anchoring of microtubules to the pSMAC/cSMAC boundary through synergy of LFA-1 and TCR signaling platforms facilitates delivery of the granule-containing vesicles directly to the clefts within the synapse (39, 40). Thus, LFA-1 accumulation within pSMAC is essential for targeting of the granules to the clefts. In the absence of LFA-1-ICAM-1 interactions, released granules may be mistargeted and may not be contained, providing an opportunity for granule contents to be diluted.
Although CTL effectively destroy other cells, they are relatively resistant to granule-mediated self destruction in vitro (45). Self protection of CTL against cytolytic granules may be explained by rapid appearance of cathepsin B on the CTL surface after TCR ligation, resulting in proteolysis of released perforin bound to the CTL membrane (46). This mechanism may be particularly effective when confined to the granule containing synaptic clefts that represent the secretory domain of the synapse, in which cathepsin B function may be optimal, as opposed to the tight membrane contact environment around the clefts (24). Thus, formation of the specialized CTL-target cell junction may contribute to a mechanism of CTL self protection from cytolytic granules.
Supplementary Material
Supporting Information
Acknowledgments
We thank T. Starr for outstanding technical assistance and Dr. Su-Yi Tseng for demonstrating the volocity (Improvision, Coventry, U.K.) software. This work was supported by National Institutes of Health Grants AI52812 (to Y.S.) and AI44931 (to M.L.D.). K.S. was supported in part by the Finnish Academy of Sciences, T.S. was supported by the National Psoriasis Foundation/USA, and V.K.T. was supported by National Institutes of Health Training Grant CA-09161.
Author contributions: N.A., K.S., T.N.S., V.K.T., M.L.D., and Y.S. designed research; N.A., K.S., T.N.S., and V.K.T. performed research; N.A., K.S., T.N.S., V.K.T., M.L.D., and Y.S. analyzed data; and N.A., M.L.D., and Y.S. wrote the paper.
Abbreviations: CTL, cytotoxic T lymphocyte(s); TCR, T cell antigen receptor; pMHC, complex of antigenic peptide with MHC protein; LFA-1, lymphocyte function-associated antigen-1; ICAM-1, intercellular adhesion molecule-1; MTOC, microtubule organizing center.
References
- 1.Lowin, B., Hahne, M., Mattmann, C. & Tschopp, J. (1994) Nature 370**,** 650-652. [DOI] [PubMed] [Google Scholar]
- 2.Berke, G. (1994) Annu. Rev. Immunol. 12**,** 735-773. [DOI] [PubMed] [Google Scholar]
- 3.Tsomides, T. J., Aldovini, A., Johnson, R. P., Walker, B. D., Young, R. A. & Eisen, H. N. (1994) J. Exp. Med. 180**,** 1283-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sykulev, Y., Joo, M., Vturina, I., Tsomides, T. J. & Eisen, H. N. (1996) Immunity 4**,** 565-571. [DOI] [PubMed] [Google Scholar]
- 5.Shaw, S., Luce, G. E., Quinones, R., Gress, R. E., Springer, T. A. & Sanders, M. E. (1986) Nature 323**,** 262-264. [DOI] [PubMed] [Google Scholar]
- 6.Somersalo, K., Anikeeva, N., Sims, T. N., Thomas, V. K., Strong, R. K., Spies, T., Lebedeva, T., Sykulev, Y. & Dustin, M. L. (2004) J. Clin. Invest. 113**,** 49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monks, C., Freiberg, B., Kupfer, H., Sciaky, N. & Kupfer, A. (1998) Nature 395**,** 82-86. [DOI] [PubMed] [Google Scholar]
- 8.Grakoui, A., Bromley, S. K., Sumen, C., Davis, M. M., Shaw, A. S., Allen, P. M. & Dustin, M. L. (1999) Science 285**,** 221-227. [DOI] [PubMed] [Google Scholar]
- 9.Potter, T. A., Grebe, K., Freiberg, B. & Kupfer, A. (2001) Proc. Natl. Acad. Sci. USA 98**,** 12624-12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dustin, M. L., Golan, D. E., Zhu, D. M., Miller, J. M., Meier, W., Davies, E. A. & van der Merwe, P. A. (1997) J. Biol. Chem. 272**,** 30889-30898. [DOI] [PubMed] [Google Scholar]
- 11.Walker, B. D., Flexner, C., Birch-Limberger, K., Fisher, L., Paradis, T. J., Aldovini, A., Young, R., Moss, B. & Schooley, R. T. (1989) Proc. Natl. Acad. Sci. USA 86**,** 9514-9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsomides, T. J., Walker, B. D. & Eisen, H. N. (1991) Proc. Natl. Acad. Sci. USA 88**,** 11276-11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotch, F., Rothbard, J., Howland, K., Townsend, A. & McMichael, A. (1987) Nature 326**,** 881-882. [DOI] [PubMed] [Google Scholar]
- 14.Valitutti, S., Muller, S., Dessing, M. & Lanzavecchia, A. (1996) J. Exp. Med. 183**,** 1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunmark, A. & Jackson, M. (1998) in MHC, eds. Fernandez, N. & Butcher, G. (Oxford Univ. Press, Oxford, U.K.), Vol. 2, pp. 53-78. [Google Scholar]
- 16.Sanchez-Madrid, F., Krensky, A. M., Ware, C. F., Robbins, E., Strominger, J. L., Burakoff, S. J. & Springer, T. A. (1982) Proc. Natl. Acad. Sci. USA 79**,** 7489-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makgoba, M. W., Sanders, M. E., Ginther Luce, G. E., Dustin, M. L., Springer, T. A., Clark, E. A., Mannoni, P. & Shaw, S. (1988) Nature 331**,** 86-88. [DOI] [PubMed] [Google Scholar]
- 18.Dransfield, I. & Hogg, N. (1989) EMBO J. 8**,** 3759-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasternack, M. S., Verret, C. R., Liu, M. A. & Eisen, H. N. (1986) Nature 322**,** 740-743. [DOI] [PubMed] [Google Scholar]
- 20.Sitkovsky, M. V. (1993) Granule Exocytosis Assay of CTL Activation, eds. Sitkovsky, M. V. & Henkart, P. A. (Birkhäuser, Boston), pp. 482-483.
