Targeting a Polyepitope Protein Incorporating Multiple Class II-Restricted Viral Epitopes to the Secretory/Endocytic Pathway Facilitates Immune Recognition by CD4+ Cytotoxic T Lymphocytes: a Novel Approach to Vaccine Design (original) (raw)
Abstract
The role of CD4+ and CD8+ cells in the generation of an effective immune response against viral infections is well established. Moreover, there is an increasing realization that subunit vaccines which include both CD4+- and CD8+-T-cell epitopes are highly effective in controlling viral infections, as opposed to those which are designed to activate a CD8+- or CD4+-T-cell response alone. One of the major limitations of epitope-based vaccines designed to stimulate virus-specific CD4+ T cells is that endogenously expressed class II-restricted minimal cytotoxic-T-lymphocyte (CTL) epitopes are poorly recognized by CD4+ CTLs. In the present study we attempted to enhance the efficiency of class II-restricted endogenous presentation of minimal class II-restricted CTL epitopes by specifically targeting a polyepitope protein to class II processing compartments through the endosomal and/or lysosomal pathway. A significantly enhanced stimulation of virus-specific CD4+-T-cell clones by antigen-presenting cells (APC) expressing the recombinant polyepitope protein targeted to the endocytic/secretory pathway was readily demonstrated in cytotoxicity assays. In addition, in vitro activation of Epstein-Barr virus- and influenza virus-specific CD4+ memory CTLs by the recombinant constructs encoding the polyepitope protein, specifically targeted to the lysosomal compartment, was also demonstrated. The enhanced stimulatory capacity of APC expressing a lysosome-targeted polyepitope protein has important implications for vaccine design.
There is now increasing evidence to suggest that both CD4+ and CD8+ T cells are critical for the generation of an effective immune response against intracellular pathogens. Although both CD4+ and CD8+ T cells recognize nonnative forms of the antigen in association with major histocompatibility complex (MHC) molecules, the presentation of antigen to these two types of T lymphocytes occurs through distinct pathways (24). In fact, the disparity in antigen presentation to these T cells is not due to processing differences but rather reflects the differences in the capacities of class I and class II molecules to bind antigenic determinants in an intracellular compartment. Indeed, earlier studies have shown that for processing and interaction with MHC class II molecules, antigen expressed de novo needs to be targeted to an endosomal or lysosomal compartment (5). There are two major pathways by which antigens are targeted to these compartments. The traditional pathway involves the phagocytosis or endocytosis of exogenous antigens, followed by degradation by acid proteases in the endosomal or lysosomal compartments (3, 8, 26, 41). On the other hand, class II-restricted presentation of endogenously synthesized proteins mainly involves membrane antigens which are thought to enter the endosomal or lysosomal pathway by internalization from the cell surface (11). Although, in certain experimental systems, cytoplasmic and nuclear proteins may also enter this endogenous pathway, generally these proteins are targets for the class I processing pathway (9, 14, 20, 27).
One of the major limitations of the epitope-based vaccines designed to stimulate virus-specific CD4+ T cells is that endogenously expressed class II-restricted minimal cytotoxic T-lymphocyte (CTL) epitopes are poorly recognized by CD4+ CTLs (2, 35, 38). Based on these observations, we reasoned that a molecular approach that directly routes these epitopes into the MHC class II pathway, such as the endocytic or lysosomal compartments, might facilitate endogenous presentation to CD4+ T cells. The lysosome-associated membrane protein (LAMP-1) and the invariant chain (Ii) are transmembrane proteins which are localized predominantly in the lysosomes and endosomes, respectively. The cytoplasmic domains of these proteins contain specific targeting signals that mediate their translocation to the specific compartments. We therefore designed a chimeric polyepitope construct capable of encoding multiple class II-restricted CTL epitopes from Epstein-Barr virus (EBV) and influenza virus linked to the cytoplasmic and/or transmembrane domains of LAMP-1 and the Ii protein, with the aim of targeting the epitopes to the endosomal and lysosomal compartments. The data presented in this study clearly demonstrate that if the endogenously synthesized polyepitope protein is targeted to the endocytic/secretory pathway, processing and presentation of all the epitopes are dramatically enhanced. More importantly, minimal epitope sequences, without any natural flanking sequences, were adequate for efficient stimulation of the virus-specific memory CTL response, a result that has important implications for epitope-based vaccine design.
MATERIALS AND METHODS
Establishment and maintenance of cell lines.
EBV-transformed lymphoblastoid cell lines (LCLs) were established from seropositive donors by exogenous virus transformation of peripheral B cells by using the B95.8 or Ag876 virus isolate (25). All cell lines were routinely maintained in RPMI 1640 containing 2 mM glutamine, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml plus 10% fetal calf serum (FCS) (growth medium).
Generation of recombinant vaccinia virus constructs.
