Productive and Nonproductive Complexes of Ku and DNA-Dependent Protein Kinase at DNA Termini (original) (raw)
Abstract
DNA-dependent protein kinase (DNA-PK) is the only eukaryotic protein kinase known to be specifically activated by double-stranded DNA (dsDNA) termini, accounting for its importance in repair of dsDNA breaks and its role in physiologic processes involving dsDNA breaks, such as V(D)J recombination. In this study we conducted kinase and binding analyses using DNA-PK on DNA termini of various lengths in the presence and absence of Ku. We confirmed our previous observations that DNA-PK can bind DNA termini in the absence of Ku, and we determined rate constants for binding. However, in the presence of Ku, DNA-PK can assume either a productive or a nonproductive configuration, depending on the length of the DNA terminus. For dsDNA greater than 26 bp, the productive mode is achieved and Ku increases the affinity of the DNA-PK for the Ku:DNA complex. The change in affinity is achieved by increases in both the kinetic association rate and reduction in the kinetic dissociation rate. For dsDNA smaller than 26 bp, the nonproductive mode, in which DNA-PK is bound to Ku:DNA but is inactive as a kinase, is assumed. Both the productive and nonproductive configurations are likely to be of physiologic importance, depending on the distance of the dsDNA break site to other protein complexes, such as nucleosomes.
One can broadly classify eukaryotic DNA repair into excision repair, mismatch repair, and double-strand break repair. Two types of double-strand break repair are homologous recombination and nonhomologous DNA end joining (NHEJ). NHEJ is the major mechanism of repairing double-stranded DNA (dsDNA) breaks during most of the cell cycle yet is the least understood type of DNA repair. Unlike single-strand breaks, which have the other strand to maintain both the physical integrity and the information content of the DNA, double-strand breaks do not. Hence, double-strand breaks have the highest potential to result in either loss of genetic information or loss of chromosomal integrity, each of which can further contribute to subsequent genomic destabilization events.
The nucleic acid and protein biochemical properties of NHEJ are largely undefined (28). Recently, we and others determined that DNA ligase IV complex is responsible for NHEJ in Saccharomyces cerevisiae (34, 39, 40). In mammalian cells, we and others also recently identified a complex of XRCC4 (X-ray cross-complementation group 4) and DNA ligase IV (9, 17). The XRCC4 stimulates the DNA ligase IV by physical association (17). This is interesting because null mutations in XRCC4 in mammalian cells result in sensitivity to ionizing radiation, especially in G1 and early S phases of the cell cycle (15). XRCC4 mutant cells that are made active for V(D)J recombination (by transfection with RAG expression vectors) are defective for both signal and coding joint formation (32, 37, 38). All of the XRCC mutants that are defective for double-strand break repair are also defective for V(D)J recombination (28). This is not surprising given that V(D)J recombination is a physiologic process of creating dsDNA breaks, which, once formed, must be repaired like pathologic dsDNA breaks.
The other three XRCC groups are XRCC5, -6, and -7 (28). XRCC5 and -6 encode the two subunits of Ku (Ku86 and Ku70, respectively) (18, 35, 36). The Ku heterodimer (Ku70-Ku86) loads onto DNA termini and diffuses in an energy-independent fashion to internal positions (10). XRCC7 is complemented by chromosomal regions that contain the gene for the DNA-dependent protein kinase (DNA-PK) (3, 23, 33), a 469-kDa protein that is activated by DNA termini (1).
The earliest work on purified DNA-PK presumed that it functioned by itself as the first (and still only) protein kinase activated by dsDNA termini (5, 26). A function suggested originally was that it served as an alarm system for exogenous (viral) or endogenous (genomic) dsDNA ends. That proposed function continues to be the most likely one (20, 21, 26). The Jackson and Dynan laboratories discovered that DNA-PK activity in vitro could be stimulated by Ku (11, 16). Gottlieb and Jackson proposed that Ku, in fact, was the DNA binding subunit for DNA-PK, implying that DNA-PK was inactive in the absence of Ku (16). They proposed that the name for the 469-kDa DNA-PK (then thought to be 350 kDa) be changed to DNA-PKCS to indicate that this is merely the catalytic subunit and that it is inactive without Ku. Recently, we found that purified, native DNA-PK can be activated by direct binding of DNA ends in the absence of Ku (41), a finding previously described for some DNA-PK phosphorylation targets by others as well (4, 11, 27, 30). We were also able to confirm Ku stimulation of DNA-PK up to eightfold (41). Hence, we retain the original name of DNA-PK. We refer to the complex that forms on DNA termini as Ku:DNA-PK:DNA or, if Ku is absent, as DNA-PK:DNA; complexes of Ku:DNA-PK do not appear to form except on DNA (41).
Our basic finding that DNA-PK can bind directly to DNA has been confirmed (19), though these authors have raised the question as to whether these results could be explained by undetectable Ku. However, our DNA-PK preparation has no detectable Ku. By Western blotting with three monoclonal antibodies and by a highly sensitive cross-linking assay standardized against the Ku Western blots, we know that Ku, if present, represents less than 1 molecule for every 110 DNA-PK molecules (41). Evidence that the DNA-PK can bind to DNA directly includes mobility shifts in which it is apparent that over 30% of the DNA-PK molecules can bind to an equimolar amount of DNA. It is clear that undetectable Ku contamination below the 0.9% level could not account for binding of 30% of the DNA-PK molecules. Results of atomic force microscopy, immunoprecipitation, and cross-linking all confirm these results (41). We have now confirmed that DNA-PK is able to maintain a Ku-independent binding at physiologic ionic strength by several independent methods (see below).
To further understand the interaction between DNA-PK and Ku at DNA termini, we have conducted a detailed study with these components, examining both the functional relationship (in the context of kinase activity) and the physical relationship. The DNA-PK kinase activity has been measured for the DNA-PK:DNA complex in the presence and absence of Ku. We find that Ku can stimulate DNA-PK activity if DNA of at least 26 bp in length is used. This 8-fold level of activation has been described previously; however, at saturating DNA concentrations, the stimulation is only about 1.6-fold. A short DNA fragment of 18 bp can fully stimulate DNA-PK (41). This size is also sufficient to bind Ku as efficiently as all larger DNA fragments. However, when this 18-bp DNA fragment is used for kinase assays, addition of Ku inhibits DNA-PK activity. In conjunction with these functional observations, we present surface plasmon resonance (SPR) measurements of Ku and DNA-PK binding with DNA. We generated the first kinetic data for Ku binding to DNA and then generated similar data for DNA-PK alone binding to DNA. Using this technique to examine the effect that Ku has on DNA-PK when binding to DNA, we find that DNA-PK is not blocked from interacting with the 18-mer DNA but rather binds to the 18-mer:Ku complex. These observations support a new model for DNA-PK and Ku interaction on DNA. In this model, Ku binds to the DNA end, changes conformation, and recruits DNA-PK to the Ku:DNA end complex. If the DNA end is long enough, the DNA-PK will be active as a kinase. If the DNA end is too short, as would be the case for breaks within internucleosomal regions, the DNA-PK kinase activity will be inhibited by being bound but in a nonproductive mode.
MATERIALS AND METHODS
Purification of DNA-PK and Ku proteins.
DNA-PK was purified from 20 liters of HeLa cells as described by Chan et al. (8). Recombinant Ku protein was expressed in a baculovirus expression system and purified by using three columns: Ni-nitrilotriacetic acid agarose with imidazole elution, dsDNA-Sepharose, and MonoQ 5/5 (41).
Oligonucleotides and DNA fragments.
