A Naturally Occurring hPMS2 Mutation Can Confer a Dominant Negative Mutator Phenotype (original) (raw)
Abstract
Defects in mismatch repair (MMR) genes result in a mutator phenotype by inducing microsatellite instability (MI), a characteristic of hereditary nonpolyposis colorectal cancers (HNPCC) and a subset of sporadic colon tumors. Present models describing the mechanism by which germ line mutations in MMR genes predispose kindreds to HNPCC suggest a “two-hit” inactivation of both alleles of a particular MMR gene. Here we present experimental evidence that a nonsense mutation at codon 134 of the hPMS2 gene is sufficient to reduce MMR and induce MI in cells containing a wild-type hPMS2 allele. These results have significant implications for understanding the relationship between mutagenesis and carcinogenesis and the ability to generate mammalian cells with mutator phenotypes.
Within the past 4 years, the genetic cause of the hereditary nonpolyposis colorectal cancer syndrome (HNPCC), also known as Lynch syndrome II, has been ascertained for the majority of kindreds affected with the disease (10). The molecular basis of HNPCC involves genetic instability resulting from defective mismatch repair (MMR). To date, six human genes that appear to participate in the MMR process, including the mutS homologs GTBP, hMSH2, and hMSH3 and the mutL homologs hMLH1, hPMS1, and hPMS2, have been identified (1a, 5, 8, 14, 17–19, 20). Germ line mutations in four of these genes (hMSH2, hMLH1, hPMS1, and hPMS2) have been identified in HNPCC kindreds (1a, 8, 10, 14, 20). Although the mutator defect that arises from the MMR deficiency can affect any DNA sequence, microsatellite sequences are particularly sensitive to MMR abnormalities (11). Microsatellite instability (MI) is therefore a useful indicator of defective MMR. In addition to its occurrence in virtually all tumors arising in HNPCC patients, MI is found in a small fraction of sporadic tumors with distinctive molecular and phenotypic properties (23).
HNPCC is inherited in an autosomal dominant fashion, so that the normal cells of affected family members contain one mutant allele of the relevant MMR gene (inherited from an affected parent) and one wild-type allele (inherited from the unaffected parent). During the early stages of tumor development, however, the wild-type allele is inactivated through a somatic mutation, leaving the cell with no functional MMR gene and resulting in a profound defect in MMR activity. Because a somatic mutation in addition to a germ line mutation is required to generate defective MMR in the tumor cells, this mechanism is generally referred to as one involving “two hits,” analogous to the biallelic inactivation of tumor suppressor genes that initiate other hereditary cancers (8, 10, 21).
In line with this two-hit mechanism, the nonneoplastic cells of HNPCC patients generally retain near-normal levels of MMR activity due to the presence of the wild-type allele. It was therefore surprising that a profound defect in MMR was found in the normal cells of two HNPCC patients. That this defect was operative in vivo was demonstrated by the widespread presence of MI in nonneoplastic cells of such patients. One of the two patients had a germ line truncating mutation of the hPMS2 gene at codon 134 (the hPMS2-134 mutation), while the other patient had a small germ line deletion within the hMLH1 gene (22). These data thus contradicted the two-hit model generally believed to explain the biochemical and biological features of HNPCC patients. The basis for this MMR deficiency in the normal cells of these patients was unclear, and several potential explanations were offered. For example, it was possible that the second allele of the relevant MMR gene was inactivated in the germ line of these patients through an undiscovered mechanism or that unknown mutations of other genes involved in the MMR process were present that cooperated with the known germ line mutation. It is clear from knockout experiments with mice that MMR deficiency is compatible with normal growth and development, supporting these possibilities (1, 2, 4). Alternatively, it was possible that the mutant alleles exerted a dominant negative effect, resulting in MMR deficiency even in the presence of the wild-type allele of the corresponding MMR gene and all other genes involved in the MMR process. To distinguish between these possibilities, we expressed the truncated polypeptide encoded by the hPMS2-134 mutation in an MMR-proficient cell line and analyzed its effect on the MMR activity of the cell. The results showed that this mutant could indeed exert a dominant negative effect, resulting in biochemical and genetic manifestations of MMR deficiency.
MATERIALS AND METHODS
Plasmids.
The full-length wild-type hPMS2 cDNA was obtained from a human HeLa cDNA library as described previously (15). An hPMS2 cDNA containing a termination codon at amino acid 134 was obtained via reverse transcriptase PCR from the patient in whom the mutation was discovered (6). The cDNA fragments were cloned into the _Bam_HI site into the pSG5 vector, which contains a simian virus 40 promoter followed by a simian virus 40 polyadenylation signal (5a). The pCAR reporter vectors described in Fig. 1 were constructed as described in references ;221 and 25 by using an episomal backbone vector.