- 21.Dustin, M. L., Rothlein, R., Bhan, A. K., Dinarello, C. A. & Springer, T. A. (1986) J. Immunol. 137**,** 245-254. [PubMed] [Google Scholar]
- 22.Dustin, M. L. (1997) J. Biol. Chem. 272**,** 15782-15788. [DOI] [PubMed] [Google Scholar]
- 23.Lyubchenko, T. A., Wurth, G. A. & Zweifach, A. (2001) Immunity 15**,** 847-859. [DOI] [PubMed] [Google Scholar]
- 24.Stinchcombe, J. C., Bossi, G., Booth, S. & Griffiths, G. M. (2001) Immunity 15**,** 751-761. [DOI] [PubMed] [Google Scholar]
- 25.Kupfer, A., Dennert, G. & Singer, S. J. (1983) Proc. Natl. Acad. Sci. USA 80**,** 7224-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger, B., Rosen, D. & Berke, G. (1982) J. Cell Biol. 95**,** 137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamata, T., Tieu, K. K., Tarui, T., Puzon-McLaughlin, W., Hogg, N. & Takada, Y. (2002) J. Immunol. 168**,** 2296-2301. [DOI] [PubMed] [Google Scholar]
- 28.Bachmann, M. F., Barner, M. & Kopf, M. (1999) J. Exp. Med. 190**,** 1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wild, M. K., Cambiaggi, A., Brown, M. H., Davies, E. A., Ohno, H., Saito, T. & van der Merwe, P. A. (1999) J. Exp. Med. 190**,** 31-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haverstick, D. M., Engelhard, V. H. & Gray, L. S. (1991) J. Immunol. 146**,** 3306-3313. [PubMed] [Google Scholar]
- 31.Negulescu, P. A., Krasieva, T. B., Khan, A., Kerschbaum, H. H. & Cahalan, M. D. (1996) Immunity 4**,** 421-430. [DOI] [PubMed] [Google Scholar]
- 32.Sedwick, C. E., Morgan, M. M., Jusino, L., Cannon, J. L., Miller, J. & Burkhardt, J. K. (1999) J. Immunol. 162**,** 1367-1375. [PubMed] [Google Scholar]
- 33.van der Merwe, P. A. (1999) J. Exp. Med. 190**,** 1371-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holdorf, A. D., Green, J. M., Levin, S. D., Denny, M. F., Straus, D. B., Link, V., Changelian, P. S., Allen, P. M. & Shaw, A. S. (1999) J. Exp. Med. 190**,** 375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dustin, M. L., Carpen, O. & Springer, T. A. (1992) J. Immunol. 148**,** 2654-2663. [PubMed] [Google Scholar]
- 36.Carpen, O., Dustin, M. L., Springer, T. A., Swafford, J. A., Beckett, L. A. & Caulfield, J. P. (1991) J. Cell Biol. 115**,** 861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faure, S., Salazar-Fontana, L. I., Semichon, M., Tybulewicz, V. L., Bismuth, G., Trautmann, A., Germain, R. N. & Delon, J. (2004) Nat. Immunol. 5**,** 272-279. [DOI] [PubMed] [Google Scholar]
- 38.Dustin, M. L. & Cooper, J. A. (2000) Nat. Immunol. 1**,** 23-29. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn, J. R. & Poenie, M. (2002) Immunity 16**,** 111-121. [DOI] [PubMed] [Google Scholar]
- 40.Poenie, M., Kuhn, J. & Combs, J. (2004) Curr. Opin. Immunol. 16**,** 428-438. [DOI] [PubMed] [Google Scholar]
- 41.Van de Wiel-van Kemenade, E., Te Velde, A. A., De Boer, A. J., Weening, R. S., Fischer, A., Borst, J., Melief, C. J. & Figdor, C. G. (1992) Eur. J. Immunol. 22**,** 1467-1475. [DOI] [PubMed] [Google Scholar]
- 42.Purbhoo, M. A., Irvine, D. J., Huppa, J. B. & Davis, M. M. (2004) Nat. Immunol. 5**,** 524-530. [DOI] [PubMed] [Google Scholar]
- 43.Faroudi, M., Utzny, C., Salio, M., Cerundolo, V., Guiraud, M., Muller, S. & Valitutti, S. (2003) Proc. Natl. Acad. Sci. USA 100**,** 14145-14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, R. E., Caulfield, J. P., Michon, J., Hein, A., Kamada, M. M., MacDermott, R. P., Stevens, R. L. & Ritz, J. (1988) J. Immunol. 140**,** 991-1002. [PubMed] [Google Scholar]
- 45.Verret, C. R., Firmenich, A. A., Kranz, D. M. & Eisen, H. N. (1987) J. Exp. Med. 166**,** 1536-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balaji, K. N., Schaschke, N., Machleidt, W., Catalfamo, M. & Henkart, P. A. (2002) J. Exp. Med. 196**,** 493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information