The Drosophila Gene for Antizyme Requires Ribosomal Frameshifting for Expression and Contains an Intronic Gene for snRNP Sm D3 on the Opposite Strand (original) (raw)
Abstract
Previously, a Drosophila melanogaster sequence with high homology to the sequence for mammalian antizyme (ornithine decarboxylase antizyme) was reported. The present study shows that homology of this coding sequence to its mammalian antizyme counterpart also extends to a 5′ open reading frame (ORF) which encodes the amino-terminal part of antizyme and overlaps the +1 frame (ORF2) that encodes the carboxy-terminal three-quarters of the protein. Ribosomes shift frame from the 5′ ORF to ORF2 with an efficiency regulated by polyamines. At least in mammals, this is part of an autoregulatory circuit. The shift site and 23 of 25 of the flanking nucleotides which are likely important for efficient frameshifting are identical to their mammalian homologs. In the reverse orientation, within one of the introns of the Drosophila antizyme gene, the gene for snRNP Sm D3 is located. Previously, it was shown that two closely linked P-element transposon insertions caused the gutfeeling phenotype of embryonic lethality and aberrant neuronal and muscle cell differentiation. The present work shows that defects in either snRNP Sm D3 or antizyme, or both, are likely causes of the phenotype.
The enzyme ornithine decarboxylase (ODC) catalyzes the first step in the synthesis of polyamines and is subject to extensive regulation (7). One mode of regulation is modulation of its half-life. At least in mammals, the protein antizyme (ODC antizyme) binds to, and inhibits, ODC (5, 8) and then targets it for degradation by 26S proteasomes through a ubiquitin-independent pathway (16, 25). Antizyme activity in insect cells has recently been reported, and insect ODC has been shown to be unstable and subject to destabilization by polyamines (12). The isolation of a partial cDNA clone (17), a genomic clone (22), and finally a full-length cDNA clone (18) has been crucial for elucidating the mechanism of antizyme synthesis. Surprisingly, translation of antizyme mRNA in at least rat, human, and Xenopus requires a specific ribosomal frameshift (9, 18, 28, 36). The amino-terminal portion is encoded by open reading frame 1 (ORF1), and the remainder is encoded by the overlapping ORF2 in the +1 reading frame. Programmed ribosomal frameshifting in the overlap region produces a chimera from the two ORFs. The frameshift efficiency is itself modulated by the concentration of polyamines in cells, resulting in an autoregulatory circuit (18, 28).
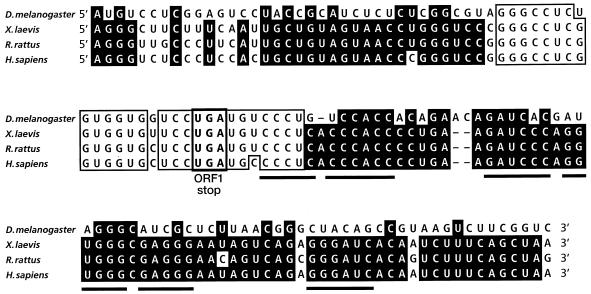
The mammalian antizyme frameshift occurs at the UCC serine codon immediately before the UGA stop codon of ORF1, most likely by occlusion of the U of UGA. A fraction of the ribosomes are switched to the +1 reading frame of ORF2 to complete synthesis of the ORF1-ORF2 product. In addition to the UCCU shift site, three _cis_-acting RNA elements contribute to the mammalian antizyme frameshift signal: the UGA stop codon of ORF1, an RNA pseudoknot 3′ of the shift site, and a partially characterized signal nested within ∼50 nucleotides (nt) 5′ of the ORF1 stop codon (18, 19).
Ribosomal frameshifting is an unusual but important mechanism of translational control of gene expression (4, 6). It plays a very important role in the expression of small genomes (viruses and retrotransposable elements). The extent to which ribosomal frameshifting is utilized in the expression of cellular genes is largely unknown. The antizyme gene is a rare example of a eukaryotic chromosomal gene regulated by translational frameshifting. Until now, no evidence to suggest that expression of antizyme in any invertebrate organism involves this kind of translational regulation has been presented.
Interestingly, a homolog of antizyme ORF2 was discovered in a Drosophila melanogaster cDNA (“guf cDNA”), and its inactivation by P-element insertions was reported to lead to embryonic lethality, deficiencies in terminal differentiation of neuronal cells, and aberrant muscle development—the gutfeeling phenotype (10, 30). On inspection of the “guf cDNA” sequence, we had a gut feeling that this interpretation was incorrect. By examining additional cDNA clones, we discovered an alternative explanation for the gutfeeling phenotype and deduced that there is a genuine D. melanogaster homolog of mammalian antizyme whose expression also involves ribosomal frameshifting.
MATERIALS AND METHODS
DNA manipulations.
The oligonucleotide primers used were GUTF1/S (5′-GCATCCGAATTCGGGCCTCTGTGGTGGTCC), GUF/A2 (5′-CGTGCCCAAGCTTAGCTTCTCCTCGGCGAACTC), SNRNP/S2 (5′-GCATCCGAATTCAAGGGACTTGGTGGGACG), GFEXN1/S (5′-GCATCCGAATTCGACGACATCAGATCACAAATTGCTG), SP6UP/S (5′-GCATCCGAATTCTCAACTTTGGGACCTGCACC), and SNRNP/A2 (5′-CGTGCCCAAGCTTGTGATTATGTGGCCCTCGGC). _Hin_dIII and _Eco_RI sites in all primers are underlined.