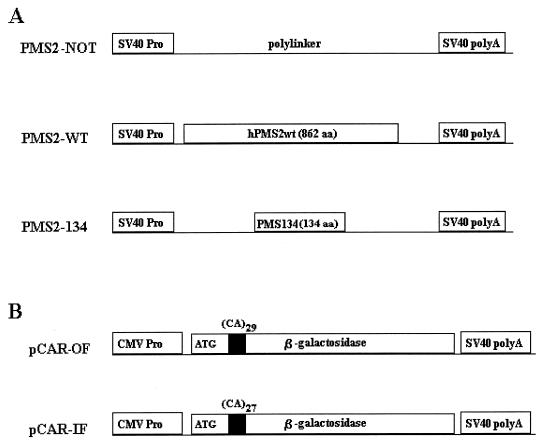
FIG. 1.
Diagrams of PMS2 expression vectors (A) and pCAR reporters (B). SV40, simian virus 40; CMV, cytomegalovirus; aa, amino acids.
Cell lines and transfection.
Syrian hamster Tk-ts13 fibroblasts were obtained from the American Type Culture Collection and cultured as described previously (12). Stably transfected cell lines expressing hPMS2 were created by cotransfection of the PMS2 expression vectors and the pLHL4 plasmid encoding the hygromycin resistance gene at a ratio of 3:1 (pCAR to pLHL4) and selected with hygromycin. Stably transfected cell lines containing pCAR reporters were generated by cotransfection of pCAR vectors together with either pNTK, encoding the neomycin resistance gene, or pLHL4. All transfections were performed with calcium phosphate as previously described (12).
β-Galactosidase assay.
At 17 days after transfection with pCAR, β-galactosidase assays were performed with 20 μg of protein in 45 mM 2-mercaptoethanol–1 mM MgCl2–0.1 M NaH2PO4–0.6 mg of chlorophenol red-β-d-galatopyranoside (CPRG; Boehringer Mannheim) per ml. The reaction mixtures were incubated for 1 h, the reactions were terminated by the addition of 0.5 M Na2CO3, and the products were analyzed by spectrophotometry at 576 nm (13). For in situ β-galactosidase staining, the cells were fixed in 1% glutaraldehyde in phosphate-buffered saline and incubated in 0.15 M NaCl–1 mM MgCl2–3.3 mM K4Fe(CN)6–3.3 mM K3Fe(CN)6–0.2% 4-chloro-3-bromo-2-indolyl-β-d-galactopyranoside (X-Gal) for 2 hours at 37°C.
Western blots.
Equal numbers of cells were lysed directly in lysis buffer (60 mM Tris [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol, 0.1 M 2-mercaptoethanol, 0.001% bromophenol blue) and boiled for 5 min. Lysate proteins were separated by electrophoresis on 4 to 12% Tris-glycine gels (for analysis of full-length hPMS2) or 4 to 20% Tris-glycine gels (for analysis of hPMS2-134). The gels were electroblotted onto Immobilon-P (Millipore) in 48 mM Tris base–40 mM glycine–0.0375% sodium dodecyl sulfate–20% methanol and blocked overnight at 4°C in Tris-buffered saline plus 0.05% Tween 20 and 5% condensed milk. The filters were probed with a polyclonal antibody generated against residues 2 to 20 of hPMS2 (Santa Cruz Biotechnology, Inc.) and a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody, with chemiluminescence (Pierce) for detection.
In vitro translation.
Linear DNA fragments containing hPMS2 and hMLH1 cDNA sequences were prepared by PCR, incorporating sequences for in vitro transcription and translation in the sense primer. A full-length hMLH1 fragment was prepared with the sense primer 5′-ggatcctaatacgactcactatagggagaccaccatgtcgttcgtggcaggg-3′ (codons 1 to 6) and the antisense primer 5′-taagtcttaagtgctaccaac-3′ (located in the 3′ untranslated region, nucleotides [nt] 2411 to 2433) and with a wild-type hMLH1 cDNA clone as the template. A full-length hPMS2 fragment was prepared with the sense primer 5′-ggatcctaatacgactcactatagggagaccaccatggagcgagctgagagc-3′ (codons 1 to 6) and the antisense primer 5′-aggttagtgaagactctgtc-3′ (located in the 3′ untranslated region, nt 2670 to 2690) and with a cloned hPMS2 cDNA as the template. A fragment encoding the amino-terminal 134 amino acids of hPMS2 was prepared with the same sense primer and the antisense primer 5′-agtcgagttccaaccttcg-3′. A fragment containing codons 135 to 862 of hPMS2-135 was generated with the sense primer 5′-ggatcctaatacgactcactatagggagaccaccatgatgtttgatcacaatgg-3′ (codons 135 to 141) and the same antisense primer as that used for the full-length hPMS2 protein. These fragments were used to produce proteins via the coupled transcription-translation system (Promega). The reaction mixtures were supplemented with [35S]methionine or unlabelled methionine, as indicated in the text. The PMS2-135 and hMLH1 proteins could not be simultaneously radiolabelled and immunoprecipitated because their similar molecular weights precluded resolution. Lower-molecular-weight bands are presumed to be degradation products and/or polypeptides translated from alternative internal methionines.
