Identification and Characterization of a Constitutively Active STAT5 Mutant That Promotes Cell Proliferation (original) (raw)
Abstract
STAT (signal transducers and activators of transcription) proteins are transcription factors which are activated by phosphorylation on tyrosine residues upon stimulation by cytokines. Seven members of the STAT family are known, including the closely related STAT5A and STAT5B, which are activated by various cytokines. Except for prolactin-dependent β-casein production in mammary gland cells, the biological consequences of STAT5 activation in various systems are not clear. We applied PCR-driven random mutagenesis and a retrovirus-mediated expression screening system to identify constitutively active forms of STAT5. By this strategy, we have identified a constitutively active STAT5 mutant which has two amino acid substitutions; one is located upstream of the putative DNA binding domain (H299R), and the other is located in the transactivation domain (S711F). The mutant STAT5 was constitutively phosphorylated on tyrosine residues, localized in the nucleus, and was transcriptionally active. Expression of the mutant STAT5 partially dispenses with interleukin 3 (IL-3) as a growth stimulant of IL-3-dependent cell lines. Further analyses of the mutant STAT5 have demonstrated that both of the mutations are required for nuclear localization, efficient transcriptional activation, and induction of IL-3-independent growth of an IL-3-dependent cell line, Ba/F3, and have indicated that a molecular basis for the constitutive activation is the stability of the phosphorylated form of the mutant STAT5.
Stimulation of cytokine receptors leads to activation of multiple signal transduction pathways, including the Ras-Raf-MEK-mitogen-activated protein kinase (MAPK) and the JAK-STAT pathways (14, 28, 34, 42, 44). The latter signaling pathway was originally found downstream of the interferon receptors and is now recognized as a common pathway downstream of most cytokine receptors. Upon stimulation with cytokines, receptor-associated JAKs are activated and phosphorylate STAT factors on tyrosine residues. The phosphorylated STAT molecules then form homo- or heterodimers through SH2-mediated interactions and translocate into nuclei to activate transcription of various target genes. Seven members of the STAT family (STAT1 through 4, -5A, -5B, and -6) are known; STAT5A and STAT5B are closely related. With the exception of STAT4 and STAT6, which were shown to be specifically activated by only one or two cytokines, interleukin 12 (IL-12) or both IL-4 and IL-13, respectively (13, 15), most of the other STATs are activated by multiple cytokines. In particular, both STAT5A and STAT5B are activated by numerous cytokines, including prolactin, IL-2, IL-3, IL-5, IL-7, granulocyte-macrophage colony-stimulating factor (GM-CSF), G-CSF, M-CSF, erythropoietin (Epo), thrombopoietin, and growth hormone (GH).
Using the receptor for the human GM-CSF as a model system, members of our group previously showed that activation of the Ras-Raf-MEK-MAPK pathway inhibits apoptosis while the region of the GM-CSF receptor, which is responsible for activation of JAK2 and STAT5 and induction of c-myc expression, plays a role in DNA synthesis (18, 41). Members of our group also demonstrated that a dominant negative STAT5 protein partially inhibited IL-3-induced cell proliferation in Ba/F3 cells, suggesting that STAT5A is involved in cell proliferation (29). A similar conclusion was reached by others who observed that IL-2 and Epo receptors defective in STAT5 activation (5, 7, 9) were also deficient in supporting a proliferative signal. In contrast, others have concluded that STAT5 is not involved in cell proliferation since mutant IL-2 (8) and Epo receptors (37) which cannot activate STAT5 are still capable of transmitting an attenuated proliferative signal. These differing interpretations of generally similar data indicate a point of controversy. Recently, mice nullizygous for STAT5A and STAT5B have been generated (24, 45). Since these mice exhibit apparently normal hematopoiesis, the contribution of STAT5 activation to proliferation or to any other biological function, such as differentiation in normal hematopoietic tissues, remains unclear.
In this paper, we report the identification and characterization of a constitutively active form of STAT5 by PCR-driven mutagenesis followed by retrovirus-mediated expression screening. The mutant STAT5 protein is constitutively phosphorylated on its tyrosine residues, is localized in the nucleus, and is transcriptionally active in the absence of growth factor stimulation. Expression of the mutant STAT5 in IL-3-dependent cells is capable of inducing cytokine-independent growth.
MATERIALS AND METHODS
Cell lines.
Mouse IL-3 (mIL-3)-dependent Ba/F3 cells were maintained in RPMI 1640 medium containing 10% fetal calf serum (FCS) and 1 ng of recombinant mIL-3 produced in silk worm/ml (26). An ecotropic retrovirus packaging cell line, BOSC23 (35), was maintained in Dulbecco’s modified Eagle’s medium containing 10% FCS and guanine phosphoribosyltransferase (GPT) selection reagents (Specialty Media). The cells were transferred into Dulbecco’s modified Eagle’s medium–10% FCS without GPT selection reagents 2 days before transfection.
Retrovirus vectors.
A retrovirus vector, pMX, was constructed as previously described (31). Its derivative pMX-puro vector harbors a simian virus 40 early promoter-driven puromycin resistance gene between the multicloning site and the 3′ long terminal repeat of the pMX vector (16). cDNAs for mouse STAT5A and STAT5B (28) were inserted into the _Eco_RI and _Not_I sites of the pMX and the pMX-puro vectors.
Production of retroviruses carrying STAT5A sequences with random mutations and infection with these viruses.