The following synthetic DNA fragments were used in this study: 16-mer, 5′-AGGCTGTGCTCAGAGG-3′ and 5′-CCTCTGAGCACAGCCT-3′; 18-mer, 5′-AGGCTGTGTCCTCAGAGG-3′ and 5′-CCTCTGAGGACACAGCCT-3′; 22-mer, 5′-AGGCTGTGTTAGCCCTCAGAGG-3′ and 5′-CCTCTGAGGGCTAACACAGCCT-3′; 26-mer, 5′-AGGCTGTGTTAAGTCGCCCTCAGAGG-3′ and 5′-CCTCTGAGGGCGACTTAACACAGCCT-3′; 30-mer, 5′-AGGCTGTGTTAAGTAGCTCGCCCTCAGAGG-3′ and 5′-CCTCTGAGGGCGAGCTACTTAACACAGCCT-3′; 35-mer, 5′-AGGCTGTGTTAAGTATCTGCGCTCGCCCTCAGAGG-3′ and 5′-CCTCTGAGGGCGAGCGCAGATACTTAACACAGCCT-3′; 59-mer, 5′-ATCAGGATGTGGTGATGCACAGTGTGATCCCTCCTCACAAAAACCGCAGGTCTTCAGTT-3′ and 5′-AACTGAAGACCTGCGGTTTTTGTGAGGAGGGATCACACTGTGCATCACCACATCCTGAT-3′; and 79-mer, 5′-GATCCTCTGAGGACACAGCCTTGTATTACTGTGCAAGACACACAATGAGCAAAAGTTACTGTGAGCTCAAACTAAAACC-3′ and 5′-GATCGGTTTTAGTTTGAGCTCACAGTAACTTTTGCTCATTGTGTGTCTTGCACAGTAATACAAGGCTGTGTACTCAGAG-3′. The complementary single-stranded oligonucleotides were annealed in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA–0.15 M NaCl by heating in boiling water for 5 min and slow cooling.
Phosphorylation assay.
Kinase assays were performed as described previously, using p53 synthetic peptide as the substrate (8, 13, 41). The phosphorylation reactions were carried out in a final volume of 20 μl with a buffer composed of 20 mM Tris-HCl (pH 7.9), 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 2 mM dithiothreitol, 0.02% Tween 20, and 10% glycerol. The concentrations of the peptide and the [γ-32P]ATP (3,000 Ci/mmol; Amersham) in the assay were 200 μM and 3.3 × 10−8 M, respectively. Time course and dose-response analysis showed that the linear range of the enzyme activity was between 5 and 20 min at 37°C and that this activity was dependent on the amount of DNA-PK in a linear manner over a 10-min time course (41). Two methods were used to separate the peptide-incorporated label from the unincorporated label. In one method, the reaction contents were applied to phosphocellulose filters and washed with acetic acid as described previously (41). In the second method, the reaction contents were separated on a sodium dodecyl sulfate (SDS)–15 to 20% polyacrylamide gel and quantitated on a PhosphorImager plate by using ImageQuaNT software.
Characterization of the purified DNA-PK and Ku proteins.
The purity of the kinase was analyzed by electrophoresis in SDS-containing gels. Following staining with Coomassie blue, no bands other than the major band migrating at approximately 470 kDa were observed. (By gel filtration, DNA-PK migrates at approximately 470 kDa, which closely matches the value computed based on the cDNA sequence of 12.5 kb.) This result indicates that the preparation was essentially devoid of contaminants and that no significant proteolysis had occurred during the purification procedure. The major band staining with Coomassie blue was also reactive with all three monoclonal antibodies tested. The specific activity of the DNA-dependent kinase in the peak fraction was determined to be 243 mol of PO4− incorporated into the p53 peptide per mg of purified protein per 10 min.
It was important to determine if the kinase preparation contained any traces of copurifying Ku. Immunoblot analysis with four anti-Ku monoclonal antibodies by using the highest sensitivity detection method did not reveal the presence of Ku (41). We have used UV cross-linking to achieve an even higher sensitivity for detection of contaminating Ku down to a level of 2 fmol (41). We detected no Ku70 or Ku86 by this method either (41). We determined that in this preparation, a maximum possible contamination with Ku, if any existed at all, would be less than 1 molecule per 110 molecules of DNA-PK (41). We therefore conclude that there is no evidence of Ku contamination and that any possible undetected contamination would have insignificant effects on the bulk activity of DNA-PK.
The Ku heterodimer was purified from HeLa cells, and recombinant Ku was purified by using a baculovirus overexpression system. The native and recombinant Ku behaved identically in DNA binding assays and DNA-PK kinase assays (41).
Protein analysis.
The proteins were analyzed for purity by electrophoresis in SDS–8% polyacrylamide. The gels were stained with Coomassie blue or transferred to a nitrocellulose membrane for Western blotting analysis using specific anti-DNA-PK and anti-Ku monoclonal antibodies as described previously (41). The immune complexes were detected with either alkaline phosphatase-conjugated (with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as substrates) or peroxidase-conjugated (enhanced chemiluminescence) anti-mouse antibodies. The protein concentration was determined by the method of Bradford (Bio-Rad), using a standard curve produced with bovine serum albumin. We independently determined the protein concentration by calculating the extinction coefficient from the amino acid composition (based on the tryptophan and tyrosine residues) and measuring the absorbance of the purified protein in 6 M guanidinium hydrochloride at 280 nm.
SPR DNA binding assays.
Binding experiments were performed on a Biacore X machine (Biacore), using basic methodology described elsewhere (14, 22). The running buffer included 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant P20. Sensorgrams involving DNA-PK with or without Ku were generated over a range of flow rates (10 to 30 μl/min) without significant change in binding characteristics. Sensorgrams of Ku binding without DNA-PK were generated over a range of flow rates (20 to 100 μl/min), and representative sensorgrams were chosen for presentation. Proteins were diluted in running buffer subsequent to injection into the Biacore unit. Each protein injection was followed by a 3- to 5-min dissociation phase in which running buffer was passed through the flow cell. All experiments were performed at least three times. A simultaneous no-oligonucleotide control run was performed for each sensorgram, and this background was subsequently subtracted from the sensorgram values. The sensorgram response units per time were evaluated by using software supplied with the instrument (Evaluation version 3.0; Biacore). To regenerate the sensor chip binding surface, a 30-s incubation with a 0.05% SDS solution was performed, which resulted in a return of the signal to pre-protein injection levels. During data analysis, sensorgrams were fitted with models considering mass transport effects when applicable.
RESULTS
Stimulation of DNA-PK activity as a function of DNA concentration in the presence or absence of Ku.
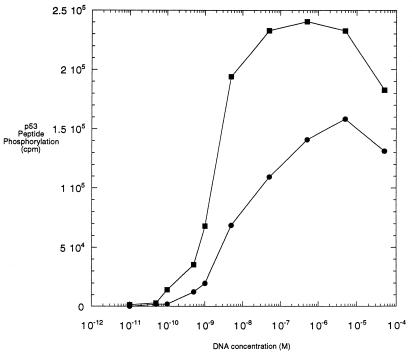
To pursue the relationship between Ku and DNA-PK, we studied how the presence or absence of Ku influences the kinase activity profile of DNA-PK as a function of DNA concentration. Using a 59-bp duplex oligonucleotide as the DNA activator, we generated a profile of kinase activity over a range of DNA concentrations under standard kinase assay conditions (7) for DNA-PK alone. The kinase activity was initially detected at a DNA concentration of 2 × 10−10 M and reached a plateau at approximately 10−6 M (Fig. 1). A similar profile was generated in the presence of a 10-fold molar excess of Ku (24 nM) over DNA-PK (2.4 nM). Under these conditions, the kinase activity was already detectable at a concentration of 6 × 10−11 M DNA, and the plateau was reached at a concentration of 5 × 10−8 M DNA (Fig. 1). Several features of these two profiles are worth noting. First, the lowest concentrations of DNA at which kinase activity was stimulated in the presence of Ku relative to its absence are within an order of magnitude of each other. Ku does not significantly shift the concentration of DNA at which DNA-PK is initially activated. However, the maximal activities observed in the two profiles are different. The maximum activity achieved by DNA-PK by itself is only 60% of the maximum activity obtained when Ku is present. The most striking feature is that in the presence of Ku, the activity reaches the plateau and also the half-maximal activity at a 20-fold lower DNA concentration. We conclude that DNA-PK alone has a significant ability to be activated by DNA and that the fold induction of DNA-PK activity by Ku depends on the DNA concentration used in the assay.
FIG. 1.
Activity of purified DNA-PK in the presence of a 59-bp DNA fragment and in the absence or presence of Ku. Reaction mixtures of 20 μl contained 1.2 × 10−9 M purified DNA-PK without (bottom curve) or with (top curve) 2.4 × 10−8 M purified Ku protein and increasing concentrations of the 59-bp DNA fragment. The probes containing Ku were preincubated with DNA for 10 min at room temperature before addition of the enzyme. The phosphorylation reaction was carried out at 37°C for 10 min as described in Materials and Methods. The curves from this experiment are representative of multiple experiments, and kinase determinations at each DNA concentration are within ±10% of the values obtained in this experiment.
Inhibition of DNA-PK activity by Ku protein.