Immunoprecipitation.
Immunoprecipitations were performed on in vitro-translated proteins by mixing the translation reaction mixtures with 1 μg of the MLH1-specific monoclonal antibody (MAB) MLH14 (Oncogene Science, Inc.), a polyclonal antibody generated to codons 2 to 20 of hPMS2 described above, or a polyclonal antibody generated to codons 843 to 862 of hPMS2 (Santa Cruz Biotechnology, Inc.) in 400 μl of EBC buffer (50 mM Tris [pH 7.5], 0.1 M NaCl, 0.5% Nonidet P-40). After incubation for 1 h at 4°C, protein A-Sepharose (Sigma) was added to a final concentration of 10% and the reaction mixtures were incubated at 4°C for 1 h. Proteins bound to protein A were washed five times in EBC and separated by electrophoresis on 4 to 20% Tris-glycine gradient gels, which were then dried and autoradiographed.
Biochemical assays for mismatch repair.
MMR activity in nuclear extracts was performed as described previously, with 24 fmol of substrate (9, 21). Complementation assays were done by adding ∼100 ng of purified MutLα or MutSα components to 100 μg of nuclear extract and adjusting the final KCl concentration to 100 mM (2a, 7, 26). The substrates used in these experiments contain a strand break 181 nt 5′ or 125 nt 3′ to the mismatch. Values represent experiments performed at least in duplicate.
RESULTS AND DISCUSSION
The MMR-proficient Syrian hamster TK-ts13 cell line (hereafter called SH cells) was cotransfected with various hPMS2 expression plasmids plus reporter constructs for assessing MMR activity. The hPMS2 expression plasmids contained the normal hPMS2 gene or the truncated hPMS2 gene identified in the patient described above (PMS2-WT and PMS2-134, respectively [Fig. 1A]). An “empty” vector devoid of hPMS2 sequences (PMS2-NOT [Fig. 1A]) served as an additional control. The reporter construct pCAR-OF (out of frame) contained a hygromycin resistance gene plus a β-galactosidase gene containing a 58-bp out-of-frame poly(C-A) tract at the 5′ end of its coding region. The reporter construct pCAR-IF (in frame) was identical except that the poly(C-A) tract was 54 bp and therefore did not disrupt the β-galactosidase reading frame (Fig. 1B). The pCAR-OF reporter would not generate β-galactosidase activity unless a frame-restoring mutation (i.e., insertion or deletion) arose following transfection.
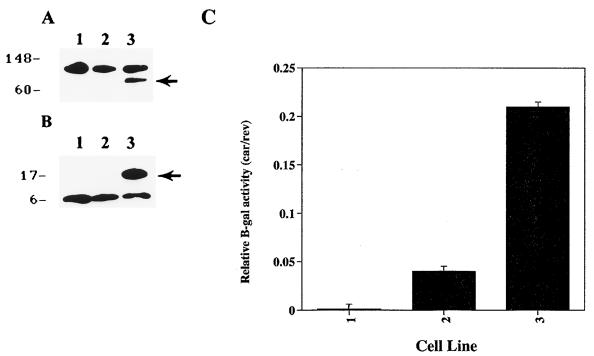
Three different transfection schemes were used to evaluate the effects of the PMS2-134 mutation on SH cells. In the first scheme, the expression vectors and the reporters were cotransfected. Pools containing greater than 100 colones were generated after selection with hygromycin for 17 days and harvested for Western blot and β-galactosidase assays. SH cells transduced with PMS2-WT and PMS2-134 synthesized the expected sizes of polypeptides, as assessed with anti-hPMS2 antibodies on Western blots (Fig. 2A and B). As expected, virtually no β-galactosidase activity was observed in SH cells transfected with the pCAR-OF reporter plus PMS2-NOT (Fig. 2C). However, SH cells transfected with PMS2-134 expressed considerable β-galactosidase activity, significantly more than those transfected with PMS2-WT (Fig. 2C). These results suggested that the truncated polypeptide encoded by the PMS2-134 construct perturbs the endogenous MMR machinery, resulting in deletions or insertions that restored the reading frame. The exact nature of these presumed deletions or insertions could not be assessed, since multiple copies of the reporter constructs were transduced under our conditions, and the wild-type β-galactosidase sequence was in great excess over the expected mutants, precluding their demonstration by direct sequencing.
FIG. 2.