_Hin_dIII and _Eco_RI sites were used to subclone PCR products in pUC19. The region overlapping intron 2 of guf1 was amplified with GUTF1/S and GUF/A2. The unknown portion of intron 1 of guf1 was amplified with primers SNRNP/S2 and GFEXN1/S. The 5′ end of guf1 was amplified with primers GUF/A2 and SP6UP/S (which is designed to prime sequences within the cloning vector). The 5′ end of guf2 was amplified with primers SP6UP/S and SNRNP/A2. Southern blotting and other general molecular biological techniques were done as described by Sambrook et al. (31). D. melanogaster cDNAs were isolated from a library constructed from 0- to 4-h embryos (2). D. melanogaster genomic clones were isolated from a cosmid library constructed from the iso-1 fly stock (35). It should be noted that the probe chosen by Salzberg et al. (30) to screen for guf cDNA clones could not have identified spliced guf1 (encoding antizyme) because it corresponds to a region inside intron 1 of that message; however, it could have identified guf2 (encoding small nuclear ribonucleoprotein [snRNP] Sm D3) clones.
In vitro transcription and translation.
CsCl-purified DNA templates were digested with _Eco_RI (NE and GUF-B) and _Bgl_II (GUF-B). One microgram of restricted DNA was used as the template for in vitro transcription with phage T7 or SP6 RNA polymerase (Promega) for NE or GUF-B, respectively, according to the manufacturer’s recommendations. After transcription, the templates were destroyed by incubation with RNase-free DNase (Promega) and the RNAs were recovered by phenol-chloroform extraction and ethanol precipitation. Transcripts were resuspended in 20 μl of 5 mM dithiothreitol containing 20 U of RNasin (Promega). One microliter of RNA was used to program a translation reaction with 10 μl of wheat germ (Promega). Then 50 mM potassium acetate was added to the reaction mixtures for optimal translation. The endogenous level of spermidine in the extract was 0.5 mM. Reaction mixtures were supplemented with spermidine, as indicated in the figure legends. [35S]methionine-labeled protein products were separated by electrophoresis through 16% polyacrylamide Tris-Tricine gels (Novex). Gels were fixed, dried under vacuum, and exposed to X-ray film. ORF1 and ORF2 for fly antizyme each contain three methionine codons. ORF1 and ORF2 for rat antizyme contain two and one methionine codons, respectively.
Northern blot analyses.
Total RNA was extracted from 0- to 24-h Canton S embryos by a standard boiling-phenol method. Twenty-five milligrams of RNA per lane was electrophoresed on a 1.2% agarose gel containing formaldehyde. Transcript sizes were determined relative to a 0.24- to 9.5-kb RNA ladder (Gibco BRL) and visualized by ethidium bromide staining. The RNA was transferred by capillary transfer to a GeneScreen Plus nylon membrane (DuPont) and cross-linked with UV light (0.12 J/cm2). [32P]dCTP-labeled antisense DNA probes were generated with a Prime-It II labeling kit (Stratagene), replacing the random primers with 25 ng of antisense primers. Exon-specific DNA templates were generated by PCR (guf1 exon 2 and 3 [and intron 2] genomic DNA, guf2 exon 1, and guf1 exon 1). Filters were hybridized in 50% formamide at 42°C and then washed at high stringency.
The Northern blot analysis presented by Salzberg et al. (30) appears to contradict our results. In their analysis, a 6-kb _Pst_I genomic fragment spanning the gutfeeling locus (including both transcripts described in this paper) was used as a probe. Major (2,100-nt) and minor (nonreproducible) (1,600-nt) transcripts were observed. We can provide an explanation for the discrepancy between the observed RNA sizes. The sizes of the RNA markers in lane 1 on the gel presented in Fig. 6 of reference 30 appear to be incorrect. The best indication of this comes from analysis of the RNA band corresponding to ribosomal protein 49 (rp49). According to the size markers on their gel, rp49 mRNA has a size of ∼1,400 nt. The real size of rp49 mRNA is ∼600 nt (26). When this is taken into consideration, the Northern blot presented by Salzberg et al. (30) is in good agreement with our findings, with the abundant 2,100-nt RNA corresponding to the antizyme mRNA and the 1,600-nt nonreproducible RNA corresponding to the snRNP Sm D3 mRNA.
RESULTS
Translational frameshifting is required for the expression of a D. melanogaster homolog of antizyme.
A computer search with the BLAST algorithm (1) for sequences similar to the mammalian antizyme frameshift site revealed a stretch of 28 nt in the 5′ untranslated region (UTR) of the “guf cDNA.” In this sequence, 26 nucleotides were identical to those that define the rat antizyme frameshift site (Fig. 1). The high degree of similarity is significant, since previous in vitro experiments suggested that such a sequence could stimulate measurable levels of ribosomal frameshifting (18) in mammals. We speculated that the presence of a potential frameshift site on the same cDNA molecule that is predicted to encode a protein with a significant homology to mammalian (and indeed all known vertebrate) antizymes was more than coincidental. The homology at a potential frameshift site is in addition to the seemingly disconnected downstream region of homology discovered previously (30). The reported 5′ UTR of the “guf cDNA” is unusually large (about 1,250 nt) and contains 23 AUG codons (Fig. 2C). These features led us to hypothesize that the reported guf cDNA clone represents an unspliced D. melanogaster antizyme clone with an intronic sequence which, when spliced out, would unite the two homologous regions of the antizyme sequences.
FIG. 1.
Nucleotide sequence comparison of antizyme frameshift sites. Black shading indicates identity among at least three sequences. Boxes with standard letters indicate identity within the most conserved region among all four sequences. Sequences involved in the formation of the UGA stop codon of ORF1 are indicated by a box with boldface letters. The stems of the vertebrate 3′ pseudoknot are underlined.
FIG. 2.