While DNA-PK clearly interacts with DNA on its own, based on the DNA-PK activation by DNA alone (Fig. 1), the nature of the stimulation by Ku is still unclear. When Ku stimulates DNA-PK, does Ku act as the binding subunit for DNA-PK or, rather, do both proteins bind DNA, with Ku acting as an allosteric effector of DNA-PK? To distinguish between these two possibilities, we used the same assay as above but with a much shorter oligonucleotide. We chose an 18-bp DNA fragment because the data from DNA footprinting demonstrate that the Ku protein covers at least this length of DNA (10). Assuming that the 18-bp DNA fragment may be entirely covered by Ku, DNA-PK will be unable to directly interact with DNA.
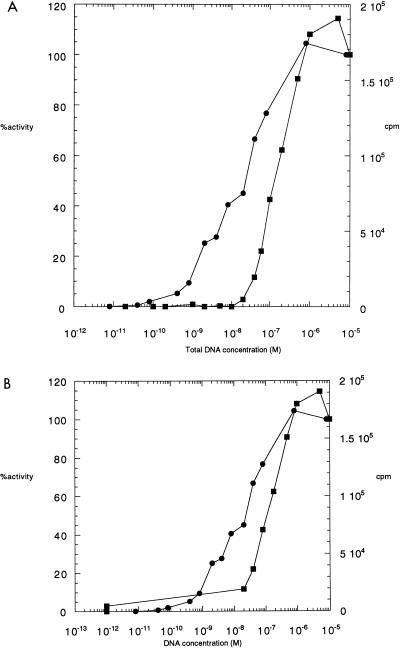
The resulting activity profiles with the smaller oligonucleotide revealed that in the presence of a 20-fold molar excess of Ku (2.4 × 10−8 M) over DNA-PK (1.2 × 10−9 M), there was an absolute inhibition of kinase activity at low DNA concentrations up until the concentration of DNA exceeded that of Ku, at 2.4 × 10−8 M DNA (Fig. 2A). In other words, only when DNA-PK was in the presence of Ku-free DNA was there any kinase activity. It is very important to note, however, that when the concentration of free DNA is corrected for the DNA bound by Ku, the kinase activity still requires a 10-fold-higher concentration of DNA compared to assays using DNA-PK and DNA alone (Fig. 2B). This finding indicates that the inhibition by Ku is not simply one of exclusion of the DNA-PK from DNA occupancy; otherwise, the two curves in Fig. 2B would be the same. Rather, it suggests that the Ku recruits the DNA-PK into a nonproductive complex.
FIG. 2.
Activity of DNA-PK in the absence or presence of Ku protein, using the 18-bp DNA fragment. (A) Reaction mixtures of 20 μl contained 1.2 × 10−9 M purified DNA-PK without (circles) or with (squares) 2.4 × 10−8 M Ku protein and increasing concentrations of the 18-bp DNA fragment. The probes containing Ku were preincubated with DNA for 10 min at room temperature before addition of the enzyme. The phosphorylation reaction was carried out for 10 min at 37°C as described in Materials and Methods. Percent activity was calculated based on an independent sample at 10−6 M of the 18-mer in the absence of Ku. The curves from this experiment are representative of multiple experiments, and kinase determinations at each DNA concentration are within ±10% of the values obtained in this experiment. (B) Same data as in panel A, but plotted as a function of the free [DNA] instead of the total [DNA] for the dsDNA fragment. The conversion from total DNA to free DNA was approximated by subtracting the total possible amount of oligonucleotide bound by Ku from the total amount of oligonucleotide added in the sample.
At higher DNA concentrations, the activity on the 18-mer DNA plus Ku finally reaches a plateau that matches that of the 18-mer without Ku. Interestingly, the 18-mer-plus-Ku kinase activity profile fails to reach the level of the 59-mer-plus-Ku kinase activity profile (Fig. 1). This effect is not due to the inability of either of the two proteins to interact with this shorter DNA fragment. The DNA-PK activity profile without addition of Ku is identical to the profile of the 59-mer oligonucleotide (Fig. 2), confirming that the 18-mer oligonucleotide can activate DNA-PK to the same extent as the 59-mer oligonucleotide. This result also indicates that the DNA-PK is recruited to the Ku:DNA complex to form a ternary complex (Ku:DNA-PK:DNA) that lacks kinase activity.
Determination of the minimal DNA length necessary for the activation of DNA-PK by Ku.
From the experiments described up to this point, it appears as if both Ku and DNA-PK must be bound to DNA simultaneously for maximal phosphorylation of the peptide substrate. Next, we designed experiments to determine the minimal length of DNA that can still activate DNA-PK if Ku is present in a molar excess over DNA. A series of synthetic dsDNA fragments with different lengths (16, 18, 22, 26, 30, and 35 bp) were tested. The ends of these fragments were blunt and uniform in sequence; the length was increased only through addition of random nucleotides in the middle of the dsDNA fragment. Thus, possible end-sequence differences that could affect kinase activity were minimized (even though no sequence specificities have been reported for DNA-PK or Ku on linear DNA).
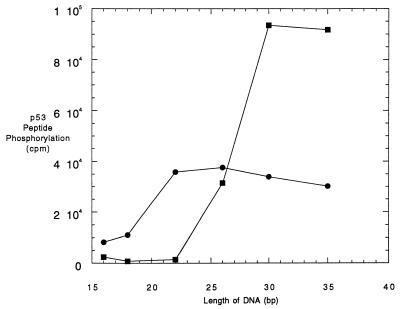
DNA-PK kinase activity was inhibited by a molar excess of Ku when DNA of 16, 18, or 22 bp in length was used (Fig. 3). These results are consistent with the data shown in Fig. 2. When 26-bp DNA was used, Ku did not inhibit the kinase activity anymore but, at the same time, did not lead to further activation. With the 30-bp DNA fragment, the expected two- to threefold additional stimulation of the kinase activity by Ku was observed. Activation of the DNA-PK by Ku was the same for the 30- or a 35-bp DNA fragment, indicating that the maximal activation had been reached with the 30-bp DNA. Hence, DNA of 30 bp in length is necessary and sufficient for the functional stability of the complex of DNA:Ku:DNA-PK.
FIG. 3.
Activity of DNA-PK in the absence and presence of Ku protein, using DNA fragments with different lengths. Reaction mixtures of 20 μl contained 1.2 × 10−9 M purified DNA-PK without (filled circles) or with (open squares) 2.4 × 10−8 M Ku protein and a DNA fragment with a length of 16, 18, 22, 26, 30, or 35 bp at a concentration of 10−8 M. The probes with Ku were incubated for 10 min at room temperature prior to the addition of DNA-PK. The phosphorylation reactions were carried out for 10 min at 37°C as described in Materials and Methods. In several experiments, kinase determinations at each DNA length were within ±10% of the values shown.
In the absence of Ku, DNA-PK was activated with these DNA fragments with different efficiencies: the shortest DNA (16 bp) activated the kinase to the lowest degree, while DNA of 22 bp and longer had higher and almost constant activity (Fig. 3). It seems that a fragment of 16 bp is too short to allow a stable interaction between the kinase and DNA. These results, together with those presented above, suggest that there is a minimal length of DNA, beginning at a 22 bp, for the activation of DNA-PK activity.
It is noteworthy that the dsDNA length necessary for half-maximal activation of the DNA:Ku:DNA-PK complex is 26 bp, whereas that for the DNA:DNA-PK complex is between 20 and 22 bp (Fig. 3). Ku binding studies indicate that it binds to 18- and 20-bp dsDNA equivalently to all larger dsDNA fragments, ranging from 18 to 130 bp for oligonucleotides and from 169 to 2,000 bp for restriction fragments or sheared salmon sperm DNA (12, 41). Both Ku (10, 12) and DNA-PK (41) appear to load onto linear dsDNA at the DNA terminus at the structural transition between single-stranded DNA and dsDNA. But Ku diffuses internally along the DNA in an ATP-independent manner (10, 41), whereas DNA-PK does not (41).
Direct analysis of DNA-PK and Ku binding to DNA by SPR.
Because the kinase assay gives a readout based on productive complexes, absence of activity could be caused by either (i) the inability of DNA-PK to bind DNA directly or (ii) DNA-PK binding to DNA and Ku, but failing to form a kinase active complex. The kinase assay cannot directly distinguish between these two possibilities. From Fig. 2B, we infer the presence of a nonproductive complex between DNA-PK, Ku, and the 18-bp oligonucleotide, based on the observed suppression by Ku of DNA-PK activity at DNA concentrations where at least half of the DNA is unbound by protein. Measuring the binding of DNA-PK to these different complexes would resolve this issue. For these studies, we use SPR. We use this approach to analyze not only the nonproductive but also the productive complex.