SH cells cotransfected with pCAR reporters and PMS2 expression vectors after 17 days of drug selection. (A) Western blots of lysates from untransfected SH cells (lane 1) or SH cells transfected with PMS2-NOT (lane 2) or PMS2-WT (lane 3). The arrow indicates the 110-kDa protein expected for hPMS2. (B) Western blots of lysates from untransfected SH cells (lane 1) or SH cells transfected with PMS2-NOT (lane 2) or PMS2-134 (lane 3). The arrow indicates the 14-kDa protein expected for hPMS134. Both A and B were probed with an antibody generated against the N terminus of hPMS2. The upper polypeptides in panel A and the lower polypeptides in panel B represent cross-reactive hamster proteins. (C) β-Galactosidase activity in lysates derived from SH cells cotransfected with PMS2-NOT (left bar), PMS2-WT (middle bar), or PMS2-134 (right bar) plus reporter plasmid. Relative β-galactosidase activities are defined as the ratio of β-galactosidase activity in cells transfected with pCAR-OF to that in cells transfected with pCAR-IF; this normalization controlled for transfection efficiency and controlled for β-galactosidase activity in the cells expressing the various PMS2 effector genes. Bars and brackets represent means and standard deviations derived from three independent experiments.
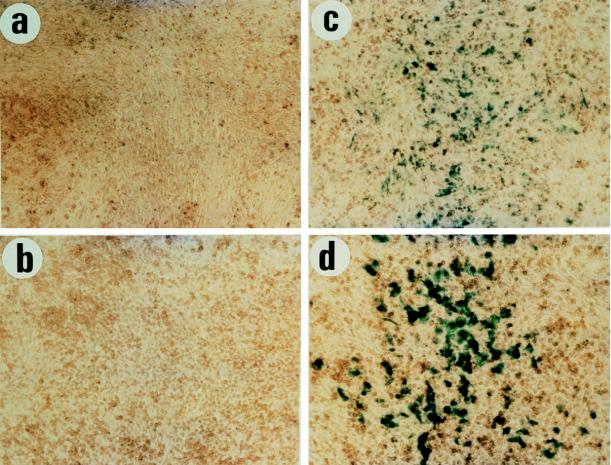
In the second scheme, SH cells were cotransfected with each of the PMS2 expression vectors plus the hygromycin resistance plasmid pLHL4. Hygromycin-resistant cultures containing more than 100 clones were pooled and expanded. These cultures were then cotransfected with pCAR-IF or pCAR-OF reporters plus a separate plasmid allowing geneticin selection. Two weeks later, the pooled cells, each containing more than 100 colonies resistant to both hygromycin and geneticin, were stained with X-Gal to assess β-galactosidase activity. As shown in Fig. 3, the cultures transfected with PMS2-134 (Fig. 3c) contained many blue cells whereas virtually no cells were blue in the cultures transfected with PMS2-NOT or PMS2-WT (Fig. 3a and b, respectively). In each case, the transfection efficiency was controlled by parallel transfections with pCAR-IF, which also served as a control for the β-galactosidase activity of cells expressing the various PMS2 effector genes, which resulted in similar β-galactosidase expression levels in all cases (an example is given in Fig. 3d).
FIG. 3.
In situ β-galactosidase activity of pooled clones of SH cells stably transduced with the PMS2-NOT (a), PMS2-WT (b), or PMS2-134 (c) expression vectors and then retransfected with pCAR-OF reporter. After 17 days of drug selection, the colonies were pooled, cultured, and stained for β-galactosidase activity. A pooled culture of PMS2-134-transduced SH cells expressing β-galactosidase from pCAR-OF is visible in panel c. The level of expression is lower, as expected, than in SH cells transfected with the pCAR-IF reporter plasmid, shown as a positive control in panel d. Each of the fields illustrated is representative of that found in triplicate experiments.
Increases in β-galactosidase activity after PMS2-134 transfection compared to PMS2-WT transfection were also observed when similar experimental protocols were applied to the MMR-proficient human embryonic kidney cell line 293. These cells were cotransfected with the pCAR-OF plus the various PMS2 effector plasmids and selected for 17 days in hygromycin. On day 17, colonies were stained with X-Gal to assess β-galactosidase activity and scored for β-galactosidase-expressing cells. As shown in Table 1, only cells expressing the PMS2-134 polypeptide expressed a detectable β-galactosidase activity. These data demonstrate a similar dominant negative effect of the hPMS2-134 protein in both rodent and human systems and validate the utility of the rodent system in these studies.
TABLE 1.