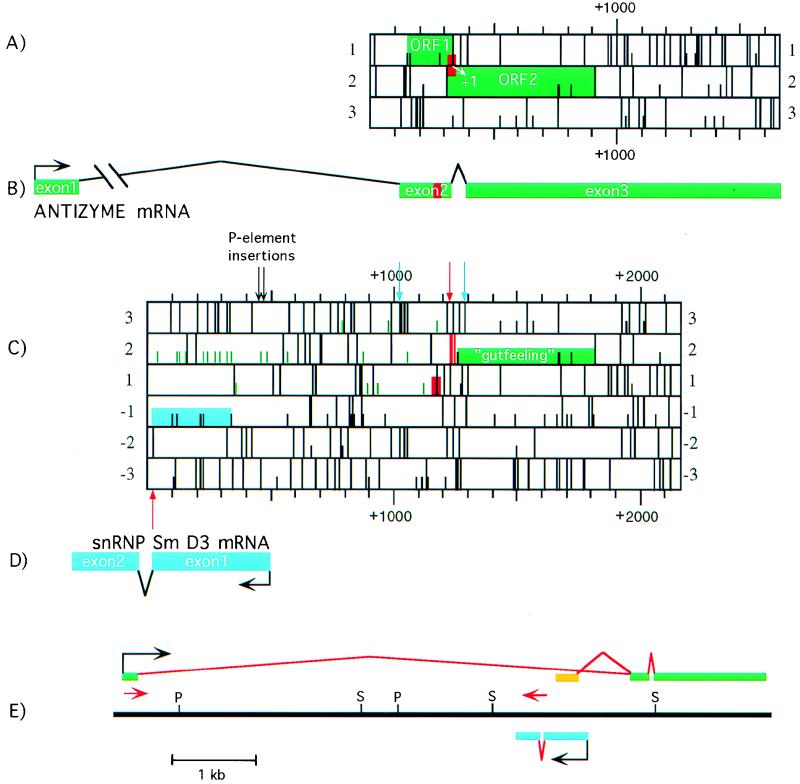
Schematic representation of the gutfeeling locus. The sequence similar to the mammalian antizyme frameshift site is indicated by a red box. Panels A to D are to the same scale, and panels A to C are positioned relative to each other. (A) D. melanogaster antizyme mRNA (guf1). Full bars represent stop codons; half bars represent AUG codons. (B) The transcriptional unit of D. melanogaster antizyme relative to guf cDNA. (C) ORF map of “guf cDNA” as defined previously (30). Green half bars designate AUG codons in the 5′ UTR. The green box indicates the gutfeeling ORF proposed previously (30). Red full bars indicate stop codons in the +1 frame situated between the region similar to the mammalian antizyme frameshift site and the downstream ORF whose product exhibits amino acid homology to vertebrate antizyme. The blue box indicates a region with high homology to sequences for S. cerevisiae and human snRNP Sm D3. Newly identified splice sites are indicated by arrows: red arrow, 5′; blue arrow, 3′. The positions of the two P-element insertions causing the gutfeeling phenotype (30) are shown by black arrows. (D) D. melanogaster snRNP Sm D3 transcriptional unit relative to “guf cDNA.” (E) Physical map of the gutfeeling locus. The exons of the antizyme mRNA are represented by green bars. The aberrant exon 1 of GUF-A is shown in orange. The exons of snRNP Sm D3 are represented by blue lines. Intervening sequences are in red. The positions of the two primers used in determining the size of intron 1 of the antizyme sequence are indicated by red arrows. P, _Pst_I; S, _Sal_I.
To search for the predicted intron, the region was amplified with PCR primers on either side. The sense primer corresponded to the guf cDNA sequence homologous to the antizyme frameshift site. The antisense primer corresponded to the guf cDNA sequence showing the highest amino acid homology to antizyme (see Materials and Methods). The PCR product obtained by amplifying genomic DNA was subcloned and sequenced. The sequence corresponded perfectly with the published “guf cDNA” sequence. In contrast, the PCR product from an embryonic (0 to 4 h) D. melanogaster cDNA library was shorter than the genomic PCR product (data not shown). We did not find a cDNA PCR product with the same size as the genomic PCR product, as would be expected from the reported “guf cDNA” sequence which is completely colinear with the genomic sequence. The sequence of the cDNA PCR product revealed the absence of 66 nt compared to the genomic sequence. The ends of the missing sequence have similarity to D. melanogaster 5′ and 3′ splice site consensus sequences (24) (Fig. 3A). Splicing out this intron (later shown to be intron 2) removes, as predicted, the four in-frame stop codons (frame 2 on Fig. 2C) between the sequence homologous to the mammalian antizyme frameshift site and the downstream sequences which are homologous to the equivalent parts of mammalian antizyme mRNA. The intron removal provides potential accessibility to these downstream sequences via a frameshifting mechanism. Furthermore, splicing removes the AUG translation initiation codon of guf cDNA proposed previously (30) (the next in-frame AUG is well past the point where the homology between guf and the antizyme sequence begins). This suggests that the initiation codon utilized is present in a different ORF.
FIG. 3.
Homology between exon-intron boundaries of the newly identified antizyme and snRNP Sm D3 transcripts and the consensus sequences for 5′ and 3′ splice sites of D. melanogaster. The invariant GU (5′ splice site) and AG (3′ splice site) are in boldface and underlined or overlined. The point of splicing is indicated by an arrow. The ratio of identical nucleotides to total nucleotides is also given.
The antizyme sequence was also amplified from Drosophila virilis genomic DNA with the same set of PCR primers. Sequencing revealed a high degree of homology between the antizyme sequences of the two flies (data not shown). The only region lacking homology corresponds to the newly discovered antizyme intron. In fact, the putative antizyme intron of D. virilis has a different size (69 nt) but has potential 5′ and 3′ consensus splice sites (Fig. 3C).