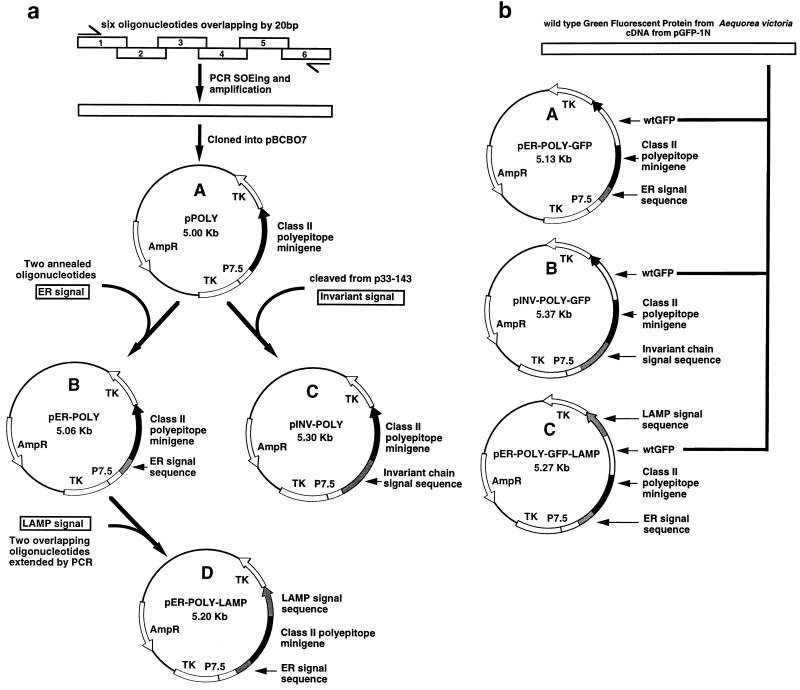
The generation of recombinant vaccinia virus constructs encoding either multiple class II-restricted epitopes as a polyepitope protein or the polyepitope protein fused to an endoplasmic reticulum (ER) (adenovirus E1A ER signal), endosomal (invariant-chain signal), or lysosomal (LAMP-1 signal) signal sequence is summarized in Fig. 1. The minigene construct which expresses multiple class II-restricted epitopes as a polyepitope protein (Fig. 1a) was designed by splicing six oligonucleotides together as described previously (37). A total of six different MHC class II-restricted CTL epitopes from EBV and influenza virus were included in this construct (Table 1). This minigene was cloned into pBCB07 (1) by using _Bam_HI and _Sal_I restriction enzymes to generate the plasmid pPOLY.
FIG. 1.
Construction of the class II polyepitope plasmids. (a) A class II polyepitope minigene was designed and constructed by splicing six overlapping oligonucleotides together by PCR and splicing by overlap extension (SOEing). The minigene was then cloned into pBCB07 to generate pPOLY. The plasmid pPOLY was then modified with DNA which coded for three different transport signal sequences. A DNA fragment flanked with appropriate restriction sites which codes for the adenovirus E1A ER signal sequence was generated by annealing two oligonucleotides together. This DNA fragment was then cloned into pPOLY at the 5′ end of the minigene to generate pER-POLY. A DNA fragment which codes for the invariant-chain signal sequence (amino acids 1 to 71) was removed from the invariant-chain cDNA in p33-143 and cloned into pPOLY at the 5′ end of the minigene to generate pINV-POLY. A DNA fragment which codes for the human LAMP-1 lysosomal signal sequence (amino acids 354 to 389) was constructed by extending two overlapping oligonucleotides. This DNA fragment was cloned into pER-POLY at the 3′ end of the minigene to generate pER-POLY-LAMP. These plasmids were subsequently used to generate four recombinant vaccinia viruses, Vacc.POLY, Vacc.ER-POLY, Vacc.INV-POLY, and Vacc.ER-POLY-LAMP, respectively, by marker rescue recombination. TK, thymidine kinase. wtGFP cDNA from the jellyfish A. victoria was cloned into the coding sequence of the class II polyepitope protein. Briefly, a DNA fragment containing the complete wtGFP cDNA except for the stop codon was removed from pGFP-1N (Clonetech) and cloned in frame at both ends into the class II polyepitope coding sequence in the plasmids pER-POLY, pINV-POLY, and pER-POLY-LAMP to generate the plasmids pER-POLY-GFP, pINV-POLY-GFP, and pER-POLY-GFP-LAMP, respectively. These plasmids were subsequently used to generate three recombinant vaccinia viruses, Vacc.ER-POLY-GFP, Vacc.INV-POLY-GFP, and Vacc.ER-POLY-GFP-LAMP, respectively, by marker rescue recombination.
TABLE 1.
CTL epitopes contained in the class II polyepitope protein
| Source (amino acids) | Cognate epitope | HLA restriction | Reference |
|---|---|---|---|
| EBV EBNA2 (280–290) | TVFYNIPPMPL | DQ2/DQ7 | 20 |
| Influenza virus HA3 (307–318) | PKYVKQNTLKLAT | DR1 | 35 |
| Influenza virus matrix (18–29) | GPLKAEIAQRLE | DR1 | 24 |
| Influenza virus HA2 (129–140) | VKILPKDRWTQH | DR11 | 24 |
| EBV BHRF1 (45–57) | TVVLRYHVLLEEI | DR4 Dw10 | 33 |
| EBV EBNA1 (515–527) | TSLYNLRRGTALA | DR1 | 17 |
To target the polyepitope protein to the endocytic/secretory pathway, the polyepitope gene in pPOLY was fused to the DNA sequences encoding either the ER, endosomal, or lysosomal signal sequence (Fig. 1a). Briefly, a synthetic DNA sequence coding for the adenovirus E1A ER signal sequence (28) was cloned into pPOLY to generate pER-POLY. A DNA fragment coding for the endosomal signal sequence (amino acids 1 to 71) was cleaved from the invariant-chain cDNA (34) and subcloned into pPOLY to generate pINV-POLY. A synthetic DNA sequence coding for the lysosomal signal sequence (amino acids 354 to 389) from human LAMP-1 (13) was constructed and cloned into the plasmid pER-POLY to generate the plasmid pER-POLY-LAMP. These plasmids were then used to generate the recombinant vaccinia viruses Vacc.POLY, Vacc.ER-POLY, Vacc.INV-POLY, and Vacc.ER-POLY-LAMP by marker rescue recombination as described previously (4).