β-Galactosidase expression of 293 clones transfected with pCAR-OF reporter construct plus PMS effector plasmidsa
| Sample | No. ofb: | |
|---|---|---|
| Blue colonies | White colonies | |
| PMS2-NOT | 0 ± 0 | 17 ± 2.7 |
| PMS2-WT | 0 ± 0 | 18 ± 4.0 |
| PMS2-134 | 15 ± 2.1 | 6 ± 2.1 |
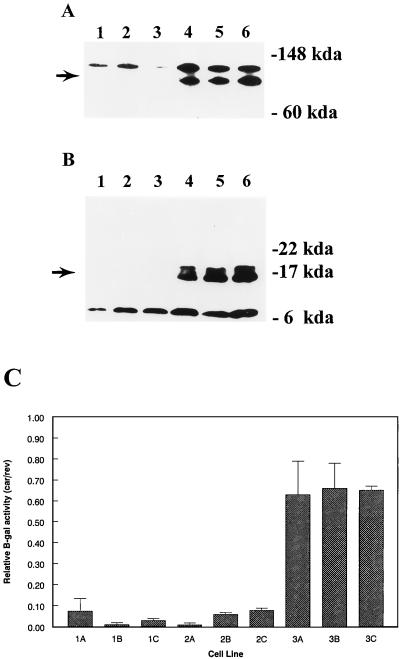
In the third scheme, SH cells were transfected with each of the PMS2 expression vectors as described for the second scheme, but individual clones, rather than pooled clones, were expanded after drug selection. Of 20 clones transfected with PMS2-WT, 5 were shown to express readily detectable levels of full-length PMS2 proteins (examples are given in Fig. 4A, lanes 4 to 6). Similar analyses of 20 PMS2-134 clones revealed four clones which expressed truncated PMS2 polypeptides of the expected size (examples are given in Fig. 4B, lanes 4 to 6). Three clones expressing full-length or truncated PMS2 proteins, as well as three randomly selected clones from PMS2-NOT-transfected cells (Fig. 4A and B, lanes 1 to 3), were chosen for further analysis. The individual clones were tested for β-galactosidase activity following cotransfection with pCAR-OF plus the pNTK plasmid, as described above for the pooled clones. As shown in Fig. 4C, each of the three clones (3A to 3C) expressing the truncated hPMS2 polypeptide yielded much higher β-galactosidase activities after transfection with pCAR-OF than did the clones expressing the full-length hPMS2 protein (2A to 2C) or no hPMS2 protein (1A to 1C).
FIG. 4.
Protein expression and β-galactosidase activity in stably transduced SH clones. (A) Western blots of lysates from clones stably transduced with PMS2-NOT (lanes 1 to 3) or PMS2-WT (lanes 4 to 6). (B) Western blots of lysates from clones stably transduced with PMS2-NOT (lanes 1 to 3) or PMS2-134 (lanes 4 to 6). The arrows indicate the polypeptide of the appropriate molecular weight. The higher- and lower-molecular-weight polypeptides in panels A and B, respectively are nonspecific proteins. (C) The clones expressing PMS2-NOT (1A to 3A), PMS2-WT (1B to 3B), or PMS2-134 (1C to 3C) were transduced with pCAR-OF or pCAR-IF reporter plasmids, and multiple subclones were selected in hygromycin plus geneticin were harvested 17 days later and assayed for β-galactosidase activity. Relative β-galactosidase activities are defined as the ratio of β-galactosidase activity in cells transduced with pCAR-OF compared to that in cells transduced with pCAR-IF. Bars and brackets represent means and standard deviations derived from three independent experiments.
The most likely explanation for the differences in β-galactosidase activity between PMS2-WT- and PMS2-134-transfected cells was that the PMS2-134 protein distrubed MMR activity, resulting in a higher frequency of mutation within the pCAR-OF reporter and reestablishing the open reading frame. To directly test the hypothesis that MMR was altered, we used a biochemical assay for MMR with the individual clones described in Fig. 4. Nuclear extracts were prepared from the clones and incubated with heteroduplex substrates containing either a /CA\ insertion-deletion or a G/T mismatch under conditions described previously (see Materials and Methods). The /CA\ and G/T heteroduplexes were used to test repair from the 3′ and 5′ directions, respectively. There was a dramatic difference between the PMS2-134-expressing clones and the other clones in these assays (Table 2). While all clones repaired substrates from the 3′ direction (/CA\ heteroduplex), cells expressing the PMS2-134 polypeptide had very little 5′ repair activity. A similar directional defect in MMR was evident with pooled clones resulting from PMS2-134 transfection or when the heteroduplex contained a 2- to 4-bp loop, examples of which are shown in Table 2. A small decrease in MMR activity was observed in the 3′ /CA\ PMS2-WT repair assays, perhaps as a result of interference in the biochemical assays by overexpression of the PMS2 protein; no significant activity was caused by PMS2-WT in the in situ β-galactosidase assays (Fig. 3; Table 1), a result more likely to reflect the in vivo condition.
TABLE 2.