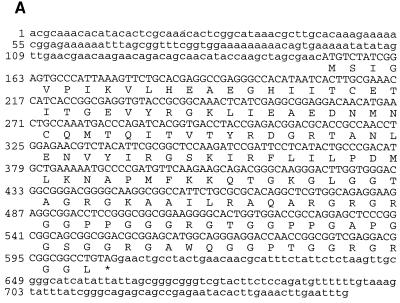
cDNA clones were isolated from the same embryonic D. melanogaster library by hybridization to probes made from the PCR products described above. Two clones, GUF-A and GUF-B, were chosen for sequencing. The 5′ end of GUF-B contained an exon (exon 1) not reported in the guf cDNA analysis reported previously (30). From the 3′ end of exon 1, the sequence jumps to nt +1015 of the published guf cDNA sequence (Fig. 2B and C). The sequence surrounding nt +1015 corresponds well to the consensus 3′ splice site (Fig. 3B). The only other intron we found that is removed to give GUF-B corresponds to the intron described in the preceding paragraph. This conclusion is supported by PCR analysis (data not shown). The first AUG codon of GUF-B initiates an ORF (ORF1) ending with the UGA stop codon of the sequence which is homologous to the vertebrate antizyme frameshift site (Fig. 2A). Sequences downstream define a second ORF (ORF2), homologous to the vertebrate antizyme gene. The two ORFs in GUF-B overlap such that they would be fused by a +1 translational frameshift, analogous to the genes encoding mammalian antizyme. The transcript represented by GUF-B is called guf1.
The unique 5′ exon in GUF-B did not correspond to any sequence in the guf cDNA, and it had no homology to vertebrate antizyme sequences. Consistent with the hypothesis that exon 1 is part of the D. melanogaster antizyme gene, all three exons were recovered on a single cosmid clone (data not shown). Additional mapping revealed that all three exons were contained in a single 8.2-kb _Hin_dIII fragment. Using primers corresponding to antizyme sequences in exon 1 and intron 1 (Fig. 2E), we determined that intron 1 is ∼6.2 kb in length. To further show that exon 1 corresponds to the 5′ end of the D. melanogaster antizyme mRNA transcript, the 5′ end of antizyme message was amplified from the 0- to 4-h-embryo cDNA library with primers within the cloning vector and exon 3. The predominant product was subcloned and sequenced. Sequence analysis of this PCR product demonstrated that D. melanogaster antizyme cDNAs contain 5′ ends beginning with exon 1 of GUF-B.
GUF-A has an unusual 5′ end. Its 5′-most sequence corresponds to nt +133 of the previously published guf cDNA. From there, GUF-A and the published sequence are colinear until nt +356 of the latter, corresponding to exon 1 of GUF-A (Fig. 2E). The other two exons of GUF-A are similar to exons 2 and 3 of GUF-B. Exon 1 of GUF-A contains numerous AUGs followed by a number of stop codons in all reading frames. This finding, combined with our analyses showing that GUF-B contains the predominant exon 1 of guf1, led us to conclude that GUF-A is an aberrant transcript of D. melanogaster antizyme.
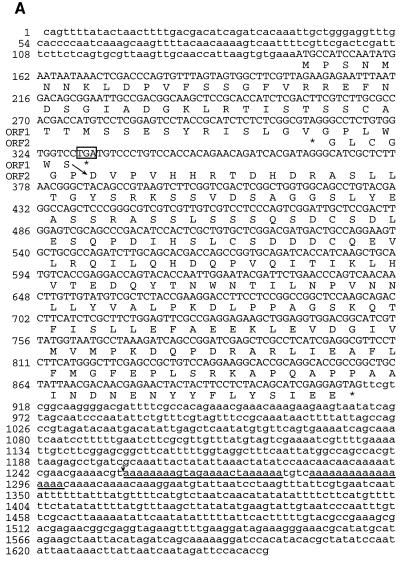
The 3′ end of GUF-B is physically close to the previously reported 3′ end of guf cDNA. GUF-A has an additional ∼350 nt at its 3′ end that are not present in GUF-B. PCR and partial sequencing analysis (data not shown) indicate that the guf1 transcript also contains these additional 350 nt. The 3′ end of GUF-B corresponds to a sequence within GUF-A containing 17 consecutive A’s (Fig. 4A), which would provide a good template for the poly(dT) primer used in generating the cDNA library. It appears that the 3′ end of GUF-B is a cDNA artifact. Interestingly, the 3′ end of the published “guf cDNA” sequence corresponds to a stretch of 24 nt, present in both GUF-B and GUF-A, that contains 20 A’s (Fig. 4A).
FIG. 4.
(A) Nucleotide sequence and predicted amino acid sequence of D. melanogaster antizyme (guf1). The predicted amino acid sequences for both ORF1 and ORF2 are given. The UGA stop codon of ORF1 is boxed. The most likely position of translational frameshifting is indicated by a diagonal arrow. The two A-rich regions (discussed in the text) within the 3′ UTR are underlined. The 3′ end of “guf cDNA” in relation to the nucleotide sequence presented here is indicated by a small vertical arrow. (B) Comparison between the amino acid sequences of D. melanogaster and D. virilis (partial) antizyme proteins and their frog, rat, and human counterparts. A black background indicates amino acid identities among at least three proteins. Boldfacing indicates amino acid similarities among at least four proteins. A symbol following the designation for D. melanogaster, D. virilis, X. laevis, or Rattus rattus indicates a PEST sequence, and the identity of the amino acids involved is indicated by the position of the corresponding symbol over the top line of that set of sequences.
ORF1 of guf1 encodes a polypeptide of 61 amino acids. By comparison, rat and Xenopus antizyme ORF1s encode polypeptides of 68 (most likely) (18) and 58 (9) amino acids, respectively, but amino acid homology between D. melanogaster and vertebrate antizyme proteins is limited and is concentrated near the C termini of the ORF1-encoded polypeptides (Fig. 4B).
If expression of D. melanogaster antizyme is similar to that of vertebrate antizyme proteins (see Discussion), +1 ribosomal frameshifting at a UCC serine codon immediately preceding the UGA stop codon of guf1 ORF1 would lead to translational fusion of ORF1 and ORF2 of guf1 to produce a polypeptide of 254 amino acids (Fig. 4A). This expected polypeptide is acidic (predicted pI = 4.78), with a predicted molecular mass of 28,282 Da. A computer search with the PROSITE algorithm identified several potential sites for posttranslational modifications, including five protein kinase C and nine casein kinase II phosphorylation sites. Seventeen amino acids encoded by ORF2 link the ORF1 polypeptide to the last 176 amino acids of the proposed gutfeeling protein. The homology of this region to vertebrate antizyme is shown in Fig. 4B. The first 64 amino acids encoded by ORF2 have no apparent homology to vertebrate antizyme and are unusual in that they contain 16 (25%) serine residues. The same region is also present in D. virilis antizyme but contains even more serines, i.e., 19. A sequence pattern termed PEST by its discoverers (27) occurs within this region of D. melanogaster antizyme and contains most of the serines (Fig. 4B). There is experimental evidence in several proteins that PEST sequences confer lability, though this is as yet unproven. The scores with the PEST algorithm for the D. melanogaster and D. virilis sequences in question are 6.8 and 21.4, respectively.