To examine the localization and expression of the class II polyepitope protein targeted to the endocytic/secretory pathway, the wild-type green fluorescent protein (wtGFP) cDNA from the jellyfish Aequorea victoria (6, 30) was cloned into the coding sequence of the polyepitope (Fig. 1b). Briefly, wtGFP cDNA was excised from the plasmid pGFP-1N (Clonetech) and subcloned into pER-POLY, pINV-POLY, and pER-POLY-LAMP to create pER-POLY-GFP, pINV-POLY-GFP, and pER-POLY-GFP-LAMP, respectively. These plasmids were then used to generate the recombinant vaccinia viruses Vacc.ER-POLY-GFP, Vacc.INV-POLY-GFP, and Vacc.ER-POLY-GFP-LAMP as described above.
The recombinant vaccinia virus constructs encoding the EBV nuclear antigen 1 (EBNA1), EBNA2, and BHRF1 antigens of EBV and a vaccinia virus construct made by insertion of the pSC11 vector alone and negative for thymidine kinase (Vacc.TK−) have been previously described (16, 21, 39). In addition, a recombinant vaccinia virus construct expressing influenza virus hemagglutinin (Vacc.HA) was also used in the study (7).
CTL clones and peptide epitopes.
The MHC class II-restricted, EBV-specific CTL clones used in this study were LC27 (EBNA2 specific; HLA DQ2/DQ7 restricted), DM2 (EBNA1 specific; HLA DR1 restricted), and SBAg1 (BHRF1 specific; HLA DR4Dw10 restricted). The specificities of these clones have been defined at the peptide epitope level: LC27 recognizes the minimal epitope TVFYNIPPMPL (residues 280 to 290) (20), DM2 recognizes the minimal epitope TSLYNLRRGTALA (residues 515 to 527) (17), and SBAg1 recognizes the minimal epitope TVVLRYHVLLEEI (residues 45 to 57) (33). These CTL clones were propagated in growth medium supplemented with recombinant interleukin-2 and MLA supernatant.
Cytotoxicity assay with recombinant vaccinia virus-infected targets.
Target cells were infected with recombinant vaccinia virus at a multiplicity of infection (MOI) of 10:1 for 1 h at 37°C. After overnight infection, cells were washed with growth medium, incubated with 51Cr for 90 min, and used as targets in a standard 5-h 51Cr-release assay (19, 25). In some experiments, monoclonal antibodies (MAb) specific for the nonpolymorphic determinants on MHC class II (IVA12) or class I (W6/32) antigens were added in the CTL assays to confirm the MHC restriction for the CTL clones.
To identify the processing pathway utilized by the class II-restricted epitopes, recombinant vaccinia virus-infected target cells were pretreated with chloroquine (20). Briefly, target cells were initially incubated in growth medium supplemented with chloroquine (80 μmol/ml) for 4 h at 37°C. Following incubation, these cells were washed and resuspended in growth medium supplemented with chloroquine (20 μmol/ml) and used as targets in a standard 5-h 51Cr release assay (25).
To verify the endogenous presentation of the epitopes through the class II pathway, two different sets of experiments were carried out. First, vaccinia virus constructs were inactivated by UV irradiation before infecting target cells (18). Briefly, these vaccinia virus constructs were treated with UV light in an open petri dish on ice with 500 mJ of short-wave UV light in a UV cross-linker apparatus (Bio-Rad, Richmond, Calif.). In the second set of experiments, unlabelled Vacc.EBNA2-infected LCL cells were mixed with 51Cr-labelled LCL cells at a ratio of 1:1 and then exposed to specific CTLs in a standard CTL assay.
Activation of memory CTL responses with recombinant vaccinia virus.
Unfractionated mononuclear cells from a donor (HLA A3, A23, B35, B44, DR1 DRW11 DQW1 DQW3 DRW52) were infected in vitro with Vacc.ER-POLY-LAMP at an MOI of 0.01:1 as described previously (21, 22, 36). After 10 days of culture in growth medium, these cells were used as polyclonal effectors in a standard 51Cr release assay against peptide-sensitized or vaccinia virus-infected autologous target cells.
Immunofluorescence assays.