MMR activity of nuclear extracts from SH clones or pooled culturesa
| Cell line | Amt of repaired substrate (fmol/15 min) | |||
|---|---|---|---|---|
| 3′ /CA\ | 3′ G/T | 5′ G/T | 3′ /CTG\ | 5′ /CTG\ |
| SH clonesb | ||||
| PMS2-NOT | ||||
| Clone A | 10.2 | 3.5 | ||
| Clone B | 12.7 | 2.9 | ||
| Clone C | 13.5 | 5.5 | ||
| PMS2-WT | ||||
| Clone A | 2.8 | 2.2 | ||
| Clone B | 5.7 | 4.8 | ||
| Clone c | 4.7 | 2.9 | ||
| PMS2-134 | ||||
| Clone A | 2.5 | 0.0 | ||
| Clone B | 2.4 | 0.0 | ||
| Clone C | 5.0 | 0.5 | ||
| Pooled cultures | ||||
| PMS2-NOT | 2.07 ± 0.09 | 2.37 ± 0.37 | 3.45 ± 1.35 | 2.77 ± 1.37 |
| PMS2-WT | 1.65 ± 0.94 | 1.86 ± 0.57 | 1.13 ± 0.23 | 1.23 ± 0.65 |
| PMS2-134 | 0.14 ± 0.2 | 0.0 ± 0.0 | 1.31 ± 0.66 | 0.0 ± 0.0 |
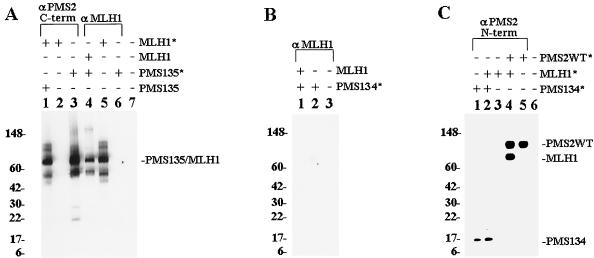
To elucidate the mechanism by which hPMS2-134 affected MMR, we analyzed the interaction between hPMS2 and hMLH1. Previous studies have shown that these two proteins dimerize to form a functionally active complex (9, 24). Proteins were synthesized in vitro with reticulocyte lysates, employing RNA generated from cloned templates (see Materials and Methods). The full-length hMLH1 and hPMS2 proteins bound to each other and were coprecipitated with antibodies to either protein, as expected (data not shown). To determine the domain of hPMS2 which bound to hMLH1, the amino terminus (codons 1 to 134, containing the most highly conserved domain among MutL proteins [16, 20]) and the carboxyl terminus (codons 135 to 862) were separately cloned and proteins were produced in vitro by coupled transcription-translation reactions. When a 35S-labelled, full-length hMLH1 protein (Fig. 5A, lane 5) was mixed with the unlabelled carboxyl-terminal hPMS2 polypeptide, a MAb to the carboxyl terminus of hPMS2 efficiently immunoprecipitated the labeled hMLH1 protein (lane 1). No hMLH1 protein was precipitated in the absence of hPMS2 (lane 2). Conversely, when the 35S-labelled carboxyl terminus of hPMS2 (lane 3) was incubated with unlabelled, full-length hMLH1 protein, an anti-hMLH1 MAb precipitated the hPMS2 polypeptide (lane 4). In the absence of the unlabelled hMLH1 protein, no hPMS2 protein was precipitated by this MAb (lane 6). The same antibody failed to immunoprecipitate the amino terminus of hPMS2 (amino acids 1 to 134) when mixed with unlabelled MLH1 protein (Fig. 5B, lane 1). This finding was corroborated by the converse experiment, in which radiolabelled hPMS134 (Fig. 5C, lane 1) was unable to coprecipitate radiolabelled MLH1 when precipitations were done with an N-terminal hPMS2 antibody (lane 2), while this antibody was shown to be able to coprecipitate MLH1 when mixed with wild-type hPMS2 (lane 4).
FIG. 5.
Immunoprecipitation of in vitro-translated hPMS2 and hMLH1 proteins. (A) Labelled (indicated by an asterisk) or unlabelled proteins were incubated with an antibody to the C terminus of hPMS2 in lanes 1 to 3 and to hMLH1 in lanes 4 to 6. Lane 7 contains a nonprogrammed reticulocyte lysate. PMS2-135 contains codons 135 to 862 of hPMS2. The major translation products of hPMS2 and hMLH1 are indicated. (B) Labelled hPMS2-134 (containing codons 1 to 134 of hPMS2) was incubated in the presence or absence of unlabelled hMLH1 plus an antibody to hMLH1 (lanes 1 and 2, respectively). Lane 3 contains lysate from a nonprogrammed reticulolysate. (C) Labelled proteins were incubated with an antibody to the N terminus of hPMS2. Lane 6 contains a nonprogrammed reticulocyte lysate. In both panels A and B, autoradiographs of immunoprecipitated products are shown.
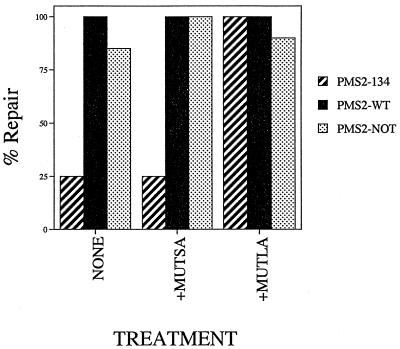
The initial steps of MMR are dependent on two protein complexes, called MutSα and MutLα (11). Since the amino terminus of hPMS2 did not mediate the binding of hPMS2 to hMLH1, it was of interest to determine whether it might instead mediate the interaction between the MutLα complex (composed of hMLH1 and hPMS2 [9]) and the MutSα complex (composed of MSH2 and GTBP ]2a]). Because previous studies have demonstrated that MSH2 and the MutL components do not associate in solution (24), we were unable to assay for direct hPMS2-134–MutSα interaction. We therefore used a different approach to address this issue and attempted to complement nuclear extracts from the various SH cell lines with MutSα or MutLα. If the truncated protein present in the PMS2-134-expressing SH cells was binding to MutSα and lowering its effective concentration in the extract, adding intact MutSα should rescue the MMR defect in such extracts. Addition of purified MutSα to such extracts had no effect (Fig. 6). In contrast, addition of intact MutLα to the extracts completely restored directional repair to the extracts from PMS2-134 cells (Fig. 6).