To introduce random mutations into the STAT5A sequence, a PCR was run for 35 cycles (1 min at 94°C, 2 min at 58°C, and 3 min at 72°C) under standard conditions described previously (31) except that the deoxynucleoside triphosphate concentration was 400 μM. The pMX-STAT5A plasmid was used as a template, and a 5′ vector primer, pMX5′ (1) (5′CCCGGGGGTGGACCATCCTCT3′), and a 3′ vector primer, pMX3′ (1) (5′CCCTTTTTCTGGAGACTAAAT3′), were used to amplify the full-length sequence of STAT5A. The average frequency of point mutations ranged from 1/600 to 1/1,200 under these conditions (data not shown). The PCR product was digested with restriction endonucleases _Eco_RI and _Not_I, and the excised 2.4-kbp STAT5A sequences harboring the PCR-generated mutations were ligated to the _Eco_RI and _Not_I sites of the pMX vector. The mixture of ligated plasmids was amplified in Escherichia coli DH5α, and plasmid DNA was prepared with a Maxi prep kit (Qiagen) to be used as a library of mutated STAT5A sequences in the pMX vector. High-titer retroviruses containing the randomly mutated STAT5A sequences were produced from the library DNA with a transient retrovirus packaging cell line, BOSC23 (35), as described previously (19). For infection, Ba/F3 cells (2 × 106) were incubated with 5 ml of the retroviruses harboring the mutated STAT5A sequence library in the presence of 10 μg of Polybrene/ml and 10 ng of mIL-3/ml. After 8 h, 5 ml of fresh growth medium was added to the culture, and the incubation was extended for 16 h. Twenty-four hours after infection, cells were washed, refed with growth medium, and allowed to grow for one more day before being subjected to selection in the absence of mIL-3 in 96-well plates. Under these conditions, the efficiency of infection of Ba/F3 cells was 20 to 40% as assessed by parallel infections with the test construct pMX-hIL-3Rα (31).
Recovery and reintroduction of the integrated retroviruses.
In order to recover retrovirally transduced STAT5A sequences, genomic DNA was isolated from each factor- independent (FI) clone, and the integrated STAT5 sequences were amplified from 10 ng of genomic DNAs by PCR with the retroviral vector primers described above. The PCR was run for 35 cycles as described above with Taq polymerase (Perkin-Elmer Cetus) under standard conditions with 100 μM deoxynucleoside triphosphates. The recovered PCR fragments were purified from the gel with QiaexII (Qiagen), digested with _Eco_RI and _Not_I, and inserted into the _Eco_RI and _Not_I sites of the pMX vector. The STAT5A sequence subcloned into the pMX vector was analyzed with Taq DyeDeoxy Terminator (Applied Biosystems) on an Applied Biosystems model 373A sequencer. The STAT5A sequences recovered from FI Ba/F3 cells were subcloned into pMX and reintroduced into parental Ba/F3 cells via retrovirus infection to confirm the ability of the integrated STAT5A sequences to induce FI in Ba/F3 cells.
Cell proliferation assay.
A cell proliferation assay was performed with cells (either bulk population or subclones) which had been stably transduced with the wild-type or the mutant STAT5 sequences and then selected by puromycin resistance, except for one experiment whose results are shown in Fig. 2. In the experiment whose results are shown in Fig. 2, we examined FI proliferation of the Ba/F3 cells in the bulk population after the transduction of STAT5 constructs via retrovirus infection (infection efficiency was 30 to 50%) without drug selection. In this particular experiment, after transducing with the wild-type and the mutant STAT5A and STAT5B sequences by using retrovirus constructs, we seeded the transduced Ba/F3 cells in the absence of mIL-3 and the growth curves were determined. Cell proliferation was quantitated either by counting the cells (see Fig. 1B and D, Fig. 2, and Fig. 6B) or by using the Alamar blue assay (see Fig. 6A) as described previously (1). For the Alamar blue assay, 2 × 104 Ba/F3 cells were suspended in 100 μl of medium and were cultured in 96-well microtiter plates for 48 h at 37°C in the presence or absence of mIL-3. Ten microliters of Alamar blue solution (Alamar Biosciences Inc.) was then added to each well, and the incubation was extended for another 24 h. The optical density was measured with an enzyme-linked immunosorbent assay microplate reader (Bio-Rad) with a test wavelength of 570 nm and a reference wavelength of 600 nm.
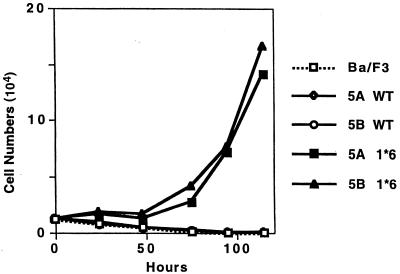
FIG. 2.
A mutant STAT5B harboring mutations homologous to those in STAT5A1*6 induces IL-3-independent growth of Ba/F3 cells. Proliferation after transduction of the sequences for the wild-type STAT5A (5A WT) or STAT5B (5B WT) or mutated STAT5A (5A 1*6) STAT5B (5B 1*6) was examined with bulk populations of Ba/F3 cells after retrovirus-mediated transduction with these constructs. The infection efficiency in this experiment was 30 to 50%. Proliferation of parental Ba/F3 cells without IL-3 is also shown.
FIG. 1.
Isolation and characterization of a constitutively active STAT5A. (A) Positions of point mutations of the mutant STAT5A. A constitutively active form of STAT5A1*6 was constructed from subclones 5-1, 5-2, and 5-3, which had been derived from FI Ba/F3 clone 5. Among these subclones, only 5-1 induced IL-3-independent growth of Ba/F3 cells. (B) Proliferation of Ba/F3 cells expressing subclone 5-1 or STAT5A1*6 in the absence of IL-3. BaF/5-1 and BaF/5A1*6 transfectants, which had been selected in the presence of puromycin and mIL-3, were cultured in the absence of mIL-3, and the cells were counted at the indicated times. The response of parental Ba/F3 cells in the presence or absence of mIL-3 is also shown. The results shown are the averages ± the standard deviations of triplicate cells. (C) Phosphorylation of STAT5A in parental Ba/F3 cells and Ba/F3 cells expressing subclone 5-1 with or without IL-3 stimulation. Stimulation with FCS had no effect on phosphorylation of STAT5A. STAT5A was immunoprecipitated by the anti-STAT5A antibody (R & D Systems) and blotted with the antiphosphotyrosine (P-Tyr) (Upstate Biotechnology Inc.) or with the anti-STAT5A antibody. (D) Long-term IL-3-independent proliferation of BaF/5A1*6 cells. BaF/5A1*6 cells were cultured in the absence of mIL-3 for the indicated periods and were counted. To exclude the possibility of autocrine growth, the growth curve of the parental Ba/F3 cells in the supernatant (BaF3 + 1*6 sup) of BaF/5A1*6 was also determined. The results shown are the averages ± the standard deviations of triplicate wells.
FIG. 6.