To examine the localization and expression of the class II polyepitope protein targeted to different compartments of the cell, HeLa cells were infected with various recombinant vaccinia viruses at an MOI of 4:1 for 1 h at 37°C. Following incubation at 37°C for 6 h, these cells were processed for immunofluorescence as described previously (21). Briefly, vaccinia virus-infected cells were fixed, permeabilized, and then incubated with rabbit anti-GFP polyclonal serum (diluted 1/400 in 1% FCS–phosphate-buffered saline [PBS]) for 1 h at room temperature. The cells were extensively washed with 1% FCS–PBS and then incubated for 1 h at room temperature with fluorescein isothiocyanate-conjugated sheep anti-rabbit immunoglobulin G (Silenus, Melbourne, Australia). The cells were washed extensively with 1% FCS–PBS and then examined under a fluorescence microscope in the presence of _n_-propyl gallate.
RESULTS AND DISCUSSION
Endogenously synthesized polyepitope protein targeted to the endocytic/lysosomal compartment facilitates presentation of multiple epitopes to CD4+ T cells.
To determine the effect of endosomal or lysosomal targeting of a polyepitope protein on the processing efficiency of class II-restricted epitopes, a panel of well-characterized CD4+ CTL clones were used as effector cells in a standard 51Cr release assay. LCLs were infected with recombinant vaccinia virus vectors encoding either the polyepitope protein (Vacc.POLY) or the polyepitope protein fused to an endosomal (Vacc.INV-POLY) or lysosomal (Vacc.ER-POLY-LAMP) targeting signal sequence. It is important to mention here that LAMP signal sequence-mediated transport of the polyepitope protein to the lysosomal compartment is dependent on efficient translocation to the secretory pathway by an ER translocation signal sequence. To ensure that the polyepitope construct is targeted to the lysosomal compartment, an N-terminal ER signal sequence was included in the construct. Target cells infected with recombinant vaccinia virus constructs encoding the polyepitope protein fused to an ER signal sequence (Vacc.ER-POLY) alone and full-length viral antigens (Vacc.EBNA2, Vacc.EBNA1, or Vacc.BHRF1) were used as controls in the assay. Targeting of the polyepitope construct to the endosomal or lysosomal pathway was based on the earlier observations that endogenous processing of antigens through the class II pathway requires translocation of these antigens to an endosomal or lysosomal compartment.
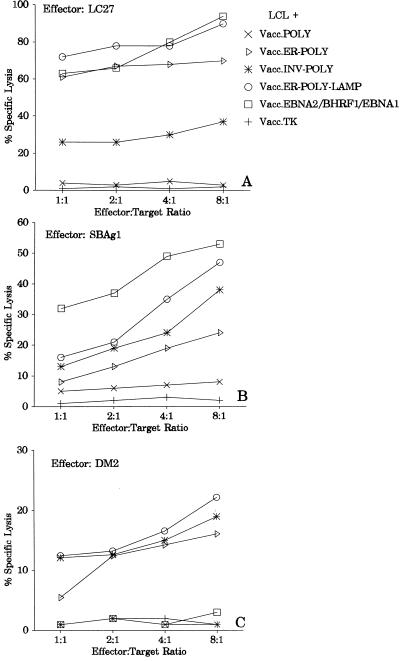
The data presented in Fig. 2 clearly demonstrate that target cells infected with Vacc.POLY were poorly recognized by all three CTL clones (LC27, SBAg1, and DM2). However, translocation of this polyepitope construct to the endosomal or lysosomal compartment significantly increased the levels of CTL lysis. The level of CTL lysis of target cells infected with Vacc.ER-POLY-LAMP was consistently higher than that of Vacc.INV-POLY-infected targets (Fig. 2). This CTL lysis was inhibited in the presence of MAb IVA12 (anti-HLA DR, DP, and DQ) (Fig. 3). Interestingly, enhanced presentation of CTL epitopes was also seen in target cells infected with Vacc.ER-POLY (Fig. 2), suggesting that translocation of minimal epitopes into the ER compartment also facilitates endogenous presentation to CD4+ T cells.
FIG. 2.
Endogenously synthesized polyepitope protein targeted to the endocytic/lysosomal compartment facilitates presentation of multiple epitopes to CD4+ T cells. Autologous LCL cells infected with different vaccinia virus recombinants (Vacc.POLY, Vacc.ER-POLY, Vacc.INV-POLY, Vacc.ER-POLY-LAMP, Vacc.EBNA2, Vacc.BHRF1, Vacc.EBNA1, and Vacc.TK−) were used as targets in standard CTL assays. CTL clones used in these assays were LC27 (EBNA2-specific), SBAg1 (BHRF1-specific), and DM2 (EBNA1-specific). LC/Ag876, SB29f, and DM/B95.8 LCLs were used as host cells for vaccinia virus constructs for LC27, SBAg1, and DM2 CTL clones, respectively. The LC/Ag876 LCL is not recognized by LC27, since this cell line is infected with a type 2 EBV which expresses a variant EBNA2 epitope sequence. SB29f LCL cells are negative for BHRF1 antigen, while the EBNA1 epitope is not endogenously processed by DM/B95.8 LCL cells, and thus these cells are not recognized by SBAg1 and DM2 CTL clones, respectively.