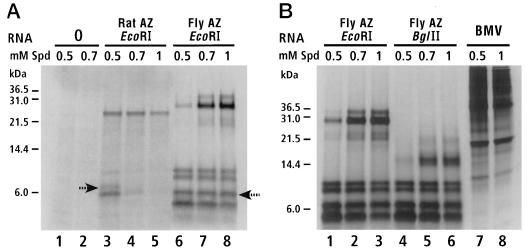
Frameshifting in decoding guf1 was tested in a wheat germ in vitro translational system with RNA transcribed from GUF-B digested with _Eco_RI. As a control, a rat antizyme transcript was also translated. The major large (>10-kDa) product from translation of guf1 had an apparent molecular mass of 28 kDa (Fig. 5), close to the predicted molecular mass (28.3 kDa) of an ORF1-ORF2 fusion (the origin of a minor protein product ∼6 kDa longer than the major product is unknown). The major large product from translation of rat antizyme RNA had an apparent molecular mass of 25 kDa (predicted molecular mass = 25.2 kDa) (Fig. 5A). To confirm that the 28-kDa protein was indeed the product of transframe translation of ORF1-ORF2, a transcript from GUF-B digested with _Bgl_II (which has a unique site within ORF2) was translated in a wheat germ extract. The size of the major large product was reduced to an apparent molecular mass of 15 kDa, as expected (predicted molecular mass, 14.7 kDa) (Fig. 5B). The large number (at least four) of small (<10-kDa) products on the gels (Fig. 5) makes it difficult to discern the termination product of ORF1 (predicted molecular masses of 6.7 kDa for fly and 7.4 kDa for rat).
FIG. 5.
In vitro translation of D. melanogaster antizyme in wheat germ extracts. Protein markers are on the left in both panels. (A) Rat and Drosophila (GUF-B) antizymes synthesized in the presence of increasing concentrations of spermidine (Spd) (0.5 to 1 mM) with products separated on a 16% Tricine gel. The products most likely to be from translational termination at the stop codon of ORF1 of rat and fly antizymes (Az) are indicated by arrows. (B) RNAs transcribed from GUF-B digested with _Eco_RI (cutting 3′ of the cloned cDNA) and _Bgl_II (cutting inside ORF2) translated in the presence of increasing concentrations of spermidine. Translation of brome mosaic virus (BMV) RNA was a control.
Addition of the polyamine spermidine increased synthesis of the 28-kDa protein from guf1 (Fig. 5). As expected (18, 28), a similar induction was seen for the rat 25-kDa antizyme protein (note that even though its absolute amount is reduced, its abundance relative to the smaller products on the gel is increased). Comparison of the products after stimulation with spermidine strongly suggests that the frameshift efficiency (in the in vitro heterologous system) of guf1 is comparable to, or greater than, that of the rat antizyme sequence in reticulocyte lysates, which is as high as 19% (18). Translation of guf1 mRNA in a rabbit reticulocyte lysate gave very similar results (data not shown), but the ubiquitous hemoglobin protein precluded visualization of the ORF1 termination products.
D. melanogaster homolog of the snRNP Sm D3 gene present in the gutfeeling locus.
A substantial portion of the 5′ UTR of the guf cDNA has a very strong homology, in the antisense orientation, to human and Saccharomyces cerevisiae snRNP Sm D3 genes (Fig. 2C), suggesting that there could be a second gene within the gutfeeling locus. To investigate this possibility, an early embryonic library was screened for cDNA clones by hybridization with a probe corresponding to the region of the published guf cDNA sequence with the highest homology to snRNP Sm D3 genes from humans and yeast. Two clones (GUF2-1 and GUF2-3) were sequenced. Both clones contained transcribed sequence in the antisense orientation relative to the guf1 transcript (Fig. 2D), indicating that they represented a distinct transcriptional unit, designated guf2. The two sequenced clones were almost identical, one having an additional 4 nt at its 5′ end. To determine if GUF2-1 and GUF2-3 are representative of the 5′ end of the guf2, the 5′ region of this mRNA was amplified (from a 0- to 4-h-embryo cDNA library), subcloned, and sequenced. The PCR clone contained several additional nucleotides at its 5′ end, compared to the cDNA clones. This data was used to define nt 1 of guf2. The first nucleotide of the guf2 transcript corresponds to nt +495 of the previously published guf cDNA sequence. The 5′ UTR of guf2 is 151 nt long. The two P-element insertions that cause the gutfeeling phenotype map to nt +28 and +47 of the 5′ UTR of guf2. Exon 1 of guf2 is 472 nt long. PCR analyses indicated that the remainder of guf2 is derived from a single exon (exon 2) and that the intervening sequence (intron 1) has a size of ∼70 nt (data not shown). guf2 cDNAs contain a single long ORF encoding a protein of 151 amino acids (Fig. 6A). The predicted protein is basic (predicted pI = 10.55) and has a molecular mass of 15,582 Da. A computer search with the BLAST algorithm revealed very high homology with snRNP Sm D3 of Homo sapiens and S. cerevisiae (Fig. 6B). The highest homology is within the first 90 amino acids of the guf2 protein (close to the size of the S. cerevisiae protein). The C-terminal half of the guf2 protein contains an arginine-glycine (R-G)-rich sequence. Similar (but shorter) R-G-rich regions are present in the C termini of human snRNP Sm D3 and snRNP Sm D1 proteins (15). Interestingly, the guf2 sequence homologous to the yeast snRNP Sm D3 sequence is located in exon 1, while the sequence encoding the R-G-rich domain is located mostly in exon 2.