Cooperation of Raf and the STAT5 mutant in supporting IL-3-independent proliferation of Ba/F3 cells. (A) IL-3-independent proliferation of BR4 clones expressing the wild-type STAT5A (W1 and W2) and STAT5A1*6 (M1 through M6). BR4 clones which express both ΔRaf-ER and either the wild-type STAT5A or the mutant STAT5A1*6 were cultured in 96-well plates (103 cells/well) in RPMI 1640 medium supplemented with 0.5% BSA in the presence or absence of increasing concentrations of ICI for 3 days and subjected to an Alamar blue assay. Growth of parental BR4 cells (BR4) and the original Ba/F3 cells (P) are shown as controls. The results shown are the averages ± the standard deviations of triplicate wells (upper panel). O.D., optical density. To examine the expression level of STAT5A and ΔRaf-ER, total lysates (10 μg) from each clone were blotted with anti-STAT5A antibody and anti-ER antibody, respectively (lower panel). (B) Proliferation of BR4 cells expressing the wild-type (BR4/W) or the mutant STAT5A1*6 (BR4/M) in the absence of serum. Cells were cultured in 24-well plates (5 × 104/well) in RPMI 1640 medium containing 0.5% BSA, with (+) or without (−) 125 nM ICI, and were counted at the indicated times. The results shown are the averages ± the standard deviations of triplicate wells.
Immunoprecipitation and Western blotting.
Immunoprecipitation, gel electrophoresis, and immunoblotting were performed according to methods described previously (46), with minor modifications. Exponentially growing cells were washed free of serum and growth factors and incubated in RPMI 1640 supplemented with 0.5% bovine serum albumin (BSA) for 12 h at 37°C. After the depletion period, cells were resuspended in the same medium at a concentration of 4 × 106/ml and were stimulated with mIL-3 or FCS at 37°C for 10 min. Cells were lysed in lysis buffer (4 × 106 cells/ml) and incubated on ice for 30 min. Cell lysates were clarified by centrifugation for 15 min at 20,000 × g before incubation with the anti-STAT5A polyclonal antibody (R & D Systems) or the anti-Flag antibody (Eastman Kodak) and protein A-Sepharose at 4°C overnight. The immunoprecipitates were washed three times with lysis buffer, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and electrophoretically transferred onto Immobilon filters (Millipore). After blocking in solution containing 3.0% BSA, the filters were probed with anti-phosphotyrosine antibody 4G10 (Upstate Biotechnology Inc.), stripped, and reprobed with the anti-STAT5 antibody to verify the loading amount. The filter-bound antibody was detected by the enhanced chemiluminescence system (Amersham).
Luciferase reporter assay.
Ba/F3 cells were transiently transfected by electroporation at 960 μF and 300 V with 10 μg of a reporter plasmid carrying a luciferase gene under the control of the β-casein promoter, 3 μg of a β-galactosidase reporter plasmid with the Rous sarcoma virus long terminal repeat promoter, and 10 μg of test DNAs at room temperature in RPMI 1640 supplemented with 10 μg of DEAE-dextran/ml. After a 12-h recovery period in the IL-3-containing medium, cells were incubated in RPMI 1640 supplemented with 0.5% BSA for 12 h, or stimulated with 10 ng of mIL-3/ml for the last 6 h before cell lysates were prepared. Cell lysates were then subjected to luciferase (Promega) and β-galactosidase (Tropix) assays. Transduction efficiency under each condition was normalized with β-galactosidase activity.
EMSA.
Nuclear extracts, prepared by the Nonidet P-40 method, were mixed with radiolabeled double-stranded oligonucleotides corresponding to the prolactin-responsive element (PRE) in the bovine β-casein promoter as described previously (46). Electrophoretic mobility shift analysis (EMSA) was performed with 8 μg of nuclear extracts in 20 μl of a reaction mixture containing 12 mM HEPES (pH 7.9), 5% glycerol, 75 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40, 1 mg of BSA/ml, 1 mg of poly(dI-dC)/ml, and 10 fmol of radioactive PRE. The mixture was incubated at room temperature for 30 min, and 5 μl was placed onto a polyacrylamide gel (0.25 × Tris-borate-EDTA). For supershift experiments, 1 μg of the anti-STAT5 polyclonal antibody (R & D Systems) was added to the reaction mixture.
Construction and expression of STAT5A-EGFP fusion molecules in NIH 3T3 cells.
A mutant green fluorescent protein (GFP), EGFP, was fused to the C termini of the wild-type STAT5A, STAT5A1, STAT5A6, and STAT5A1*6 with pEGFP-N1 (Clontech), and the _Eco_RI-_Not_I fragment encoding the STAT5-GFP fusion molecule was inserted into the pMX vector. The resulting constructs (pMX-STAT5A-EGFP, pMX-STAT5A1-EGFP, pMX-STAT5A6-EGFP, and pMX-STAT5A1*6-EGFP) were expressed in NIH 3T3 cells via retrovirus infection as described previously (19). Two days after infection, the NIH 3T3 cells were observed under a fluorescence microscope (IX 70; Olympus).
Stability of phosphorylated wild-type and mutant STAT5 proteins in Ba/F3 transfectants expressing Flag-tagged wild-type and mutant STAT5A.
To distinguish the endogenous and the transduced STAT5 molecules, the Flag tag was added to the C termini of the wild-type and the mutant STAT5s. The sequences for the Flag-tagged STAT5s were inserted into the _Eco_RI and _Not_I sites of the pMX-neo vector with a construct by Wang et al. (47), and Ba/F3 stable transfectants expressing the Flag-tagged STAT5s were established after gene transduction via retrovirus infection followed by selection in the presence of 1 mg of G418 (Gibco-BRL)/ml. The Ba/F3 transfectants expressing the STAT51*6-Flag were cultivated in the absence of IL-3 after the establishment of transfectants because the expression of the STAT51*6-Flag decreased over time in the media containing IL-3, probably due to a growth-inhibitory effect of the STAT51*6-Flag when stimulated with IL-3 (30). The stable transfectants were deprived of mIL-3 for 12 h in the presence of 10% FCS, stimulated with 10 ng of IL-3/ml for 30 min, washed with medium, and cultured for various time periods. The cell lysates were prepared before IL-3 stimulation and 0, 0.5, 1, 2, 4, and 8 h after the stimulation. The cell lysates were subjected to immunoprecipitation and Western blotting to examine the stability of the phosphorylated STAT5 molecules.
Establishment of Ba/F3 transfectants expressing both an inducible form of Raf, ΔRaf-ER, and either the wild-type or the mutant STAT5A.