To accurately measure protein-DNA interactions with SPR, we performed the studies in a buffer containing 150 mM salt to approximate the ionic strength within the nucleus and also to prevent any nonspecific interactions between the proteins and the supporting matrix. A recent study was unable to detect DNA-PK kinase at salt concentrations of 100 mM and above in the absence of Ku (19). However, we have conducted a study of the kinase activity as a function of monovalent salt concentration by using a kinase assay that distinguishes between incorporated and unincorporated phosphate more accurately than that used by Hammersten and Chu (19). Though we find that DNA-PK activity decreases with increasing salt concentration, we find that there is vigorous kinase activity that is 258-fold above background at 150 mM salt and over 1,000-fold above background at 100 mM salt (Table 1). This demonstrates that DNA-PK can in fact phosphorylate peptides under physiologic conditions and that binding studies under these conditions are physiologically relevant.
TABLE 1.
DNA-PK kinase activity as a function of monovalent salt in the absence of Ku
| Conditiona | Phosphorylated peptide (total counts)b | Fold increase over control counts |
|---|---|---|
| 1 | 6.2 × 103 | 1 |
| 2 | 21 × 106 | 3,416 |
| 3 | 6.4 × 106 | 1,032 |
| 4 | 1.6 × 106 | 258 |
Direct examination of Ku binding to DNA by SPR.
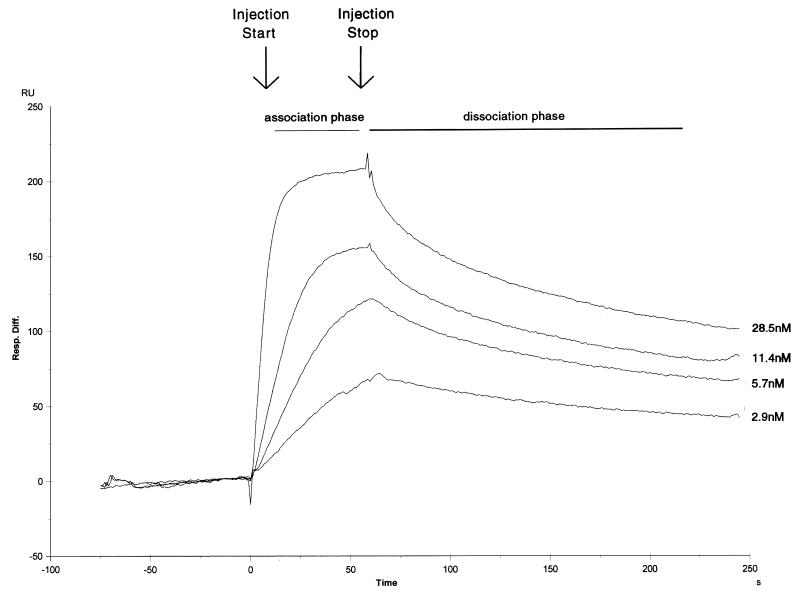
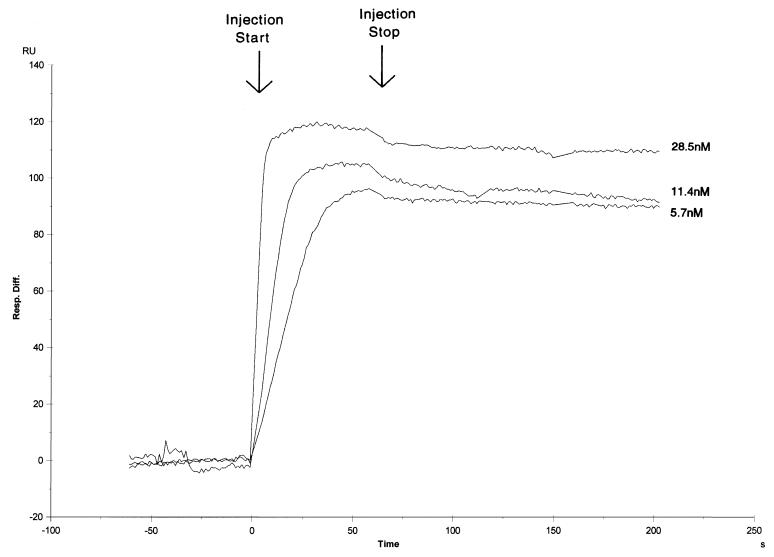
We first examined the kinetics of Ku binding to the 18- and 35-bp double-stranded oligonucleotides (Fig. 4 and 5) to determine the stability of the Ku:DNA complex. Low levels of double-stranded oligonucleotide were loaded onto separate streptavidin-precoated sensor chips to give a maximum Ku binding response of between 100 and 200 response units. Purified recombinant Ku was then passed over the DNA-dextran matrix at varied concentrations close to the published equilibrium dissociation constant (2, 29). In the sensorgram shown in Fig. 4, four different concentrations of Ku were examined in the context of the 18-bp oligonucleotide (the bulk refractive index changes have been removed from the curves). Ku shows both a high association rate and a low dissociation rate. Using a 1:1 Langmuir fitting program, we calculated the equilibrium dissociation constant to be 5.9 × 10−10 M, which is very close to our own (unpublished) and to previously published data obtained in assays using filter binding or mobility shift techniques (2, 29). The calculated kinetic rate of association in our measurements is 2.3 × 107 M−1 s−1, while the calculated kinetic rate of dissociation is 1.4 × 10−2 s−1.
FIG. 4.
Sensorgram of Ku protein binding to a sensor chip loaded with the 18-bp oligonucleotide. Single runs of four Ku concentrations (28.5, 11.4, 5.7, and 2.9 nM) are shown.
FIG. 5.
Sensorgram of Ku protein binding to a sensor chip loaded with the 35-bp oligonucleotide. Single runs of three Ku concentrations (28.5, 11.4, and 5.7 nM) are shown.
The sensorgrams of Ku binding with the 35-bp oligonucleotide are shown in Fig. 5. Three different concentrations of Ku were used under identical conditions of Ku binding to the 18-bp oligonucleotide. Interestingly, Ku binding under these conditions shows a higher affinity for the DNA. Difficulties in fitting the curves precluded precise measurements of the kinetic constants. However, the kinetic rate of dissociation is at least 10-fold lower than that for Ku binding to the 18-bp oligonucleotide, while the kinetic rate of association is equal to or higher than that for Ku binding to the 18-bp oligonucleotide. Though Ku appears to bind to both oligonucleotides in a stable fashion, there appears to be a difference in degree of stability. We are currently pursuing further studies regarding this difference.
Direct examination of DNA-PK binding by SPR.
Having used SPR to establish the stability of Ku binding, we examined the binding of DNA-PK. Direct binding of DNA-PK to DNA in the absence of Ku has already been established in assays using UV cross-linking (41). However, attempts in our laboratory at examining DNA-PK binding using quasi-equilibrium methods, such as filter binding based assays or gel shift assays, have not been successful. To measure DNA-PK binding, we created a sensor chip with a high concentration of 35-bp oligonucleotide (as reflected by a higher magnitude of response units).
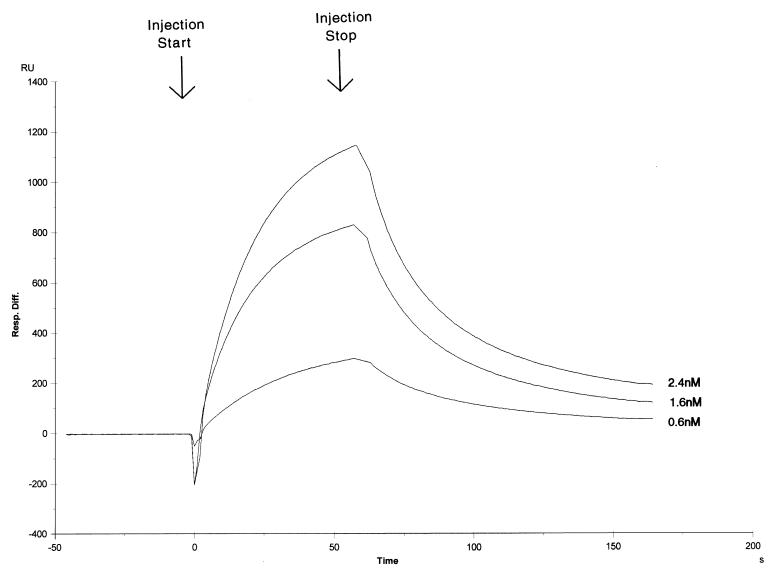
In Fig. 6, the sensorgram of three different DNA-PK concentrations are shown. DNA-PK has both extremely high association and dissociation rates to give an equilibrium dissociation constant of 3.1 × 10−9 M. However, the calculated kinetic rate of association is 1.5 × 107 M−1 s−1, while the calculated kinetic rate of dissociation is 0.048 s−1. These extremely high association and dissociation rates explain why it was difficult to measure DNA-PK binding by using the electrophoretic mobility shift (in which rapid debinding results in loss of the complex) or filter binding (in which rapid debinding results in loss of the complex in the washing step). Similar results were achieved with DNA-PK binding to the 18-bp oligonucleotide (data not shown).