FIG. 6.
(A) Nucleotide sequence and predicted amino acid sequence of D. melanogaster snRNP Sm D3. (B) Sequence alignment between D. melanogaster snRNP Sm D3 protein and snRNP Sm D3 from H. sapiens and S. cerevisiae. Dark background indicates amino acid identities between at least two proteins. Boldfacing indicates amino acid similarities between at least two proteins.
Expression analysis of guf1 and guf2 transcripts.
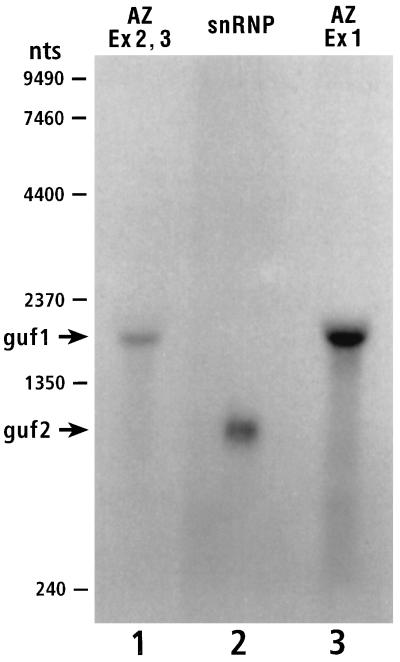
cDNA analyses led us to predict two transcripts within the gutfeeling locus, one of 750 nt (guf2) and the other of 1,650 nt (guf1). Northern blot analysis was performed with probes for each of the two transcripts (guf1 and guf2). The probe for guf1 (exon 1 or exons 2 and 3) revealed an RNA of 1,800 nt (Fig. 7, lanes 1 and 3). The probe for guf2 (exon 1) revealed a single RNA species with an apparent size of 950 nt (Fig. 7, lane 2). The sizes corresponded well to the deduced sizes of the two transcripts [allowing for 150 to 200 nt of poly(A) tails]. A Northern blot study presented by Salzberg et al. (30) appears to disagree with our analysis. However, upon more careful consideration, the two sets of data are in good agreement (for details see Materials and Methods).
FIG. 7.
Northern blot analysis of guf1 and guf2 transcripts. Shown is a Northern blot of embryonic (0- to 24-h) RNA probed with DNA fragments corresponding to exons (Ex) 2 and 3 of the antizyme sequence (Az) (guf1) (lane 1), exon 1 of the snRNP Sm D3 sequence (guf2) (lane 2), and exon 1 of the antizyme sequence (lane 3).
The guf1 transcript is quite abundant. While screening the D. melanogaster cDNA library for antizyme cDNA clones, we found a multitude of positive clones, at least 2 orders of magnitude more than for guf2. In addition, during the writing of this paper, the D. melanogaster expressed sequence tag sequencing project generated no fewer than 18 clones (from a 0- to 24-h-embryo cDNA library) corresponding to the guf1 transcript. High levels of antizyme message in mammalian tissues were also reported (17). No clones corresponding to the previously published “guf cDNA” clone or to guf2 were present in the data bank.
DISCUSSION
Antizyme in Drosophila: utilization of programmed frameshifting.
The discovery of intron 2 (in guf1) demonstrated that the presence of two regions of guf cDNA with homology to mammalian antizyme cDNA is not merely coincidental. Splicing out intron 2 removes the only potential start codon for independent initiation of ORF2. It also removes the blocking in-frame stop codons to leave unencumbered overlapping ORF1 and ORF2. This permits a major expansion to the theory that this mRNA encodes a homolog of mammalian antizyme. Evidence that this sequence causes efficient regulated programmed frameshifting directly comparable to that of its mammalian counterpart is provided by the in vitro translation experiments which yielded a transframe protein of the expected size that was responsive to polyamine concentration. These results, combined with our knowledge of the expression of mammalian antizyme, lead us to conclude that the expression of D. melanogaster antizyme involves programmed, polyamine-regulated, translational frameshifting.
Programmed frameshifting is known to occur in single-celled eukaryotes (yeast and protozoa) and in Xenopus but has not previously been discovered in any intermediate organism. The finding of antizyme programmed frameshifting in Drosophila provides the opportunity to study the evolution of this recoding event, which involves a transitory alteration of the rules of readout of the genetic code. Studies on this evolutionary aspect will be reported elsewhere, but the degree of conservation of shift site sequences is considered below. Notably, however, the 3′ mRNA message feature that is an important stimulator, a pseudoknot, for the recoding signal in mammalian systems is not recognizable in Drosophila, and the identification of its presumed alternative is of major interest.
Of the three known RNA elements that stimulate antizyme frameshifting in mammalian cells, the other two are present in guf1. One of these is the UGA stop codon of ORF1. It is interesting that even though in vitro translation experiments have shown that the other two stop codons (UAG and UAA) are almost as effective in stimulating frameshifting in decoding mammalian antizyme sequences (18), so far all eukaryotic antizyme genes have UGA as the stop codon of ORF1. Another stimulatory element, a sequence immediately 5′ of the UGA stop codon, is also likely to be present in the D. melanogaster antizyme sequence. Of 18 nucleotides 5′ of the UGA stop codon, 16 are identical for guf1 and the rat antizyme sequence (with one of the two mismatches being an antizyme polymorphism) (Fig. 1). This level of 5′ sequence homology is striking and provides a clear indication that this region plays an important role in antizyme frameshifting. The apparent absence of an RNA pseudoknot 3′ of the UGA stop codon of guf1 is puzzling. The nucleotides within the stems of this pseudoknot are absolutely conserved among all known vertebrate antizyme sequences (Fig. 1 and unpublished data). Despite the apparent absence of a 3′ pseudoknot, there is nucleotide sequence homology between this region of guf1 and vertebrate antizyme sequences (Fig. 1). Perhaps some other RNA structure is present in this region of guf1. Indirect evidence supports such a hypothesis. Comparison of D. melanogaster and D. virilis antizyme nucleotide sequences reveals that the 53 nt between the UGA of the putative frameshift site and intron 2 (a region most likely to contain a stimulatory 3′ RNA structure) are completely conserved between the two species, even though the conservation of the rest of the known exonic sequences is only 75%. Computer programs predict several stem loops in this region, but the relevance of any of them to translational frameshifting is unknown. Even though additional frameshift-stimulatory elements in the Drosophila antizyme sequence are not obvious, there is a strong indication that there is more to the guf1 frameshift site than the 28-nt region homologous to the mammalian antizyme frameshift site. Results presented previously (18, 19) indicate that the 28 nt alone cannot stimulate more than 3% frameshifting in vitro. Our data for the in vitro translation of guf1 strongly suggests that the frameshift efficiency in that system is much higher than 3%, thus implying the existence of additional frameshift-stimulatory signals in Drosophila antizyme mRNA.