We first attempted to establish Ba/F3 transfectants expressing an inducible form of Raf, ΔRaf-ER (40). This protein consists of the catalytic domain of human c-Raf-1 (ΔRaf) and the hormone-binding domain of the estrogen receptor (ER). Addition of β-estradiol or its analog ICI 182 780 (ICI) (33) to cells expressing ΔRaf-ER leads to immediate activation of the Raf-MAPK cascade. We introduced ΔRaf-ER with a retrovirus construct harboring the G418 resistance gene and established several clones in the presence of 1 mg of G418 (Gibco-BRL)/ml. We chose two clones (BR4 and BR6) in which the expression of ΔRaf-ER and the activation of MAPK by ICI were confirmed by Western blot analysis with anti-Raf antibodies (Signal Transduction Lab) and the MAPK assay with the myelin basic protein as a substrate (data not shown). We then introduced pMX-puro-STAT5A or pMX-puro-STAT5A1*6 into BR4 cells via retrovirus infection and isolated several clones as well as bulk cells expressing the transduced STAT5A sequences in the presence of 1 μg of puromycin/ml.
RESULTS
Identification of mutant STAT5s which induce FI growth of Ba/F3 cells.
We attempted to identify constitutively active forms of STAT5A by PCR-mediated mutagenesis followed by a high-efficiency retrovirus expression screening system (19, 32). Briefly, random mutations were introduced into the STAT5A sequence by PCR, and the mutated STAT5A sequences were expressed in an mIL-3-dependent cell line (Ba/F3) with retrovirus vector pMX (31) and a transient retrovirus packaging cell line, BOSC23 (35). Ba/F3 cells infected with these viruses were then screened for IL-3-independent growth, and 13 IL-3-independent clones were isolated. The integrated STAT5A-coding sequences in FI clones were recovered by PCR with vector primers. To confirm the ability of the mutant forms of STAT5A to induce IL-3-independent proliferation of Ba/F3 cells, the PCR fragments were subcloned into pMX, reintroduced into Ba/F3 cells, and analyzed for their ability to confer FI growth. The recovered STAT5A sequences from six FI clones but not the other seven FI clones efficiently reproduced the FI phenotype in Ba/F3 cells. Clone 5 showed the best phenotype and was chosen for further experiments. Several point mutations were found in the STAT5A sequence recovered from Ba/F3 FI clone 5; three independent PCR-recovered DNA subclones designated 5-1, 5-2, and 5-3 are shown in Fig. 1A. Of these subclones, only 5-1 induced FI when reintroduced into naive Ba/F3 cells (Fig. 1B and data not shown). The other two clones (5-2 and 5-3) had several point mutations in common with subclone 5-1 (i.e., point mutations 1 and 6); however, 5-2 and 5-3 also harbored additional point mutations, including a nonsense mutation, which were probably generated during the recovery of the integrated STAT5A sequence by PCR (Fig. 1A). Common point mutation 1 was located in the STAT transactivation domain and caused an amino acid change from serine to phenylalanine (S711F). Point mutation 6, on the other hand, was located upstream of the putative DNA binding domain and resulted in an amino acid change from histidine to arginine (H299R). In the Ba/F3 stable transfectant expressing the mutant STAT5A (5-1), the mutant STAT5A was constitutively phosphorylated at tyrosine residues (Fig. 1C), but constitutive activation of JAK2 and MAPK were not observed (data not shown).
Two mutations were required for constitutive activation of STAT5A.
To clarify which of the point mutations observed in subclone 5-1 was responsible for IL-3-independent growth of Ba/F3 cells, we constructed retroviruses encoding STAT5A with each single point mutation alone and assessed their ability to promote IL-3-independent proliferation of Ba/F3 cells. The sequences for these STAT5A mutants were subcloned into the pMX-puro retrovirus vector, and Ba/F3 cells transduced with these constructs (infection efficiency was 30 to 50%) were selected with 1 μg of puromycin/ml for a week. The STAT5A mutants harboring either mutation 1 (STAT5A1) or 6 (STAT5A6) alone could not support long-term growth of Ba/F3 cells (data not shown). Since the original subclone 5-1 gave rise to the FI phenotype, we reconstructed the double mutant STAT5A1*6 to examine the effect of combining the two point mutations. Interestingly, expression of the double mutant STAT5A1*6 in Ba/F3 cells induced IL-3-independent proliferation in Ba/F3 stable transfectants (BaF/5A1*6) (Fig. 1B) and was able to support long-term growth of the transfectants in the absence of IL-3 (Fig. 1D). In Fig. 1B, continuous proliferation of the BaF/5A1*6 transfectant, which is similar to that of the Ba/F3 transfectant expressing the original subclone 5-1 (BaF/5-1), is shown. Expression of STAT5A1*6 also induced FI growth of other IL-3-dependent cell lines, such as TF-1 (20) and OTT-1 (12) (data not shown). In contrast, introduction of the wild-type STAT5A never gave rise to FI growth (Fig. 2). To exclude the possibility that the growth of BaF/5A1*6 cells was mediated through an autocrine mechanism, we examined whether growth-promoting activity was present in the supernatant of BaF/5A1*6 cells. As shown in Fig. 1D, the supernatant of BaF/5A/1*6 cells did not have any growth-promoting activity for the parental Ba/F3 cells.
The same mutations in STAT5B have similar biological effects.
STAT5B is 95% identical to STAT5A at the amino acid level (22, 23, 28), and although expression of STAT5B has been shown to be critical in the GH-GH receptor system (45), little is known regarding the functional difference between STAT5A and STAT5B in hematopoietic cells. Since the amino acid sequences surrounding the mutations (mutations 1 and 6) were conserved in STAT5A and STAT5B but not in other STAT proteins, we introduced the same mutations into STAT5B and examined the ability of the mutant STAT5B to induce FI. As shown in Fig. 2, expression of the mutant forms of STAT5A or STAT5B rendered Ba/F3 cells FI with the same efficiency, suggesting functional similarity between STAT5A and STAT5B. This result also confirms the efficacy of these mutations in the constitutive activation of STAT5 molecules.
Characterization of the mutant STAT5A molecules.