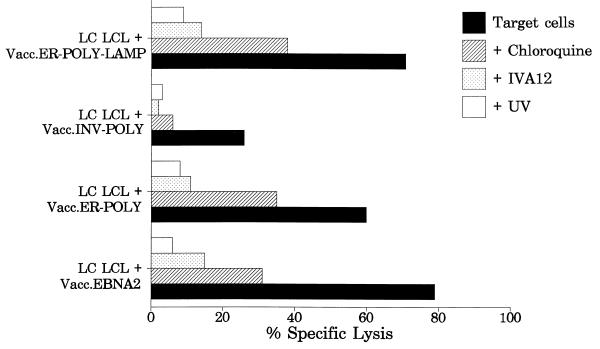
FIG. 3.
Effect of chloroquine and UV treatment of vaccinia virus constructs on CTL recognition of target cells. Recombinant vaccinia virus-infected LC/Ag876 LCL cells were pretreated with chloroquine (for details, see Materials and Methods) and then exposed to LC27 CTL. To determine whether presentation of the EBNA2 epitope requires de novo endogenous synthesis of the viral protein and is not exogenously processed, LC/Ag876 LCL cells infected with UV-inactivated Vacc.EBNA2, Vacc.ER-POLY, Vacc.INV.POLY, and Vacc.ER-POLY-LAMP were exposed to the LC27 clone. In addition, vaccinia virus-infected LC/Ag876 LCL cells were also exposed to LC27 CTLs with or without the addition of MAb IVA12 (anti-HLA DR, DP, and DQ) (1:20-diluted ascites). An effector-to-target ratio of 4:1 was used in the assay.
One of the surprising results from these experiments relates to the endogenous presentation of the EBNA1 epitope. Earlier studies have shown that although EBNA1 includes potential CTL epitopes which can be presented by class II molecules, these epitopes are not endogenously processed and presented by virus-infected cells. The data presented in Fig. 2C show that the translocation of the EBNA1 epitope to the endosomal/lysosomal compartment significantly improved the endogenous presentation of this epitope. Consistent with our earlier observations, this epitope is not endogenously processed by target cells infected with either Vacc.EBNA1 or Vacc.POLY.
Earlier studies have shown that conventional class II antigen presentation requires assembly of the peptide epitope with class II molecules in an acidified vacuolar compartment and is therefore sensitive to lysosomotropic agents that disrupt acidification of endosomes (5, 12, 24). We therefore tested whether the presentation of the CTL epitopes, targeted to the endocytic/secretory compartments, was dependent on this pathway by preventing acidification of the endosomal compartment with chloroquine treatment. Treatment of Vacc.ER-POLY-, Vacc.INV-POLY-, and Vacc.ER-POLY-LAMP-infected target cells with chloroquine significantly reduced the level of CTL lysis by the clone LC27 (Fig. 3). A similar effect on the presentation of other CTL epitopes was also seen following treatment of target cells with chloroquine (data not shown).
To exclude the possibility that the observed CTL recognition is due to peptides present in the vaccinia virus stocks, target cells were infected with vaccinia virus constructs that had been inactivated by irradiation with UV light. As shown in Fig. 3, target cells infected with UV-treated vaccinia virus were not lysed by the CTLs. Moreover, premixing of 51Cr-labelled target cells with recombinant vaccinia virus-infected cells showed no lysis of target cells (data not shown). Thus, de novo synthesis of the CTL epitopes from the polyepitope transcripts was required for processing and presentation of this epitope.
Activation of virus-specific memory CTL response by Vacc.ER-POLY-LAMP.
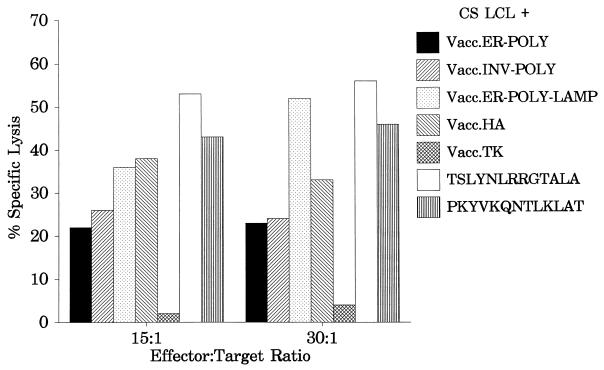
The results presented above clearly demonstrate that processing of endogenously synthesized class II-restricted epitopes can be significantly enhanced by targeting a polyepitope protein to the lysosomal compartment. A series of experiments were carried out to determine whether this polyepitope construct could be used to stimulate a memory CTL response in vitro from peripheral blood lymphocytes. Unfractionated mononuclear cells from an HLA DR1-positive donor were infected with Vacc.ER-POLY-LAMP at an MOI of 0.01:1 for 10 days, and the resulting polyclonal CTLs were then used as effectors against peptide-sensitized or vaccinia virus-infected autologous LCLs in a standard CTL assay. The data presented in Fig. 4 clearly demonstrate that Vacc.ER-POLY-LAMP was capable of activating CTL responses to both EBV and influenza virus epitopes. Consistent with the data presented in Fig. 2C, a strong EBNA1-specific CTL response was noticed following in vitro stimulation with Vacc.ER-POLY-LAMP (Fig. 4). It is important to mention here that in vitro stimulation with Vacc.EBNA1 or Vacc.POLY failed to stimulate an EBNA1-specific CTL response from the HLA DR1-positive donor (data not shown). These results suggest that direct LAMP-1-mediated lysosomal targeting of the EBNA1 epitopes could be used for stimulating CD4+-CTL responses in a vaccine designed to control EBV-associated diseases.