FIG. 6.
Complementation of MMR activity in transduced SH cells. Lysates from pooled clones stably transduced with PMS2-NOT, PMS2-WT, or PMS2-134 were complemented with purified MutSα or MutLα MMR components by using the 5′G/T heteroduplex substrate. The values are presented as the percentage of repair activity in each case compared to that in lysates complemented with both purified MutLα and MutSα components to normalize for repair efficiency in the different lysate backgrounds. The values shown represent the average of at least three different determinations.
The results described above lead to several conclusions. First, expression of the amino terminus of hPMS2 results in an increase in MI, consistent with a replication error (RER) phenotype. That this elevated MI is due to MMR deficiency was proven by evaluation of extracts from stably transduced cells. Interestingly, the expression of PMS134 resulted in a polar defect in MMR, which was observed only with heteroduplexes designed to test repair from the 5′ direction (no significant defect in repair from the 3′ direction was observed in the same extracts). Interestingly, cells deficient in hMLH1 also have a polar defect in MMR, but in this case the defect preferentially affects repair from the 3′ direction (3). It is known from previous studies with both prokaryotes and eukaryotes that the separate enzymatic components mediate repair from the two different directions. Our results, in combination with those of Drummond et al. (3), strongly suggest a model in which 5′ repair is dependent primarily on hPMS2 while 3′ repair is dependent primarily on hMLH1. It is easy to envision how the dimeric complex between PMS2 and MLH1 might set up this directionality. The combined results also demonstrate that a defect in directional MMR is sufficient to produce a RER+ phenotype.
We anticipated that the dominant negative function of the PMS2-134 polypeptide resulted from its binding to MLH1 and consequent inhibition of MutLα function. This hypothesis was based in part on the fact that the most highly conserved domain of the PMS2 gene is located in its amino terminus and that the only known biochemical partner for PMS2 is MLH1. Our binding studies revealed, however, that the carboxyl terminus of PMS2, rather than the highly conserved amino terminus, actually mediated binding to MLH1. This result is consistent with those recently obtained with Saccharomyces cerevisciae, in which the MLH1-interacting domain of PMS1 (the yeast homolog of human PMS2) was localized to its carboxyl terminus (19a). Our add-back experiments additionally showed that the hPMS2-134 mutant was not likely to mediate an interaction with the MutSα complex (Fig. 6). The best explanation at present to explain the various observations made here is that the hPMS134 polypeptide does not inhibit the initial steps in MMR but, rather, interacts with and inhibits a downstream component of the pathway, perhaps a nuclease required for repair from the 5′ direction.
The demonstration that the hPMS134 mutation can confer a dominant negative MMR defect to transfected cells helps to explain the phenotype of the kindred in which this mutant was discovered. Three individuals from this kindred, a father and his two children, were found to carry the mutation. Both children exhibited MI in their normal tissues, and both developed tumors at an early age, while the father had no evidence of MI in his normal cells and was completely healthy at age 35. The only difference known to us with respect to the MMR genes in this family is that the father’s mutant allele was expressed at lower levels than the wild-type allele as assessed by sequencing of reverse transcriptase-PCR products of RNA from lymphocytes. The children expressed both alleles at approximately equal levels (reference 22 and unpublished observations). We suspect that the dominant negative attribute of the hPMS2-134 mutant will only be manifest when it is present at sufficient concentrations (at least equimolar), thus explaining the absence of MMR deficiency in the father. The reason for the differential expression of the hPMS2-134 allele in this kindred is not clear, although imprinting is a possibility. Hopefully, the ascertainment of additional, larger kindreds with such mutations will facilitate the investigation of this issue.
Finally, the ability to inactivate endogenous MMR of cells through the introduction of the hPMS2-134 protein may have some practical value. In particular, it suggests a way to make other eucaryotic cells MMR deficient. Notable in this regard is that the human hPMS2-134 mutant affected MMR activity in hamster cells. Although MMR deficiency can be generated by knockouts of MMR genes in mice, gene deletion strategies to create MMR deficiency are impractical in the germ line of other animals and in most somatic cells. Transgenosis with an hPMS2-134 mutant gene could prove useful in such circumstances and might facilitate the production of highly diverse agricultural and livestock products.
ACKNOWLEDGMENTS
We thank Luigi Grasso for his technical assistance.