FIG. 6.
Sensorgram of DNA-PK protein binding to a sensor chip loaded with the 35-bp oligonucleotide. Single runs of three DNA-PK concentrations (2.4, 1.6, and 0.6 nM) are shown.
DNA-PK binding to the Ku:35-bp complex.
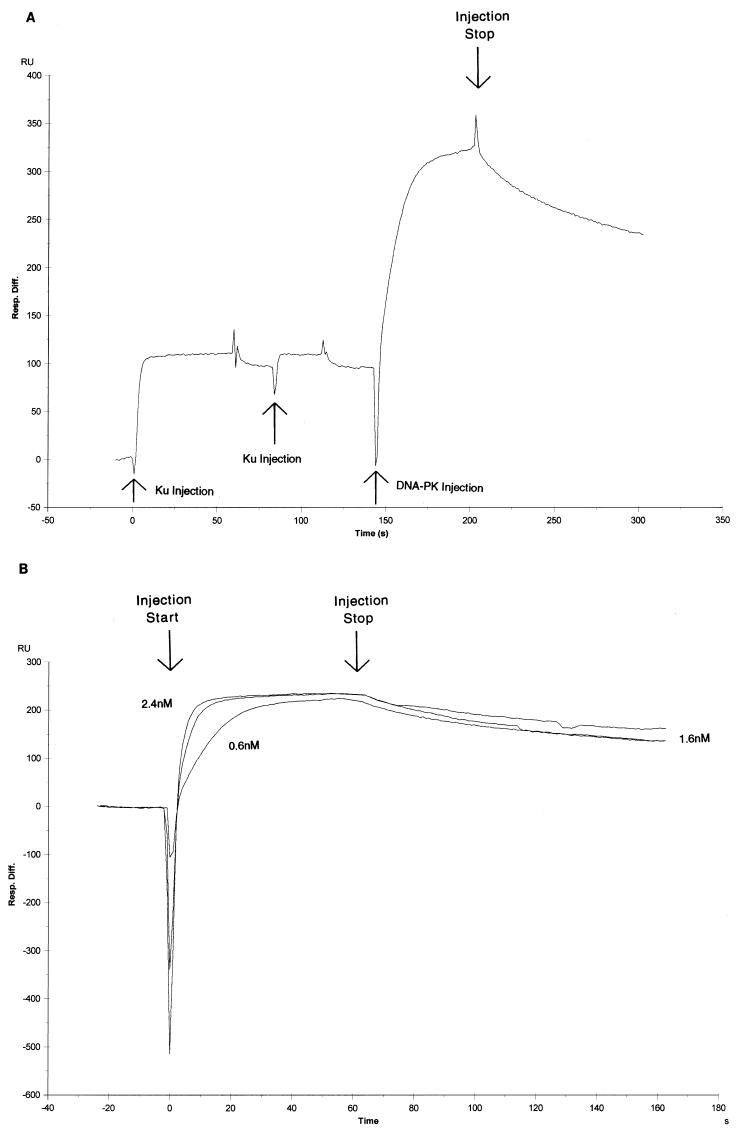
The preincubation of Ku with the 35-bp oligonucleotide in the kinase assay demonstrated that Ku shifts the half-maximal activity to lower DNA concentrations (Fig. 1). This finding suggests that Ku stabilizes DNA-PK binding in the DNA complex. To evaluate the effect of Ku on the stability of DNA-PK association with DNA, we preincubated Ku with a chip containing a low level of the 35-bp oligonucleotide. Immediately after attaining saturation of the DNA with Ku, we injected DNA-PK. An example of this series of injections is shown in the sensorgram in Fig. 7A: two consecutive Ku injections followed by an injection of DNA-PK. Multiple Ku injections are used to ensure that saturation of the DNA is achieved. This is followed by a large rise and fall in response units due to the injection and subsequent dissociation of DNA-PK.
FIG. 7.
Binding of DNA-PK with the Ku:35-bp DNA fragment complex. (A) Example of a complete sensorgram of sequential Ku (28.5 nM) and DNA-PK (2.4 nM) injections onto a sensor chip loaded with the 35-bp oligonucleotide. (B) Sensorgram of the DNA-PK binding portion of the sensorgram in panel A. Single runs of three DNA-PK concentrations (2.4, 1.6, and 0.6 nM) are shown.
In Fig. 7B, we show three different concentrations of the DNA-PK association and dissociation part of the sensorgram. The analysis of these sensorgrams is less reliable due to the poorer fit of the modeling program because of the complexity of the macromolecular interactions. However, it is obvious from the curves that the kinetic rate of dissociation is much lower than with DNA-PK alone. We calculate that the kinetic rate of dissociation is 5 × 10−3 M−1 s−1. The kinetic rate of association appears not to be significantly affected; we calculate it to be about 10-fold higher (1.4 × 108 M−1 s−1) than with DNA-PK alone. The difference between the rate of dissociation of DNA-PK alone and DNA-PK with Ku illustrates that the primary effect of Ku is to stabilize the DNA-PK on the DNA, though Ku does have some effect on the fast association rate of DNA-PK. These findings are completely consistent with the kinase assay results in Fig. 1. Furthermore, the striking qualitative difference between DNA-PK binding with and without Ku conclusively demonstrates that there is no Ku contamination in the DNA-PK stock; otherwise, the binding profiles would be identical.
Previous work has shown that DNA-PK and Ku do not associate in the absence of DNA (41). To confirm this in the more sensitive SPR system, we immobilized Ku on an nitrilotriacetic acid-containing chip through a C-terminal histidine tag engineered on each of the polypeptides. We find no significant binding when DNA-PK is introduced under conditions similar to those in the studies described above (data not shown).
DNA-PK binding to the Ku:18-bp complex.
As mentioned above, the findings in Fig. 2 suggest that DNA-PK and Ku enter into a nonproductive complex for kinase activity when incubated with the 18-bp oligonucleotide. However, the other interpretation of these results is that Ku simply prevents DNA-PK from contacting DNA. We used a similar technique as with the 35-bp oligonucleotide in Fig. 7 to analyze whether or not DNA-PK bound to a Ku:18-bp complex.