FIG. 4.
Activation of virus-specific memory CTL response by the polyepitope protein targeted to the lysosomal compartment. Polyclonal effectors from an HLA DR1-positive donor were generated by restimulating peripheral blood lymphocytes with Vacc.ER-POLY-LAMP. Target cells were autologous LCLs either infected with recombinant vaccinia virus constructs or sensitized with the indicated peptide (10 μmol/ml). Effector/target ratios of 15:1 and 30:1 were used in the assay.
Detection of polyepitope protein targeted to the endocytic/secretory pathway.
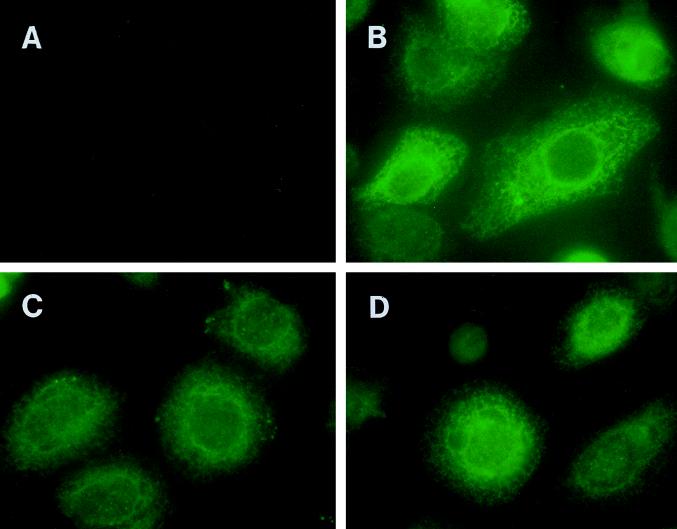
To examine the localization and expression of the class II polyepitope protein targeted to the endocytic/secretory pathway, wtGFP was incorporated into different polyepitope constructs. A standard immunofluorescence assay was used to detect these chimeric molecules in recombinant vaccinia virus-infected HeLa cells by using a GFP-specific antibody (Fig. 5). It is important to mention here that our earlier attempts to detect the wtGFP by direct fluorescence were unsuccessful due to a rapid loss of fluorescence at 37°C (10, 29). A distinct pattern of staining, restricted primarily to the ER, was seen in cells infected with Vacc.ER-POLY (Fig. 5B), while cells infected with Vac.ER-POLY-LAMP and Vacc.INV-POLY showed fluorescence in vesicles which resemble lysosomes in size and perinuclear localization (Fig. 5C and D). These results are consistent with the previously described endosomal/lysosomal localization of Ii protein and LAMP-1.
FIG. 5.
Localization of polyepitope protein targeted to the endocytic/secretory pathway by immunofluorescence. HeLa cells were infected with Vacc.TK− (A), Vacc.ER-POLY-GFP (B), Vacc.INV-POLY-GFP (C), or Vacc.ER-POLY-GFP-LAMP (D) at an MOI of 4:1 for 6 h at 37°C. Following incubation, these cells were processed for immunofluorescence as described in Materials and Methods. Cells were initially labelled with wtGFP-specific antibody and then incubated with fluorescein isothiocyanate-conjugated sheep anti-rabbit immunoglobulin G. The cells were examined under a fluorescence microscope in the presence of _n_-propyl gallate.
This study clearly illustrates that targeting signal sequences can be utilized to direct multiple class II-restricted CTL epitopes into the endosomal and lysosomal compartments. This approach not only preferentially translocates the polyepitope protein to these compartments but also enhances endogenous presentation of CTL epitopes. Furthermore, this strategy was successfully used to activate a virus-specific memory CTL response from peripheral blood lymphocytes. These observations have a number of important implications for the endogenous presentation of MHC class II-restricted epitopes and for vaccine design in general. First, the use of a polyepitope protein targeted to the lysosomal compartment overcomes the problems associated with the use of oncogenic viral antigens as vaccines. Second, multiple epitopes from different viruses can be included in these constructs to specifically stimulate CD4+ T cells, avoiding any requirement for multiple recombinant whole-protein sequences (31, 32). This might be important in settings where an antibody response results in enhanced infection. For example, dengue virus infection is augmented when nonneutralizing antibodies complex with virus (15). Another important implication of these results relates to the ability of the LAMP-1-targeted polyepitope protein to activate a CD4+-CTL response to cytoplasmic or nuclear antigens that are not normally targeted to the class II pathway. Indeed, the data presented in this study showed that although the EBNA1 antigen is poorly immunogenic, LAMP-1-mediated targeting of the EBNA1 CTL epitope significantly enhanced the activation of the EBNA1-specific CTL response in vitro. These results indicate that the latter approach might be used to enhance the EBNA1-specific response in vivo to control EBV-associated malignancies where EBV latent gene expression is restricted to this antigen. Recent studies have shown that immunization with the papillomavirus E7 antigen fused to LAMP-1 induced potent E7-specific antitumor immunity (23, 40).