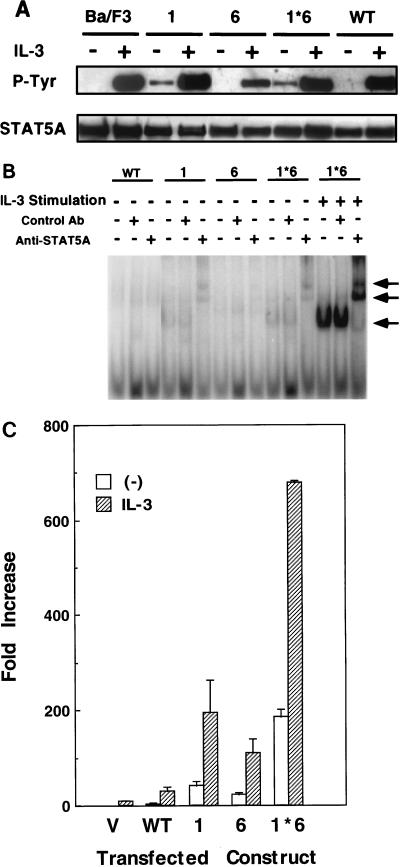
To examine the mechanistic basis of STAT5A activation by the two mutations, we characterized the biochemical properties of the mutant STAT5As expressed in bulk populations of puromycin-selected Ba/F3 cells transduced with sequences for wild-type STAT5A, STAT5A1, STAT5A6, or STAT5A1*6. In cells depleted of mIL-3 and serum for 12 h, tyrosine phosphorylation was detected on STAT5A1 and STAT5A1*6 but not on the wild-type or STAT5A6 protein. The level of phosphorylation was, however, much weaker than that resulting from IL-3 stimulation (Fig. 3A). Consistent with these results, STAT5A1 and STAT5A1*6, but not the wild-type STAT5A or STAT5A6, exhibited weak but significant DNA binding activity without IL-3 stimulation, as assessed by EMSA (Fig. 3B). Interestingly, in a transactivation assay in which Ba/F3 cells were cotransfected with various STAT5 constructs and a STAT5-responsive luciferase reporter plasmid (46), all three mutant STAT5As were able to induce luciferase activity in the absence of IL-3 stimulation (Fig. 3C). Transactivation by the mutant STAT5A1*6 was strongest, and its constitutional transcriptional activity was even stronger than that of the wild-type STAT5A stimulated by IL-3 (Fig. 3C).
FIG. 3.
Characterization of the mutant STAT5As. (A) Tyrosine phosphorylation of STAT5A in Ba/F3 cells expressing the wild-type (WT) or the mutant STAT5As (1, 6, and 1*6) with (+) and without (−) IL-3 stimulation for 10 min. (B) DNA binding activities, without IL-3-stimulation, were examined by EMSA of the nuclear extracts derived from Ba/F3 cells expressing the wild-type STAT5A (WT) and the mutant STAT5As (1, 6, and 1*6). As a positive control, nuclear extracts were also prepared from Ba/F3 cells expressing the mutant STAT5A1*6 after stimulation with 10 ng of IL-3/ml for 10 min. The EMSA complexes were confirmed to contain STAT5A by supershift analysis with anti-STAT5A antibody. Arrows indicate the positions of the STAT5A bands as well as the supershifted complexes. Ab, antibody. (C) Transcriptional activation by the mutant STAT5As. Luciferase activities in the lysates of Ba/F3 cells transfected with the pMX vector (V), the sequence for the wild-type STAT5A (WT), and the sequence for the mutant STAT5As (1, 6, and 1*6) with or without IL-3 stimulation are shown. Transactivation observed in the control experiment (V) was derived from endogenous STAT5A and -5B. The results shown are the averages + the standard deviations of three independent experiments.
Subcellular localization of the wild-type and the mutant STAT5As was also assessed by using STAT5-EGFP fusion constructs (Fig. 4). While the wild-type STAT5A-EGFP as well as STAT5A1-EGFP and STAT5A6-EGFP did not show any particular localization in most NIH 3T3 cells (except for a few in which STAT5-GFP proteins localized to the nucleus) (Fig. 4A to C), the mutant STAT5A1*6-EGFP localized to the nucleus in most NIH 3T3 cells (Fig. 4D).
FIG. 4.
Nuclear localization of the mutant STAT5A. The fusion molecules STAT5A-EGFP (A), STAT5A1-EGFP (B), STAT5A6-EGFP (C), and STAT5A1*6-EGFP (D) were expressed in NIH 3T3 cells and observed under a fluorescence microscope. The photographs were taken with an automatic microscopic camera system (PM30; Olympus). Bars, 30 μm.
Stability of the phosphorylated forms of the wild-type and the mutant STAT5As.
To further elucidate the molecular basis for the activation of the mutant STAT5s, we established Ba/F3 transfectants expressing Flag-tagged wild-type STAT5A, STAT5A1, STAT5A6, and STAT5A1*6. These cells were deprived of mIL-3 for 12 h and stimulated with 10 ng of mIL-3/ml for 30 min, and the tyrosine phosphorylation was chased for 8 h. As shown in Fig. 5, phosphorylation of STAT5A1*6 after IL-3 stimulation was much stronger and more stable than that of the wild-type STAT5A, STAT5A1, and STAT5A6, whose tyrosine phosphorylation started to decrease 2 h after IL-3 stimulation and returned to the basal level in 4 h. Significant tyrosine phosphorylation of the Flag-tagged STAT5 was observed only in the cells expressing the Flag-tagged STAT51*6 before IL-3 stimulation (Fig. 5, lower right panel, first lane). The result shown in Fig. 5 is inconsistent with the result shown in Fig. 3A, where STAT5A1*6 as well as STAT5A1 mutants showed constitutive phosphorylation. This inconsistency may be due to the addition of the Flag-tag to STAT5 in the experiment whose results are shown in Fig. 5.
FIG. 5.
Stability of the phosphorylated forms of the wild-type and the mutant STAT5As. Phosphorylation of the wild-type and the mutant STAT5As was examined in the transfectants (bulk population) expressing the wild-type STAT5A-Flag (wild type), STAT5A1-Flag (1), STAT5A6-Flag (6), and STAT5A1*6 (1*6). The cells were depleted of IL-3 for 12 h (−), stimulated with 10 ng of mIL-3/ml for 10 min, and cultured in the absence of mIL-3 for the indicated time periods, and lysates were prepared from the cells. The cell lysates were immunoprecipitated (IP) by the anti-Flag antibody and then blotted with 4G10 (α pY) or the anti-STAT5A antibody (α STAT5A). Arrows indicate positions of phosphorylated STAT5A-Flag.