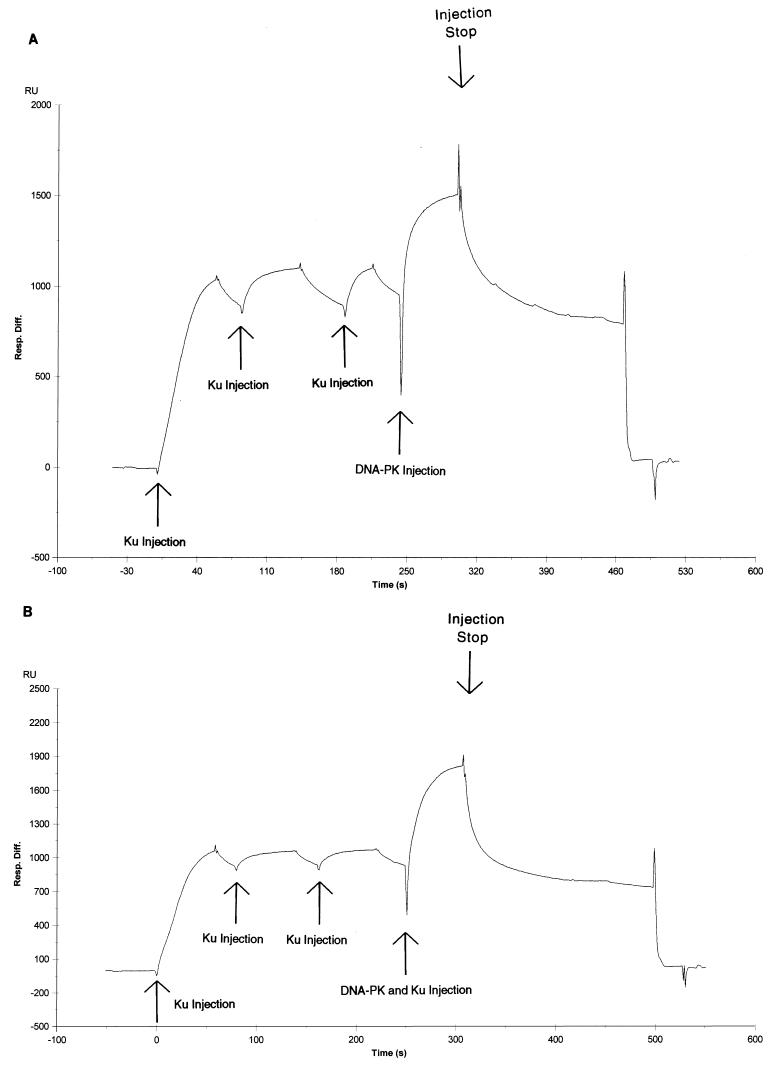
In Fig. 8, two sensorgrams for the DNA-PK:Ku:18-bp oligonucleotide are shown. In Fig. 8A, three consecutive injections with Ku are made before purified DNA-PK is injected. Under these conditions, Ku does achieve saturation; however, due to its weaker stability on the 18-bp oligonucleotide than on the 35-bp oligonucleotide, slightly more Ku dissociates from the DNA before the DNA-PK is injected. Despite this, a robust signal is achieved when DNA-PK is injected. This signal is comparable in intensity (900 response units [RU] versus 1,200 RU on chips with similar DNA amounts) to the signal seen with DNA-PK binding to the 35-bp oligonucleotide alone (Fig. 7). If Ku was blocking DNA-PK binding on the 18-bp oligonucleotide, one would expect to see a much lower signal given that Ku was saturated on the chip prior to DNA-PK injection. Though the nature of these conditions precludes our analysis of kinetic constants, it appears from the shape of the sensorgram that the complex is very unstable. However, the magnitude of the signal exceeds the maximum possible signal if DNA-PK was binding to the free DNA at saturating conditions. In SPR studies of DNA-PK at similar low DNA concentrations (data not shown), we find that the DNA-PK-generated signal is much lower than saturation. Thus, the signal that we observe in Fig. 8A comes from weak direct binding between DNA-PK and the Ku:18-bp oligonucleotide. However, to ensure that DNA-PK is binding to DNA complexed with Ku and not just free DNA, we injected a mixture of DNA-PK and Ku in Fig. 8B. The concentration of Ku is the same as for the previous saturating injections, while the concentration of DNA-PK is the same as that injected in the assay represented in Fig. 8A. We find that the ensuing signal is higher than the signal for DNA-PK alone. This finding demonstrates that DNA-PK and Ku were not competing for the small fraction of sites that were vacated after the last saturating Ku injection. We conclude that DNA-PK can form a weak nonproductive complex with Ku and short pieces of DNA. This corroborates the functional data in Fig. 2, where we find that Ku has a weak inhibitory effect on the kinase activity independent of its binding to DNA.
FIG. 8.
Binding of DNA-PK with the Ku:18-bp DNA fragment complex. (A) Complete sensorgram of sequential Ku (28.5 nM) and DNA-PK (2.4 nM) injections onto a sensor chip loaded with the 18-bp oligonucleotide. (B) Complete sensorgram of sequential Ku (28.5 nM) and DNA-PK (2.4 nM)–Ku (28.5 nM) injections onto a sensor chip loaded with the 18-bp oligonucleotide.
DISCUSSION
Using kinase assays, we show that DNA-PK has strong kinase activity independent of Ku over a wide range of DNA concentrations. Using SPR, we find that DNA-PK has high association and dissociation rates for binding DNA termini. Combining these methodologies, we find that Ku:DNA can stabilize DNA-PK in its active or productive configuration as a kinase. The productive DNA-PK in complex with Ku:DNA (called DNA-PK:Ku:DNA or the ternary complex) is shifted in half-maximal kinase activity to lower DNA concentrations. Furthermore, we find that on short DNA fragments, DNA-PK and Ku can complex on DNA in a nonproductive mode. We functionally determine the minimum DNA terminus length required for a productive complex. Based on these quantitative and qualitative insights, we propose a model for the order of events at DNA termini when DNA-PK and Ku are both present (see below).
DNA-PK interactions with DNA in the absence of Ku.
We have studied DNA-PK interaction with DNA in both its affinity for DNA and its activation by DNA in the absence of Ku. The kinase profiles yield some interesting data (Fig. 1). First, the maximum activities for the two profiles differ only marginally. The maximal activity in the presence of Ku is 1.6-fold higher than the maximal activity observed in absence of Ku. This finding confirms that DNA-PK has at least a functionally significant DNA binding domain. While previous studies demonstrated that DNA-PK could bind DNA termini directly (41), it was unclear whether this binding could activate the kinase function to the same extent that Ku does. Second, the DNA-PK activity alone on DNA never reaches the activity achieved with Ku. This small effect (1.6-fold) may be an allosteric one. Third, the kinase activity in the presence of Ku never falls to that of the activity without Ku. This means that the preference of DNA-PK for Ku:DNA versus free DNA is large. We discuss this aspect further below.
The sensorgram of DNA-PK binding to the 35-bp oligonucleotide presented in Fig. 6 defines the DNA binding affinities of DNA-PK. These are the first measurements of DNA-PK binding to DNA in equilibrium. This experiment clearly defines DNA-PK as a protein capable of binding to DNA alone under physiologic conditions. It must be stressed that there can be no Ku contamination that might be responsible for this binding activity because the DNA-PK and Ku complex has a completely different binding profile (Fig. 7). The extremely high association and dissociation rates explain much of the behavior of DNA-PK in different kinase assay conditions (see below).
The productive complex of DNA-PK and Ku.
In the presence of Ku, the concentration of DNA required to give maximum kinase activity drops 100-fold, from 5 μM to 50 nM. This profound effect could explain why previous studies (7) showed such large increases in stimulation by Ku. At concentrations of DNA between 1 to 10 nM, a large difference in activity is seen, but at higher concentrations this difference disappears.
Though the kinase profile with Ku spans only a range of 2 logs in DNA concentration, the kinase profile without Ku is much broader, spanning at least 3 logs. The binding study of DNA-PK alone (Fig. 6) clearly shows why the DNA-PK profile is so spread out. With extremely high rates of association and dissociation, the stability of DNA-PK alone on DNA is very poor. At low concentrations of DNA, the kinase may not be activated for sufficient time to allow the kinase steps of peptide binding and phosphorylation to take place. At high concentrations of DNA, the overall occupancy of the DNA binding site may be sufficient for efficient kinase activity. Though it shifts the plateau of maximum activity, Ku does not significantly change the concentration of DNA at which activity begins. This could be due to the similar binding affinity constants that Ku and DNA-PK possess, which indicates that the two proteins begin interaction with DNA at similarly low DNA concentrations.
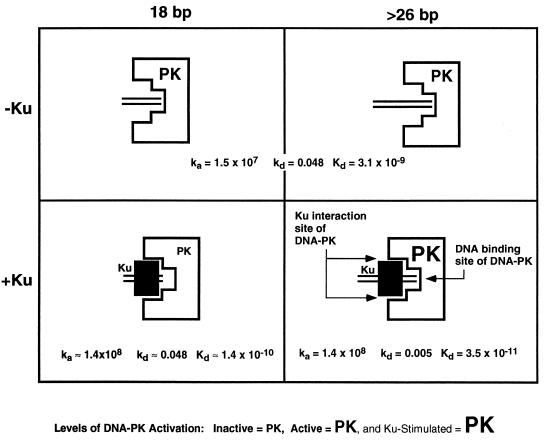
As mentioned, a final interesting feature of the kinase profiles is that the plateau of kinase activity with Ku does not decrease to the plateau without Ku at high DNA concentrations (Fig. 1). At the highest DNA concentration, the concentration of free DNA is 1,000 times greater than the concentration of Ku-bound DNA. It is surprising that the Ku effect (higher maximum activity) is not diluted out. This clearly indicates that DNA-PK has a much higher preference for Ku-bound DNA. A comparison of the sensorgrams of DNA-PK alone (Fig. 6) and DNA-PK with Ku on the 35-bp oligonucleotide (Fig. 7) illustrates why. In a mixed population of free DNA and Ku:DNA complexes, DNA-PK will very quickly find and form stable complexes with the Ku bound DNA. Because of its very high association (1.5 × 107 M−1 s−1) and dissociation rates (0.048 s−1) DNA-PK will encounter and dissociate from many free DNA molecules in a short period of time. However, when DNA-PK encounters Ku-bound DNA, the dissociation rate changes dramatically (5 × 10−3 s−1). In a very short period of time, DNA-PK can search through a large excess of free DNA to find Ku:DNA complexes. Once there, the stability of DNA-PK is 100 times greater (Fig. 9, _Kd_s at top right versus bottom right).