ACKNOWLEDGMENTS
This work was supported by grant from the Co-operative Research center for Vaccine Technology and The National Health and Medical Research Council (NHMRC). R.K. is supported by a R. Douglas Wright Fellowship from NHMRC.
REFERENCES
- 1.Andrew M E, Boyle D B, Coupar B E, Whitfeld P L, Both G W, Bellamy A R. Vaccinia virus recombinants expressing the SA11 rotavirus VP7 glycoprotein gene induce serotype-specific neutralizing antibodies. J Virol. 1987;61:1054–1060. doi: 10.1128/jvi.61.4.1054-1060.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bikoff E K. Formation of complexes between self-peptides and MHC class II molecules in cells defective for presentation of exogenous protein antigens. J Immunol. 1992;149:1–8. [PubMed] [Google Scholar]
- 3.Blum J S, Cresswell P. Role for intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci USA. 1988;85:3975–3979. doi: 10.1073/pnas.85.11.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle D B, Coupar B E, Both G W. Multiple-cloning-site plasmids for the rapid construction of recombinant poxviruses. Gene. 1985;35:169–177. doi: 10.1016/0378-1119(85)90169-6. [DOI] [PubMed] [Google Scholar]
- 5.Braciale T J, Morrison L A, Sweetser M T, Sambrook J, Gething M J, Braciale V L. Antigen presentation pathways to class I and class II. Immunol Rev. 1987;98:95–114. doi: 10.1111/j.1600-065x.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- 6.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 7.Coupar B E, Andrew M E, Both G W, Boyle D B. Temporal regulation of influenza hemagglutinin expression in vaccinia virus recombinants and effects on the immune response. Eur J Immunol. 1986;16:1479–1487. doi: 10.1002/eji.1830161203. [DOI] [PubMed] [Google Scholar]
- 8.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 9.Cresswell P. Invariant chain structure and MHC class II function. Cell. 1996;84:505–507. doi: 10.1016/s0092-8674(00)81025-9. [DOI] [PubMed] [Google Scholar]
- 10.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–445. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 11.Eager K B, Hackett C J, Gerhard W U, Bennink J, Eisenlohr L C, Yewdell J, Ricciardi R P. Murine cell lines stably expressing the influenza virus hemagglutinin gene introduced by a recombinant retrovirus vector are constitutive targets for MHC class I- and class II-restricted T lymphocytes. J Immunol. 1989;143:2328–2335. [PubMed] [Google Scholar]
- 12.Fleisher G R, Collins M, Fager S. Humoral immune response in infectious mononucleosis. Late emergence of anti-EA (R) and the effects of corticosteroid therapy. J Adolesc Health Care. 1985;6:424–428. doi: 10.1016/s0197-0070(85)80046-2. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, Viitala J, Matteson J, Carlsson S R. Cloning of cDNAs encoding human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Comparison of their deduced amino acid sequences. J Biol Chem. 1988;263:18920–18928. [PubMed] [Google Scholar]
- 14.Germain R N. Immunology. The ins and outs of antigen processing and presentation. Nature. 1986;322:687–689. doi: 10.1038/322687a0. [DOI] [PubMed] [Google Scholar]
- 15.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 16.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. Localization of Epstein-Barr virus cytotoxic T-cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanna R, Burrows S R, Moss D J. Immune regulation in Epstein-Barr virus-associated diseases. Microbiol Rev. 1995;59:387–405. doi: 10.1128/mr.59.3.387-405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna R, Burrows S R, Moss D J, Silins S L. Peptide transporter (TAP-1 and TAP-2)-independent endogenous processing of Epstein-Barr virus (EBV) latent membrane protein 2A: implications for cytotoxic T-lymphocyte control of EBV-associated malignancies. J Virol. 1996;70:5357–5362. doi: 10.1128/jvi.70.8.5357-5362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna R, Burrows S R, Suhrbier A, Jacob C A, Griffin H, Misko I S, Sculley T B, Rowe M, Rickinson A B, Moss D J. EBV peptide epitope sensitization restores human cytotoxic T cell recognition of Burkitt’s lymphoma cells. Evidence for a critical role for ICAM-2. J Immunol. 1993;150:5154–5162. [PubMed] [Google Scholar]
- 20.Khanna R, Burrows S R, Thomson S A, Moss D J, Cresswell P, Poulsen P, Cooper L. Class I processing defective Burkitt’s lymphoma cells are recognised efficiently by CD4+ EBV-specific CTL. J Immunol. 1997;157:3619–3625. [PubMed] [Google Scholar]
- 21.Khanna R, Jacob C A, Burrows S R, Kurilla M G, Kieff E, Misko I S, Moss D J. Expression of Epstein-Barr virus nuclear antigens in anti-IgM-stimulated B cells following recombinant vaccinia infection and their recognition by human cytotoxic T-cells. Immunology. 1991;74:504–510. [PMC free article] [PubMed] [Google Scholar]