Cooperation between the mutant STAT5s and the Raf-1 protein.
Although both mutant STAT5A and STAT5B induced IL-3-independent growth of Ba/F3 cells, these cells still required FCS for proliferation. This observation indicates that an additional signal(s) supplied by the serum is required to support cell proliferation. Since previous results suggested that both the JAK-STAT and the Raf-MAPK pathways are necessary to stimulate cell proliferation (18, 29), we reasoned that the signal missing under the serum-free conditions may be complemented by the activation of the Raf-MAPK pathway. To test this possibility, we expressed an inducible form of Raf, ΔRaf-ER (40), together with the wild-type or mutant STAT51*6 in Ba/F3 cells.
The biological effects of coexpression of ΔRaf-ER and STAT5A were assessed both in clones (Fig. 6A) and in bulk cultures (Fig. 6B) of drug-selected Ba/F3 cells transduced with ΔRaf-ER and either the wild-type or mutant STAT5A. A Ba/F3 clone expressing only ΔRaf-ER, designated BR4, did not exhibit IL-3-independent growth in response to ICI treatment. However, BR4 subclones expressing relatively higher levels of the mutant STAT5A1*6 (M2, M3, M4, and M6) proliferated without IL-3 and serum in the presence of ICI (in a dose-dependent manner), while clones (M1 and M5) which expressed lower levels of the mutant STAT5A1*6 failed to proliferate (Fig. 6A). On the other hand, expression of the wild-type STAT5A at levels much higher than that observed for clones expressing the highest levels of mutant STAT5A (compare W1 and W2 with M2, M3, M4, and M6) did not proliferate in response to ICI (Fig. 6A). To address the possibility of clonal differences among the infected BR4 subclones, we also tested bulk cultures of puromycin-selected cells for ICI-dependent, IL-3-independent growth (Fig. 6B). A bulk population of BR4 cells transduced with the sequence for the mutant STAT5A1*6 followed by selection with puromycin proliferated in the presence of ICI. However, ICI did not stimulate the proliferation of BR4 cells and BR4 cells expressing the wild-type STAT5A. Similar results were obtained in the experiment with another ΔRaf-ER-transduced Ba/F3 clone, BR6 (data not shown). These results showed that activation of neither the Raf-MAPK pathway nor the mutant STAT5A by itself can support proliferation of Ba/F3 cells under serum-free conditions but that activation of the two together induces cytokine- and serum-independent proliferation of Ba/F3 cells.
Activation of Raf had no effect on DNA binding activity of the mutant STAT5A.
To test the direct effects of the activation of the Raf-MAPK pathway on the DNA binding activity of the wild-type and the mutant STAT5A, we examined nuclear extracts prepared from BR4, W1, and M6 cells in an EMSA with the PRE as a probe (46) (Fig. 7). Nuclear extract from M6 cells contained DNA binding activity, albeit at a lower level than that observed in IL-3-stimulated extracts, which was supershifted by anti-STAT5A antibody. Activation of ΔRaf-ER by ICI had no detectable effect on DNA binding activity of the double mutant, M6, suggesting that activation of the Raf-MAPK pathway is not important for DNA binding activity of the mutant STAT5. Similar results were obtained when bulk selected cells were used for the experiment (data not shown).
FIG. 7.
Raf activation did not increase the DNA binding capability of either the wild-type or the mutant STAT5A. Nuclear extracts from the BR4 clones expressing the wild-type STAT5A (W1) and the mutant STAT5A (M6) and the parental BR4 cells were tested for DNA (PRE) binding by EMSA. The arrows indicate bands showing DNA binding and bands supershifted by the anti-STAT5 antibody. I, stimulated with ICI; 3, stimulated with IL-3. The M6 cells showed weak but significant DNA binding which was supershifted by the anti-STAT5A antibody but not by the control antibody (Ab).
DISCUSSION
Using PCR-driven random mutagenesis followed by retrovirus-mediated expression screening, we identified a constitutively active form of STAT5 harboring two point mutations. In addition to being constitutively phosphorylated at tyrosine residues, the mutant STAT5A1*6 is capable of binding a target DNA sequence, stimulating transcription of a reporter plasmid, and supporting FI proliferation of IL-3-dependent cell lines (summarized in Table 1). Constitutive activation of the mutant STAT5A is also associated with nuclear translocation of the molecule, as demonstrated by the experiment in which the mutant STAT5A1*6-EGFP fusion molecule was expressed in NIH 3T3 cells. The observation that a constitutively active STAT5A mutant promotes cell proliferation is consistent with the previous observation that a dominant negative form of STAT5 partially inhibits IL-3-driven cell proliferation (29). However, the property of the mutant molecule does not necessarily reflect that of the wild-type molecule. For instance, while v-abl and bcr-abl are highly mitogenic, there is no evidence to suggest that c-abl is required for proliferation (38). Thus, it is possible that the constitutively active mutation detected in STAT5A is a gain-of-function mutation. Therefore, whether STAT5 activation is physiologically involved in cell proliferation is still open to question.
TABLE 1.
Summary of biological characteristics of the wild-type and mutant STAT5As
| Protein | TP and DNA bindinga | Transcriptional activationb | Nuclear localizationc | FId |
|---|---|---|---|---|
| STAT5A (wild type) | − | − | − | − |
| STAT5A1 | + | + | − | − |
| STAT5A6 | − | + | − | − |
| STAT5A1*6 | + | ++ | + | + |
Previous studies of cytokine receptor signaling have defined at least two pathways important for proliferation: the JAK-STAT and the Raf-MAPK pathways (18, 41). It was also shown that serum by itself weakly stimulated the Raf-MAPK pathway (18). On the other hand, in sharp contrast to the mutant STAT5, the constitutively active form of MPL which was able to stimulate both the JAK-STAT and the Raf-MAPK pathways could induce FI growth even in the absence of FCS (32). From these results, we predicted that the mutant STAT5A would collaborate with the Raf-MAPK pathway to support cell growth. To test this possibility, we transduced either the wild-type STAT5A or the mutant STAT5A1*6 in BR4 cells expressing the ΔRaf-ER construct. As expected, estrogen-induced Raf activation synergized with expression of the mutant STAT5A1*6, but not with the wild-type STAT5A, to induce cell proliferation in the absence of both IL-3 and serum.