Rat antizyme is a short-lived protein (8). It contains a PEST sequence associated with proteins that rapidly turn over (27). Xenopus and rat antizymes also contain PEST sequences (9) (Fig. 4B). The predicted Drosophila antizyme protein contains a PEST sequence within a region with no apparent homology to vertebrate antizymes (Fig. 4B). It should be noted that the PEST sequences of rat, Xenopus, and Drosophila antizymes are located in different regions of the protein, and so their significance is not clear. However, the presence of the PEST sequence may indicate that Drosophila antizyme, like rat antizyme, is also a short-lived protein.
snRNP Sm D3 gene of Drosophila: a nested gene.
Our analysis revealed a second gene in the gutfeeling locus. This gene has a transcriptional unit (guf2) that is oriented in a direction opposite to that of the transcriptional unit guf1. This second gene, with two exons, is entirely within intron 1 of guf1 (Fig. 2E). This nested gene organization, where one gene exists within an intron of another gene (on the opposite strand), is unusual; however, several such examples have been described (reference 21 and references therein). This gene encodes a protein that has very high homology to snRNP Sm D3 proteins from other organisms. snRNPs U1, U2, U4/U6, and U5 are essential for pre-mRNA splicing (23, 34). Two classes of snRNP proteins exist. The class Sm includes proteins common to all four snRNPs. The other class includes proteins that are specific to each snRNP particle. snRNP Sm D3 is one of the proteins common to all snRNPs (14). On the basis of the amino acid homology (Fig. 6B), we conclude that the guf2 product is the same as D. melanogaster snRNP Sm D3.
R-G-rich regions.
The C terminus of D. melanogaster snRNP Sm D3 contains an R-G-rich region (Fig. 6). A search revealed a number of proteins in the public data bank that contain R-G-rich regions, including fibrillarin (Schizosaccharomyces pombe), NAB2 (S. cerevisiae), FMR-1 (H. sapiens), GAR1 (S. cerevisiae), EWS (H. sapiens), nucleolin (Xenopus laevis), heterogeneous ribonuclear particle protein A1.b (X. laevis), glycine-rich RNA-binding protein GRP1A (Sinapis alba), GAM1 (S. cerevisiae), basic fibroblast growth factor (Rattus norvegicum), Epstein-Barr virus nuclear antigens 1 and 2 (H. sapiens), and ALY (Mus musculus). The role of the R-G-rich sequences in these proteins is not entirely clear. Many, but not all, of these R-G-rich regions contain the RGG RNA binding box (3, 11). It is thought that RGG box regions bind to RNA through a non-sequence-specific mechanism. Even though snRNP Sm D3 from Drosophila contains two RGG repeats, the human homolog contains none in its R-G-rich region. In fact, not all proteins we have identified that contain R-G-rich regions are thought to bind RNA (some are transcription factors with no known RNA binding motifs).
The only feature common to all these proteins that contain R-G-rich sequences is that they all are located in the nucleus. This raises the possibility that, at least in some proteins, the R-G-rich region could be a nuclear localization sequence (NLS) or be involved in some aspect of nuclear localization (for nucleolin and NLS-binding protein (NSR1), regions other than the R-G-rich one have been implicated as NLSs [32, 37]). The feature that unites all NLSs is their richness in basic amino acids (33), and the R-G-rich region would most likely satisfy this criterion. The presence of an RNA binding motif and NLS in the same region of a protein is not unprecedented. Previously published work (13) has shown that for the majority of nuclear proteins for which an NLS and DNA- or RNA-binding domain have been determined, the two are either overlapping or flanking. Perhaps the best indication that the R-G-rich region might be involved in nuclear localization comes from an analysis of fibrillarin proteins from different species. Fibrillarin is needed for pre-rRNA processing and is located in the nucleus (20). All known fibrillarins from eukaryotic species contain an R-G-rich region in their N termini. A genuine homolog of fibrillarin appears to exist in several Methanococcus (archaeon) species (GenBank accession no. X73987, X73988, and 2127901). As our hypothesis predicts, archaeon fibrillarin proteins (which do not have to be transported across a nuclear membrane) do not have the R-G-rich region, even though they have extensive similarity to the rest of their eukaryotic homologs.
gutfeeling.
Since both P-element insertions responsible for the gutfeeling mutant phenotype map to the 5′ UTR of D. melanogaster snRNP Sm D3 mRNA and therefore would cause a severe gene disruption, we propose that defective expression of snRNP Sm D3 is a likely contributor to the gutfeeling phenotype. However, since both P elements are also in intron 1 of the antizyme gene homolog, they could disrupt its splicing or possibly its transcription and so contribute to the phenotype. Perhaps snRNP Sm D3 and antizyme contribute to different aspects of the gutfeeling phenotype. This could be determined by identifying and analyzing point mutations in each of the genes for the two proteins.