- 22.Khanna R, Jacob C A, Burrows S R, Moss D J. Presentation of endogenous viral peptide epitopes by anti-CD40 stimulated human B cells following recombinant vaccinia infection. J Immunol Methods. 1993;164:41–49. doi: 10.1016/0022-1759(93)90274-b. [DOI] [PubMed] [Google Scholar]
- 23.Lin K Y, Guarnieri F G, Staveley O F, Levitsky H I, August J T, Pardoll D M, Wu T C. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 24.Morrison L A, Braciale V L, Braciale T J. Antigen form influences induction and frequency of influenza-specific class I and class II MHC-restricted cytolytic T lymphocytes. J Immunol. 1988;141:363–368. [PubMed] [Google Scholar]
- 25.Moss D J, Misko I S, Burrows S R, Burman K, McCarthy R, Sculley T B. Cytotoxic T-cell clones discriminate between A- and B-type Epstein-Barr virus transformants. Nature. 1988;331:719–721. doi: 10.1038/331719a0. [DOI] [PubMed] [Google Scholar]
- 26.Neefjes J J, Stollorz V, Peters P J, Geuze H J, Ploegh H L. The biosynthetic pathway of MHC class II but not class I molecules intersects the endocytic route. Cell. 1990;61:171–183. doi: 10.1016/0092-8674(90)90224-3. [DOI] [PubMed] [Google Scholar]
- 27.Nuchtern J G, Biddison W E, Klausner R D. Class II MHC molecules can use the endogenous pathway of antigen presentation. Nature. 1990;343:74–76. doi: 10.1038/343074a0. [DOI] [PubMed] [Google Scholar]
- 28.Persson H, Jornvall H, Zabielski J. Multiple mRNA species for the precursor to an adenovirus-encoded glycoprotein: identification and structure of the signal sequence. Proc Natl Acad Sci USA. 1980;77:6349–6353. doi: 10.1073/pnas.77.11.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pines J. GFP in mammalian cells. Trends Genet. 1995;11:326–327. doi: 10.1016/s0168-9525(00)89092-7. [DOI] [PubMed] [Google Scholar]
- 30.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 31.Rowell J F, Ruff A L, Guarnieri F G, Staveley O, Lin X, Tang J, August J T, Siliciano R F. Lysosome-associated membrane protein-1-mediated targeting of the HIV-1 envelope protein to an endosomal/lysosomal compartment enhances its presentation to MHC class II-restricted T cells. J Immunol. 1995;155:1818–1828. [PubMed] [Google Scholar]
- 32.Sanderson S, Frauwirth K, Shastri N. Expression of endogenous peptide-major histocompatibility complex class II complexes derived from invariant chain-antigen fusion proteins. Proc Natl Acad Sci USA. 1995;92:7217–7221. doi: 10.1073/pnas.92.16.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt C W, Misko I S. The ecology and pathology of Epstein-Barr virus. Immunol Cell Biol. 1995;73:489–504. doi: 10.1038/icb.1995.79. [DOI] [PubMed] [Google Scholar]
- 34.Strubin M, Berte C, Mach B. Alternative splicing and alternative initiation of translation explain the four forms of the Ia antigen-associated invariant chain. EMBO J. 1986;5:3483–3488. doi: 10.1002/j.1460-2075.1986.tb04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweetser M T, Morrison L A, Braciale V L, Braciale T J. Recognition of pre-processed endogenous antigen by class I but not class II MHC-restricted T cells. Nature. 1989;342:180–182. doi: 10.1038/342180a0. [DOI] [PubMed] [Google Scholar]
- 36.Thomson, S., S. Elliott, M. Sherritt, K. W. Sproat, B. E. Coupar, A. A. Scalzo, C. A. Forbes, A. Ladhams, X. Y. Mo, R. Tripp, P. C. Doherty, D. J. Moss, and A. Suhrbier. Recombinant polyepitope vaccines for the delivery of multiple CD8+ cytotoxic T cell epitopes. J. Immunol., in press. [PubMed]
- 37.Thomson S A, Khanna R, Gardner J, Burrows S R, Coupar B, Moss D J, Suhrbier A. Minimal epitopes expressed in a recombinant polyepitope protein are processed and presented to CD8+ cytotoxic T cells: implications for vaccine design. Proc Natl Acad Sci USA. 1995;92:5845–5849. doi: 10.1073/pnas.92.13.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss S, Bogen B. MHC class II-restricted presentation of intracellular antigen. Cell. 1991;64:767–776. doi: 10.1016/0092-8674(91)90506-t. [DOI] [PubMed] [Google Scholar]
- 39.White C A, Cross S M, Kurilla M G, Kerr B M, Schmidt C, Misko I S, Khanna R, Moss D J. Recruitment during infectious mononucleosis of CD3+CD4+CD8+ virus-specific cytotoxic T cells which recognise Epstein-Barr virus lytic antigen BHRF1. Virology. 1996;219:489–492. doi: 10.1006/viro.1996.0277. [DOI] [PubMed] [Google Scholar]
- 40.Wu T C, Guarnieri F G, Staveley O F, Viscidi R P, Levitsky H I, Hedrick L, Cho K R, August J T, Pardoll D M. Engineering an intracellular pathway for major histocompatibility complex class II presentation of antigens. Proc Natl Acad Sci USA. 1995;92:11671–11675. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yewdell J W, Bennink J R. Brefeldin A specifically inhibits presentation of protein antigens to cytotoxic T lymphocytes. Science. 1989;244:1072–1075. doi: 10.1126/science.2471266. [DOI] [PubMed] [Google Scholar]