This work was supported by National Cancer Institute grants CA35494, CA62924 (to B.V.), and CA71544 (to S.J.L.) and by NIGMS grant GM45190 (to P.M.). B.V. and P.M. are Investigators of the Howard Hughes Medical Institute.
ADDENDUM IN PROOF
Another hPMS2 mutation giving rise to an apparent dominant form of microsatellite instability has recently been described (Oncogene **15:**2877–2881, 1997).
REFERENCES
- 1.Baker S M, Bronner C E, Zhang L, Plug A W, Robatez M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, Bradley N, Flavell R A, Liskay R M. Male defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 1a.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag R J, Godwin A R, Ward D C, Nordenskjold M, Fishel R, Kolodner R, Liskay R M. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 2.de Wind N, Dekker M, Berns A, Radman M, Riele H T. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–300. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 2a.Drummond J T, Li G M, Longley M J, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 3.Drummond J T, Anthoney A, Brown R, Modrich P. Cisplatin and adriamycin resistance are associated with MutLα and mismatch repair deficiency in an ovarian tumor cell line. J Biol Chem. 1996;271:9645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 4.Edelmann W, Cohen P E, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chagnatti R, Pollard J W, Kolodner R D, Kucherlapati R. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 5.Fishel R, Lescoe M, Rao M R S, Copeland N J, Jenkins N A, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;7:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5a.Green S, Issemann I, Sheer E. A versatile in vivo eucaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton S R, Liu B, Parsons R E, Papadopoulos N, Jen J, Powell S M, Krush A J, Berk T, Cohen Z, Tetu B, Kinzler K W, Vogelstein B. The molecular basis of Turcot’s syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 7.Holmes J, Clark S, Modrich P. Strand specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, Guan X Y, Zhang J, Meltzer P S, Yu J W, Kao F T, Chen D J, Cerosaletti K M, Fournier R E K, Todd S, Lewis T, Leach R J, Naylor S L, Weissenbach J, Mecklin J P, Jarvlnen J A, Petersen G M, Hamilton S R, Green J, Jass J, Watson P, Lynch H T, Trent J M, de la Chapelle A, Kinzler K W, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 9.Li G-M, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human mutL homologs. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Parsons R, Papadopoulos N, Nicolaides N C, Lynch H T, Watson P, Jass J R, Dunlop M, Wyllie A, Peltomaki P, de la Chapelle A, Hamilton S R, Vogelstein B, Kinzler K W. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 11.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 12.Nicolaides N C, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via MYB binding in the 5′ flanking region of the human c-myb gene. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolaides N C, Correa I, Casadevall C, Travali S, Soprano K J, Calabretta B. The Jun family members, c-JUN and JUND, transactivate the human c-myb promoter via an Ap1 like element. J Biol Chem. 1992;267:19665–19672. [PubMed] [Google Scholar]
- 14.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter C J, Dunlop M G, Hamilton S R, Petersen G M, de la Chapelle A, Vogelstein B, Kinzler K W. Mutations of two PMS homologs in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaides N C, Kinzler K W, Vogelstein B. Analysis of the 5′ region of PMS2 reveals heterogenous transcripts and a novel overlapping gene. Genomics. 1995;29:329–334. doi: 10.1006/geno.1995.9997. [DOI] [PubMed] [Google Scholar]
- 16.Nicolaides N C, Carter K C, Shell B K, Papadopoulos N, Vogelstein B, Kinzler K W. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 17.Nicolaides N C, Palombo F, Kinzler K W, Vogelstein B, Jiricny J. Molecular cloning of the N-terminus of GTBP. Genomics. 1996;31:395–397. doi: 10.1006/geno.1996.0067. [DOI] [PubMed] [Google Scholar]
- 18.Palombo F, Hughes M, Jiricny J, Truong O, Hsuan J. Mismatch repair and cancer. Nature. 1994;367:417. doi: 10.1038/367417a0. [DOI] [PubMed] [Google Scholar]
- 19.Palombo F, Gallinari P, Iaccarino I, Lettleri T, Hughes M A, Truong O, Hsuan J J, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 19a.Pang Q, Prolla T A, Liskay R M. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papadopoulos N, Nicolaides N C, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter C J, Dunlop M G, Hamilton S R, Petersen G M, de la Chapelle A, Vogelstein B, Kinzler K W. Mutation of a mutL homolog is associated with hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 21.Parsons R, Li G M, Longley M J, Fang W H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 22.Parsons R, Li G M, Longley M, Modrich P, Liu B, Berk T, Hamilton S R, Kinzler K W, Vogelstein B. Mismatch repair deficiency in phenotypically normal human cells. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 23.Perucho M. Cancer of the microsattelite mutator phenotype. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 24.Prolla T A, Pang Q, Alani E, Kolodner R A, Liskay R M. MLH1, PMS1, and MSH2 interaction during the initiation of DNA mismatch repair in yeast. Science. 1994;264:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 25.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 26.Su S S, Lahue R S, Au K G, Modrich P. Mispair specificity of methyl directed DNA mismatch corrections in vitro. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]