FIG. 9.
Summary of Ku and DNA-PK interactions with long and short DNA, depicting the different complexes that DNA-PK can form with DNA (parallel lines) in the presence or absence of Ku (filled rectangle). Depending on the length of the DNA, DNA-PK can be active or inactive for kinase activity in the presence of Ku. In the lower right panel, DNA-PK is larger, representing an increase in activity when it is productively associated with Ku.
The nonproductive complex of DNA-PK and Ku.
While DNA-PK can clearly bind DNA, it is unclear whether this activity is used when Ku is present, given that Ku is present in the cell in approximately a fivefold molar excess and given that Ku has a tighter equilibrium binding constant (1, 39a). The direct interaction of DNA-PK and DNA could account for the difference in phenotype for V(D)J recombination between cells lacking Ku (which fail to form both signal and coding joints) versus those lacking DNA-PK (which fail to form coding joints but still form signal joints) (25, 35–37). The data presented in Fig. 2 provide direct evidence that the DNA binding by Ku alone (without DNA-PK contact of the DNA) is not sufficient to activate the kinase activity. In kinase profiles with an oligonucleotide 18 bp in length, Ku inhibits kinase activity. This inhibition appears to involve two different phenomena. At DNA concentrations lower than the Ku concentration, there is an absolute inhibition of kinase activity. At DNA concentrations above the Ku concentration, the kinase activity is four- to fivefold less than expected for the concentration of free DNA, unbound by Ku (Fig. 2B). Only when the ratio of free to Ku-bound DNA reaches 100 to 1 does this inhibition disappear.
There are two potential mechanisms for Ku inhibition of kinase activity. Ku could directly compete for the DNA that DNA-PK requires for kinase activity. Ku could also be a noncompetitive inhibitor by binding to DNA-PK and preventing it from productively interacting with DNA.
Our studies using the kinase assay and the SPR analysis indicate that there must be some extent of inhibition due to generation of nonproductive complexes. When the total DNA that Ku could bind has been subtracted from the kinase profile, the kinase activity is still less than that seen in the profile without Ku (Fig. 2B). Because Ku can bind only one DNA oligomer at a time (41), it cannot further change the free DNA concentration. Therefore, in order for Ku to inhibit, it must be interacting with DNA-PK. However, this complex is clearly not as stable as a complex with longer DNA. Challenging the nonproductive complex with more DNA appears to lead to some DNA-PK binding to free DNA, thereby resulting in kinase activity. If the nonproductive complex were extremely stable, we would not see activity regardless of how much free DNA was added. In contrast, the productive complex remains intact, despite a 1,000-fold excess of free DNA (see above) (Fig. 1). Thus, the nonproductive complex appears to be considerably less stable than the productive complex.
The sensorgrams in Fig. 7 and 8 establish that DNA-PK forms a strong complex with Ku and the 35-bp oligonucleotide whereas it forms a weaker complex with the Ku:18-bp oligonucleotide. Thus, the observations from both the kinase activity experiments and the SPR experiments indicate that two complexes exist: a transient, nonproductive complex that occurs on short DNA fragments, and a stable, kinase-active complex that occurs on longer DNA fragments.
Functional footprinting of DNA:DNA-PK and DNA:Ku:DNA-PK.
The relationship between Ku and DNA-PK is further defined by determining the minimum length of DNA required for kinase activity (Fig. 3). Several groups have footprinted Ku bound on DNA (2, 16, 24). In this study, we define the functional footprint of DNA-PK:DNA and of Ku:DNA-PK:DNA. Between 22 and 26 bp, the Ku:DNA-PK:DNA complex begins to have significant kinase activity. The length of DNA may be critical in determining with which DNA lesions this complex can interact. This may be of greater importance when considering that the internucleosomal distance is 20 to 80 bp. A double-strand break in this region may leave only a very short stretch of DNA for recognition. The lengths of DNA for DNA-PK binding and activation defined here may be helpful in considering dsDNA breaks in such regions (Fig. 9).
Physiologic functions of DNA-PK independent of Ku.
We have examined the activities of DNA-PK without Ku previously (41) and in this study. We have conducted several assays to check for the presence of Ku and found no evidence of Ku contamination, based on results of Western blot analyses, cross-linking assays, selective immunoprecipitation assays, atomic force microscopy, electrophoretic mobility shift assays, and now SPR analysis. The fraction of the DNA-PK molecules that bind DNA in the absence of Ku is nearly 2 orders of magnitude higher than the upper limit of any possible contamination.
Recently, one group has claimed that DNA-PK in the absence of Ku has no kinase activity in salt conditions that approximate those in the nucleus (19). However, we find clear evidence of kinase activity by DNA-PK in the absence of Ku (Table 1). Furthermore, the study by Hammarsten and Chu (19) shows that the kinase activity of DNA-PK is inhibited by the presence of Ku when a 22-bp DNA fragment is used as the activator. The authors, however, offer no explanation as to the nature of the inhibition or how this phenomenon might influence the physiologic activity of DNA-PK. Our studies examine the interaction between DNA-PK and Ku in the context of short DNA fragments in a much more detailed manner by examining both a functional characteristic (kinase activity) and structural characteristic (binding affinity constants) of the complex. Our experiments show that while DNA-PK in the presence of Ku on short DNA fragments has no kinase activity, there does exist a complex of slightly lower stability of DNA-PK and Ku on the DNA. Our much more extensive study provides a picture that includes both productive and nonproductive complexes at physiologic ionic strength and establishes the profile of lengths over which the transition from nonproductive to productive occurs.
Model for the interaction of DNA-PK with Ku and DNA.
Based on the data that we have derived, we propose the following. First, DNA-PK binds and is activated as a kinase by binding to DNA termini, regardless of DNA length (Fig. 9, top right and left). Second, if Ku is present, it can stimulate DNA-PK by associating with DNA-PK and lowering its kinetic dissociation constant and by increasing its association constant (Fig. 9, lower right). Third, if the DNA is too short (<26 bp), then Ku, DNA-PK, and DNA may associate, but the DNA is too short to stimulate DNA-PK (Fig. 9, lower left).
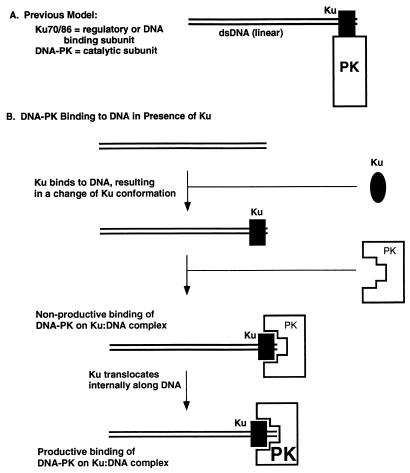
We have incorporated these observations into a model for the function of Ku and DNA-PK (Fig. 10B), which is compared with the model proposed by Gottlieb and Jackson (16) (Fig. 10A). Ku is present in the cell in excess over DNA-PK. Ku also binds to DNA termini with an affinity higher than that of DNA-PK alone. Upon binding to DNA termini, Ku must change conformation. This is the only explanation for why free Ku and free DNA-PK fail to associate but bound Ku associates with DNA-PK (41). After Ku binds to the terminus, DNA-PK binds to the Ku:DNA binary complex. We propose that initially, DNA-PK binds to the Ku:DNA complex in a nonproductive mode, until Ku diffuses internally along the DNA terminus to permit direct DNA-PK:DNA association to yield a Ku:DNA-PK:DNA ternary complex.
FIG. 10.
Schematic model for the assembly and activity of the Ku:DNA-PK complex with a dsDNA end. (A) Previous model (16) for DNA-PK; (B) model based on data from this study, where Ku and DNA-PK bind separately and sequentially. The model depicts Ku with an oval when it is free and a rectangle when it is bound to DNA. This is based on the fact that free Ku does not associate with DNA-PK to any detectable extent, whereas bound Ku does associate with DNA-PK. The level of kinase activity of DNA-PK is reflected by the size of the letters “PK” as in Fig. 9.