Although it is generally accepted that both Myc and Ras-Raf are required for cell proliferation, the mutant STAT5A in concert with Raf was apparently able to stimulate cell proliferation without significant induction of the Myc protein. The doubling time of proliferation driven by the mutant STAT5A and Raf (24 h), however, is longer than that of IL-3-driven proliferation (12 h). One possible explanation for this difference is inadequate expression of c-myc in Ba/F3 cells expressing the mutant STAT5A and/or ΔRaf-ER. Since the expression of c-myc is tightly correlated with proliferation in many systems and is efficiently induced by IL-3, it will be interesting to test whether ectopic Myc expression can complement activation of STAT5A and Raf to yield maximal proliferation.
The same two point mutations in the corresponding region of STAT5B also induced IL-3-independent proliferation of Ba/F3 cells. This result further substantiates the significance of these mutations. In order to investigate how these two mutations constitutively activate STAT5, we constructed mutant STAT5A molecules possessing each mutation (mutation 1 or 6). Mutation 1, but not 6, was by itself able to induce constitutive phosphorylation of STAT5A and constitutive binding of STAT5A to the target DNA sequence. However, for efficient transcriptional activation, nuclear localization, and cell growth, both mutations were required (summarized in Table 1). The fact that mutation 1 (S711F) results in constitutive DNA binding activity is reminiscent of the behavior of C-terminally truncated STAT5s (2, 27, 47) that exhibited delayed dephosphorylation and sustained DNA binding. The phenotype displayed by proteins with these different alterations in the C-terminal region may thus be mechanistically related, but it is important to note that these C-terminal truncations resulted in an overall dominant negative phenotype. Alternatively, mutation 1 may somehow facilitate dimerization of STAT5 because of its close proximity to the C-terminal tyrosine (Y694 of STAT5A or Y699 of STAT5B), which becomes phosphorylated in response to cytokine stimulation and mediates STAT5 dimerization (10). These two possibilities are not mutually exclusive, because dimerization of STAT5 should facilitate DNA binding. On the other hand, it is mysterious that the N-terminal mutation 6 (H299R) is also capable of inducing constitutive transcriptional activity although this mutant lacks constitutive phosphorylation at tyrosine residues and did not show DNA binding activity without IL-3 stimulation. This discrepancy may be explained by the difference in the sensitivities of the assay systems (luciferase assay versus Western blotting and EMSA). We hypothesized that mutation 6 was involved in stabilization of the phosphorylated form of STAT5 since it was reported that the N-terminal part of STAT1 was required for its dephosphorylation (43). In addition to inhibiting dephosphorylation, mutation 6 may also interfere with other mechanisms of down-regulation, such as ubiquitin-proteasome-mediated degradation (17) or proteasome- and p53-dependent protein masking (39); these mechanisms were shown to be involved in regulation of activated STAT1 or STAT3 and STAT5, respectively. To test this hypothesis, we expressed Flag-tagged wild-type STAT5 and mutant STAT5s. However, STAT5A6 did not show any difference in the stability of phosphorylated STAT5A after IL-3 stimulation, and both mutations 1 and 6 are required for the stability of the phosphorylated STAT5. The extremely high degree of transcriptional activation by the mutant STAT51*6 after IL-3 stimulation (Fig. 3C) is probably due to the stability of the phosphorylated STAT5A1*6 (Fig. 5). In summary, the present data strongly indicate that the mechanism of constitutive phosphorylation and activation of the mutant STAT5A1*6 is the stability of the phosphorylated mutant STAT5A1*6.
Constitutive activation of STATs in patients’ leukemic cells has been reported by several groups (11, 48). STAT activation has also been demonstrated in transformed cells, such as human T-cell leukemia virus type 1-infected cells (25) and cells expressing v-src (3), v-abl (6), or bcr-abl (4). While activation of STATs is usually associated with concomitant activation of JAKs or other kinases, we observed phosphorylation of mutant STAT5 without detectable activation of JAKs. Recently, a TEL-JAK2 fusion protein has been implicated in leukemogenesis in three patients with leukemia (21, 36). Lacronique et al. have also shown that the TEL-JAK2 fusion protein can induce constitutive activation of STAT5 in Ba/F3 cells (21). Therefore, it would be interesting to test whether mutation of STAT5 could also be involved in leukemogenesis. Experiments with patients’ leukemic cells and transgenic mice expressing the STAT5A1*6 mutant are in progress.
We have shown in this paper the mechanism by which the mutant STAT5 is activated. To clarify whether STAT5 activation is involved in cell proliferation as well as in other biological processes, such as differentiation, it is necessary to identify the target gene(s) whose expression can promote cellular proliferation.
ACKNOWLEDGMENTS
Tetsuya Nosaka, Kazuhide Misawa, Atsushi Miyajima, and Toshio Kitamura were partly supported by the Ministry of Education, Science and Culture of Japan. DNAX Research Institute is supported by the Schering-Plough Corporation. The Department of Hemopoietic Factors was supported in part by Chugai Pharmaceutical Company Ltd.
We thank James Ihle for the STAT5-Flag plasmid construct.