Knockout experiments have demonstrated that the snRNP Sm D3 gene is an essential gene in S. cerevisiae (29). It is reasonable to assume that the same is true for D. melanogaster. How disruption of snRNP Sm D3, a protein which is an integral part of eukaryotic spliceosomes, would result in specific deficiencies of neuron and/or muscle differentiation is not clear. Whatever the mechanism, it is most likely nonspecific. For example, it is possible that mutations disrupting zygotic snRNP Sm D3 function coupled with gradual dilution of the pool of maternally supplied protein would initially and differentially affect the splicing of a subset of mRNAs, one or more of which are required for neuron and/or muscle cell differentiation. The possible role of antizyme in the gutfeeling phenotype has already been discussed at length previously (30).
The discovery that Drosophila antizyme is apparently regulated by translational frameshifting suggests that this regulatory event is much more common than previously thought. It is in fact very likely that translational frameshifting is involved in the regulation of antizyme in all animals expressing this protein.
ACKNOWLEDGMENTS
We thank Norma Wills for kind help with the translation experiments and Senya Matsufuji for discussions.
K.S. is a Developmental Biology Training Grant (5T32HD07491-03) recipient. A.L. has a JFRA-657 award. R.F.G. is an investigator from the Howard Hughes Medical Institute. This work was also supported by a grant (RO1-GM48152) from the National Institutes of Health to J.F.A.
REFERENCES
- 1.Altchul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown N H, Kafatos F C. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 3.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 4.Farabaugh P J. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 5.Fong W F, Heller J S, Canellakis E S. The appearance of an ornithine decarboxylase inhibitory protein upon the addition of putrescine to cell cultures. Biochim Biophys Acta. 1976;23:456–465. doi: 10.1016/0304-4165(76)90054-4. [DOI] [PubMed] [Google Scholar]
- 6.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi S. Multiple mechanisms for the regulation of mammalian ornithine decarboxylase. In: Hayashi S, editor. Ornithine decarboxylase: biology, enzymology, and molecular genetics. New York, N.Y: Pergamon Press; 1989. pp. 35–45. [Google Scholar]
- 8.Heller J S, Fong W F, Canellakis E S. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci USA. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichiba T, Matsufuji S, Miyazaki Y, Hayashi S-I. Nucleotide sequence of ornithine decarboxylase antizyme cDNA from Xenopus laevis. Biochim Biophys Acta. 1995;1262:83–86. doi: 10.1016/0167-4781(95)00062-l. [DOI] [PubMed] [Google Scholar]
- 10.Kania A, Salzberg A, Bhat M, D’Evelyn D, He Y, Kiss I, Bellen H J. P-element mutations affecting embryonic peripheral nervous system development in Drosophila melanogaster. Genetics. 1996;139:1663–1678. doi: 10.1093/genetics/139.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koguchi K, Murakami Y, Hayashi S. Involvement of antizyme-like regulatory protein in polyamine-caused repression of ornithine decarboxylase in insect-derived Trichoplusia ni 5 cells. Biochim Biophys Acta. 1997;1353:291–296. doi: 10.1016/s0167-4889(97)00033-5. [DOI] [PubMed] [Google Scholar]
- 13.LaCasse E C, Lefebvre Y A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmeier T, Foulaki K, Lührmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990;18:6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmeier T, Parker V, Hermann H, Lührmann R. cDNA cloning of the Sm proteins D2 and D3 from human small nuclear ribonucleoproteins: evidence for a direct D1-D2 interaction. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Coffino P. Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol Cell Biol. 1993;13:2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsufuji S, Miyazaki Y, Kanomoto R, Kameji T, Murakami Y, Baby G T, Fujita K, Ohno T, Hayashi S. Analyses of ornithine decarboxylase antizyme mRNA with a cDNA cloned from rat liver. J Biochem. 1990;108:365–371. doi: 10.1093/oxfordjournals.jbchem.a123207. [DOI] [PubMed] [Google Scholar]
- 18.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S-I. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsufuji, S. Personal communication.
- 20.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–935. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 21.McNabb S, Greig S, Davis T. The alcohol dehydrogenase gene is nested in the outspread locus of Drosophila melanogaster. Genetics. 1996;143:897–911. doi: 10.1093/genetics/143.2.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki Y, Matsufuji S, Hayashi S. Cloning and characterization of a rat gene encoding ornithine decarboxylase antizyme. Gene. 1992;113:191–197. doi: 10.1016/0378-1119(92)90395-6. [DOI] [PubMed] [Google Scholar]
- 23.Moore M J, Query C C, Sharp P A. Slicing of precursors to mRNA by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 24.Mount S M, Burks C, Hertz G, Stromo G D, White O, Fields C. Splicing in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteosome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 26.O’Connell P O, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 28.Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc Natl Acad Sci USA. 1994;91:3959–3963. doi: 10.1073/pnas.91.9.3959. . (Author’s correction, 91:9195.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy J, Zheng B, Rymond B C, Woolford J L., Jr Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzberg A, Golden K, Bodmer R, Bellen H J. gutfeeling, a Drosophila gene encoding an antizyme-like protein, is required for late differentiation of neurons and muscles. Genetics. 1996;144:183–196. doi: 10.1093/genetics/144.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schmidt-Zachmann M S, Nigg E A. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- 33.Silver P A. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 34.Steitz J A, Black D L, Gerke V, Parker K A, Krämer A, Frendewey D, Keller W. Functions of the abundant U-snRNPs. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Berlin, Germany: Springer; 1988. pp. 115–154. [Google Scholar]
- 35.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 36.Tewari D S, Qian Y, Thornton R D, Pieringer J, Taub R, Mochan E, Tewari M. Molecular cloning and sequencing of a human cDNA encoding ornithine decarboxylase antizyme. Biochim Biophys Acta. 1994;14:293–295. doi: 10.1016/0167-4838(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 37.Yan C, Mélèse T. Multiple regions of NSR1 are sufficient for accumulation of a fusion protein within the nucleolus. J Cell Biol. 1993;123:1081–1091. doi: 10.1083/jcb.123.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]