This model would explain the inhibitory effect of Ku on DNA-PK activity on short DNA fragments. On short DNA, the DNA-PK:Ku:DNA interaction results in inhibition (Fig. 2) because the configuration is that of the nonproductive complex (Fig. 9, lower left). In these cases, the DNA is not long enough to activate the DNA-PK because Ku is occupying too much of the DNA length. We do not know if there is any direct contact between the DNA and DNA-PK at this step in the nonproductive configuration. On longer DNA, this association results in activation (Fig. 1) because the configuration is that of the active complex (Fig. 9, lower right).
It seems likely that DNA-PK does not recognize DNA alone initially if Ku is present; rather it binds to some part of Ku or both Ku and DNA in the Ku:DNA complex. Figure 1 shows that once the plateau is reached, a 100-fold additional excess of Ku-free DNA does not change the level of kinase activity. If DNA-PK initially made contact with DNA, the plateau in the Ku-stimulated curve should eventually fall back down to that in the Ku-absent case as the Ku:DNA complexes are progressively diluted out by the free DNA. The fact that this does not happen in the kinase assays indicates that if Ku is present, it recruits DNA-PK to the DNA. The extent of this recruitment is reflected by the higher kinetic on rate of DNA-PK binding to DNA when Ku is absent than when it is present. The fact that Ku affects both the on and off kinetic rates supports an initial Ku or, at least, Ku:DNA contact. Otherwise, if the DNA were the only initial contact, then only the off rate would be affected.
Once on the DNA, a second aspect to the Ku:DNA-PK:DNA interaction is that the kinetic off rate of DNA-PK from the DNA is lower when Ku is present than when it is absent. Hence, Ku both recruits and stabilizes the binding of DNA-PK to the DNA terminus.
Based on our previous biochemical atomic force microscopy study (41) and the biochemical work presented here, the model for DNA-PK and Ku at the terminus proposes that Ku takes up a position internal to the terminus relative to DNA-PK (Fig. 10B). Neither we nor others (6) have observed movement of DNA-PK to internal positions from the DNA terminus, whereas Ku is able to do this (10, 31, 41).
We believe that the nonproductive mode may be of key importance for end occupancy by Ku and DNA-PK. This may permit the further configuration of proteins and the DNA ends themselves along the path toward resolution. Upon movement of the complex internally, perhaps after moving nucleosomes further internally, the kinase is activated by the DNA end, and this may actually cause the complex to eventually disassociate. There are some data suggesting that the productive mode of the complex phosphorylates itself, ultimately leading to its inactivation (7). Hence, the nonproductive mode may be a critical step in nonhomologous DNA end joining.
Concluding remarks.
These studies help define the functional relationships of Ku and DNA-PK on relevant lengths of DNA. These studies further illustrate that DNA-PK can have kinase activity in the absence of Ku. In the mammalian cell nucleus, the ratio of Ku to DNA-PK may be about 5 to 1 (1). At this ratio, we predict that Ku will initially bind, translocate internally, and recruit DNA-PK; DNA-PK could then bind DNA directly and be activated.
The DNA lengths examined here cover those relevant in DNA damage. In the internucleosomal regions, dsDNA breaks would leave regions that are in many cases shorter than 30 bp in length from the DNA end to the nearest nucleosome. The studies here will be useful as we consider the order of events at a dsDNA break.
ACKNOWLEDGMENTS
We thank Robert Tracy and Yunmei Ma for comments on the manuscript. We are indebted to Shirley Demer at Biacore for advice.
This work was supported by NIH grants to M.R.L., who is a Leukemia Society of America Scholar and who is the Rita & Edward Polusky Basic Cancer Research Professor.
REFERENCES
- 1.Anderson C W, Carter T H. The DNA-activated protein kinase-DNA-PK. In: Jessberger R, Lieber M R, editors. Molecular analysis of DNA rearrangements in the immune system. Heidelberg, Germany: Springer-Verlag; 1996. pp. 91–112. [Google Scholar]
- 2.Blier P R, Griffith A J, Craft J, Hardin J A. Binding of Ku protein to DNA. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 3.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 4.Brush G, Anderson C W, Kelly T J. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during SV40 DNA replication. Proc Natl Acad Sci USA. 1994;91:12520–12524. doi: 10.1073/pnas.91.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter T H, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cary R B, Peterson S R, Wang J, Bear D G, Bradbury E M, Chen D J. DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci USA. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan D W, Lees-Miller S P. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 8.Chan D W, Mody C H, Ting N S, Lees-Miller S P. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem Cell Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]
- 9.Critchlow S, Bowater R, Jackson S P. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 10.deVries E, van Driel W, Bergsma W G, Arnberg A C, van den Vliet P C. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J Mol Biol. 1989;208:65–78. doi: 10.1016/0022-2836(89)90088-0. [DOI] [PubMed] [Google Scholar]
- 11.Dvir A, Stein L Y, Calore B L, Dynan W S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 12.Falzon M, Fewell J, Kuff E L. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 13.Finnie N J, Gottlieb T, Blunt T, Jeggo P, Jackson S P. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher R J, Fluach M, Casa-Finet J, Erckson J W, Kondoh A, Bladen S, Fishewr C, Watson D, Papas T. Real-time DNA binding measurements of the ETSL recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994;3:257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaccia A, Weinstein R, Hu J, Stamato T D. Cell cycle-dependent repair of double-strand DNA breaks in a gamma-ray-sensitive Chinese hamster cell. Somatic Cell Mol Genet. 1985;11:485–491. doi: 10.1007/BF01534842. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb T, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–42. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 17.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson T E, Mann M, Lieber M R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 18.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson S P. The recognition of DNA damage. Curr Opin Genet Dev. 1996;6:19–25. doi: 10.1016/s0959-437x(96)90005-2. [DOI] [PubMed] [Google Scholar]
- 21.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 22.Jonsson U, Fajerstam L, Roos H, Ronnberg J, Sjolander S, Stenberg E, Stahlberg R, Urbaniczky C, Ostlin H, Malmqvist M. Surface plasmon resonance and microfluidics for real time biospecific interaction analysis. BioTechniques. 1991;11:520–527. [PubMed] [Google Scholar]
- 23.Kirschgessner C, Patel C, Evans J, Cuomo C, Fried L, Carter T, Oettinger M, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1185. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 24.Knuth M W, Gunderson S I, Thompson N E, Strasheim L A, Burgess R R. Purification and characterization of proximal sequence element-binding protein 1, a transcription activating protein related to Ku and TREF that binds the proximal sequence element of the human U1 promoter. J Biol Chem. 1990;265:17911–17920. [PubMed] [Google Scholar]
- 25.Kulesza, P., and M. R. Lieber. DNA-PK is essential only for coding joint formation in V(D)J recombination. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 26.Lees-Miller S P, Chen Y-R, Anderson C W. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber M R, Grawunder U, Wu X, Yaneva M. Tying loose ends: roles of Ku and DNA-dependent protein kinase in the repair of double-strand breaks. Curr Opin Genet Dev. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- 29.Mimori T, Hardin J A. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 30.Pan Z, Amin A A, Gibbs E, Niu H, Hurwitz J. Phosphorylation of the p34 subunit of human single-stranded-DNA-binding protein in cyclin A-activated G1 extracts is catalyzed by cdk-cyclin A complex and DNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:8343–8347. doi: 10.1073/pnas.91.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang D, Yoo S, Dynan W S, Jung M, Dritschilo A. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997;57:1412–1415. [PubMed] [Google Scholar]
- 32.Pergola F, Zdzienicka M Z, Lieber M. V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol Cell Biol. 1993;13:3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Loss of the catalytic subunit of DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of S. cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smider V, Rathmell W K, Lieber M, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant mammalian cells by Ku. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 36.Taccioli G, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 37.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 38.Taccioli G E, Rathbun G, Shinkai Y, Oltz E M, Cheng H, Whitmore G, Stamato T, Jeggo P, Alt F W. Activities involved in V(D)J recombination. Curr Top Microbiol Immunol. 1992;182:107–114. doi: 10.1007/978-3-642-77633-5_13. [DOI] [PubMed] [Google Scholar]
- 39.Teo S H, Jackson S P. Identification of S. cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.West, R. B., M. Yaneva, and M. R. Lieber. Unpublished data.
- 40.Wilson T E, Grawunder U, Lieber M R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 41.Yaneva M, Kowaleski T, Lieber M R. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]