REFERENCES
- 1.Ahmed S A, Gogal R, Jr, Walsh J E. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods. 1994;170:211–224. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 2.Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 3.Cao X, Tay A, Guy G R, Tan Y H. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlesso N, Frank D A, Griffin J D. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damen J E, Wakao H, Miyajima A, Krosl J, Humphries R K, Cutler R L, Krystal G. Tyrosine 343 in the erythropoietin receptor positively regulates erythropoietin-induced cell proliferation and Stat5 activation. EMBO J. 1995;14:5557–5568. doi: 10.1002/j.1460-2075.1995.tb00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danial N N, Pernis A, Rothman P B. Jak-STAT signaling by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 7.Friedmann M C, Migone T, Russell S M, Leonard W J. Different interleukin 2 receptor b-chain tyrosines couple to at least two signaling pathways and synergistically mediate interleukin 2-induced proliferation. Proc Natl Acad Sci USA. 1996;93:2077–2082. doi: 10.1073/pnas.93.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii H, Nakagawa Y, Schindler U, Kawahara A, Mori H, Gouilleux F, Groner B, Ihle J N, Minami Y, Miyazaki T, Taniguchi T. Activation of Stat5 by interleukin 2 requires a carboxyl-terminal region of the interleukin 2 receptor b chain but is not essential for the proliferative signal transmission. Proc Natl Acad Sci USA. 1995;92:5482–5486. doi: 10.1073/pnas.92.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobert S, Chretien S, Gouilleux F, Muller O, Pallard C, Dusanter-Fourt I, Groner B, Lacombe C, Gisselbrecht S, Mayeux P. Identification of tyrosine residues within the intracellular domain of the erythropoietin receptor crucial for STAT5 activation. EMBO J. 1996;15:2434–2441. [PMC free article] [PubMed] [Google Scholar]
- 10.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse J-F, Capiod J-C, Delobel J, Weber-Nordt R, Dusanter-Fourt I, Dreyfus F, Groner B, Prin L. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 12.Hawley T S, McLeish W A, Hawley R G. Establishment of a novel factor-dependent myeloid cell line from primary cultures of mouse bone marrow. Cytokine. 1991;3:60–71. doi: 10.1016/1043-4666(91)90011-2. [DOI] [PubMed] [Google Scholar]
- 13.Hou J, Schindler U, Henzel W J, Ho T C, Brasseur M, McKnight S L. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 14.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson N G, Szabo S J, Weber-Nordt R M, Zhong Z, Schreiber R D, Darnell J, Jr, Murphy K M. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami Y, Miura T, Bissonnette R, Hata D, Khan W N, Kitamura T, Maeda-Yamamoto M, Hartman S E, Yao L, Alt F W, Kawakami T. Bruton’s tyrosine kinase regulates apoptosis and JNK/SAPK kinase activity. Proc Natl Acad Sci USA. 1997;94:3938–3942. doi: 10.1073/pnas.94.8.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim T K, Maniatis T. Regulation of interferon-g-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita T, Yokota T, Arai K, Miyajima A. Suppression of apoptotic death in hematopoietic cells by signalling through the IL-3/GM-CSF receptors. EMBO J. 1995;14:266–275. doi: 10.1002/j.1460-2075.1995.tb07000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan G P. Efficient screening of retroviral cDNA expression libraries. Proc Natl Acad Sci USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao Y-F, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 21.Lacronique V, Boureux A, Della Valle V, Poirel H, Tran Quang C, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 22.Lin J X, Mietz J, Modi W S, John S, Leonard W J. Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. J Biol Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 23.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Robinson G W, Wagner K-U, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 25.Migone T-S, Lin J-X, Cereseto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Constitutive activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–83. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 26.Miyajima A, Schreurs J, Otsu K, Kondo A, Arai K, Maeda S. Use of the silkworm, Bombyx mori, and an insect baculovirus vector for high-level expression and secretion of biologically active mouse interleukin-3. Gene. 1987;58:273–281. doi: 10.1016/0378-1119(87)90382-9. [DOI] [PubMed] [Google Scholar]
- 27.Moriggl R, Gouilleux-Gruart V, Jähne R, Berchtold S, Gartmann C, Liu X, Hennighausen L, Sotiropoulos A, Groner B, Gouilleux F. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol Cell Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mui A L-F, Wakao H, O’Farrell A, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mui A L F, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425–2433. [PMC free article] [PubMed] [Google Scholar]
- 30.Nosaka, T., K. Ikuta, and T. Kitamura. Unpublished data.
- 31.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Applications of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 32.Onishi M, Mui A L F, Morikawa Y, Cho L, Kinoshita S, Nolan G P, Gorman D M, Miyajima A, Kitamura T. Identification of an oncogenic form of the thrombopoietin receptor MPL using retrovirus-mediated gene transfer. Blood. 1996;88:1399–1406. [PubMed] [Google Scholar]
- 33.Osborne C K, Coronado-Heinsohn E B, Hilsenbeck S G, McCue B L, Wakeling A E, McClelland R A, Manning D L, Nicholson R I. Comparison of the effects of a pure steroidal anti-estrogen with those of tamoxifen in a model of human breast cancer. J Natl Cancer Inst. 1995;87:746–750. doi: 10.1093/jnci/87.10.746. [DOI] [PubMed] [Google Scholar]
- 34.O’Shea J J. Jaks, STATs, cytokine signal transduction, and immunoregulation: are we there yet? Immunity. 1997;7:1–11. doi: 10.1016/s1074-7613(00)80505-1. [DOI] [PubMed] [Google Scholar]
- 35.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peeters P, Raynaud S D, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Fusion of TEL, the ets-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 37.Quelle F W, Wang D, Nosaka T, Thierfelder W E, Stravopodis D, Weinstein Y, Ihle J N. Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol Cell Biol. 1996;16:1622–1631. doi: 10.1128/mcb.16.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raitano A B, Whang Y E, Sawyers C L. Signal transduction by wild-type and leukemogenic Abl proteins. Biochim Biophys Acta. 1997;1333:201–216. doi: 10.1016/s0304-419x(97)00023-1. [DOI] [PubMed] [Google Scholar]
- 39.Rayanade R J, Patel K, Ndubuisi M, Shama S, Omura S, Etlinger J D, Pine R, Sehgal P B. Proteasome- and p53-dependent masking of signal transducer and activator of transcription (STAT) factors. J Biol Chem. 1997;272:4659–4662. doi: 10.1074/jbc.272.8.4659. [DOI] [PubMed] [Google Scholar]
- 40.Samuels M L, Weber M J, Bishop J M, McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human Raf-1 protein kinase. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato N, Sakamaki K, Terada N, Arai K, Miyajima A. Signal transduction by the high-affinity GM-CSF receptor: two distinct cytoplasmic regions of the common beta subunit responsible for different signaling. EMBO J. 1993;12:4181–4189. doi: 10.1002/j.1460-2075.1993.tb06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindler C, Darnell J., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 43.Shuai K, Liao J, Song M M. Enhancement of antiproliferative activity of gamma interferon by the specific inhibition of tyrosine dephosphorylation of Stat1. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniguchi T. Cytokine signaling through nonreceptor protein tyrosine kinases. Science. 1995;268:251–255. doi: 10.1126/science.7716517. [DOI] [PubMed] [Google Scholar]
- 45.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S-H, Ram P, Waxman D J, Davey H W. Requirement of Stat5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber-Nordt R M, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]