Scaffold Attachment Region-Mediated Enhancement of Retroviral Vector Expression in Primary T Cells (original) (raw)
Abstract
We have studied retroviral transgene expression in primary human lymphocytes. Our data demonstrate that transgene expression is high in activated primary CD4+ T cells but significantly decreased in mitotically quiescent cells. Incorporation of a DNA fragment from the scaffold attachment region (SAR) of the human beta interferon gene into the vector improved transgene expression, particularly in quiescent cells. The SAR element functioned in an orientation-dependent manner and enhanced expression of Moloney murine leukemia virus- and murine embryonic stem cell-based vectors. Clonal analysis of transduced T cells showed that the SAR sequence did not confer position-independent expression on a transgene but rather prevented the decrease of expression when cells became quiescent. The SAR sequence also enhanced transgene expression in T cells generated from retrovirally transduced CD34-enriched hematopoietic progenitor-stem cells in a SCID-hu thymus-liver mouse model. We have used the SAR-containing retroviral vector to express the RevM10 gene, a _trans_-dominant mutant of the human immunodeficiency virus type 1 (HIV-1) Rev gene. Compared to a standard retroviral vector, the SAR-containing vector was up to 2 orders of magnitude more efficient in inhibiting replication of the HIV-1 virus in infected CD4+ peripheral blood lymphocyte populations in vitro. This is the first demonstration that SAR elements can be used to improve retroviral vector expression in human primary T cells.
Peripheral blood lymphocytes (PBLs) have been used as cellular targets for gene therapy applications of immune disorders including SCID-ADA deficiency and HIV disease (1, 4, 34). At present, retroviral vectors are the gene transfer modality of choice mainly because the integration of retrovirally transduced genes into the chromosome of the target cells supports persistent transgene expression (reviewed in reference 17). Protocols for efficient gene marking of PBLs have been developed (6, 35), but little is known about the regulation of transgene expression in primary T cells. In vivo, the majority of circulating PBLs are in a resting state and genes carried by standard retroviral vectors based on the Moloney murine leukemia virus (Mo-MuLV) (24) or the murine embryonic stem cell virus (MESV) (14) are not efficiently expressed in quiescent primary T cells (26). The factors that control transgene expression in primary T cells are not known but may render retrovirus-based gene therapy approaches inefficient against certain diseases, including human immunodeficiency virus (HIV) disease (26).
Scaffold attachment regions (SARs), also referred to as matrix attachment regions (MARs), are DNA sequences that bind with high affinity to isolated nuclear scaffolds or nuclear matrices in vitro (reviewed in reference 2). SAR elements are several hundred base pairs long and are AT rich (≈70%). Although cloned SAR and MAR elements share common structural features, no consensus sequence has been identified (5). SARs have been located upstream of, downstream of, and within genes (introns), suggesting that they may represent functionally distinct classes (2). It is thought that SAR elements define boundaries of independent chromatin domains encompassing all required _cis_-regulatory elements for coordinated expression of the genes within the domain (2). SAR elements can enhance expression of heterologous genes in in vitro transfection experiments (19, 20, 28) and in transgenic mice (22, 32). In some instances, it has been reported that SAR elements can confer position-independent expression to a linked transgene (19, 22).
In an attempt to improve gene expression in resting primary T cells we have inserted into a retroviral vector the SAR sequence derived from the human beta interferon (IFN-β–SAR) gene (20). The SAR-containing vectors were expressed at significantly higher levels than were the control vectors in transduced PBLs, as well as in T cells generated from transduced hematopoietic stem-progenitor cells (HSPC) in SCID-hu thymus-liver mice. Additionally, we have shown that the enhanced RevM10 expression in vectors with the SAR element leads to a greater reduction of HIV replication in vitro, demonstrating the utility of a SAR sequence for developing improved vectors for the gene therapy of HIV disease.
MATERIALS AND METHODS
Construction of recombinant retroviral vectors and retrovirus-producing cells.
The retroviral vectors LMiLy and MESV-MiLy (Fig. 1) have been described previously (26). The 800-bp _Hin_dIII-_Bam_HI IFN-β–SAR fragment from the pCL plasmid (23) was inserted in reverse orientation into the _Nhe_I site in the 3′ long terminal repeat (LTR) of the LNCX retroviral vector (24). Subsequently, the _Cla_I-_Xba_I fragment spanning the SAR sequence and a portion of the 3′ LTR was excised from LNCX-SAR and inserted into LMiLy to create the LMiLy2S vector (Fig. 1). In the LMiLyS vector the 800-bp IFN-β–SAR fragment was inserted (blunt) into the _Cla_I site of the LMiLy. MESV-MiLy2S and MESV-MiLy2S-F were generated by inserting (blunt) an _Hin_dIII-_Eco_RI IFN-β–SAR fragment from pCL into the _Nhe_I site of the 3′ LTR of the MESV-MiLy vector. There is no difference between the _Hin_dIII-_Bam_HI and the _Hin_dIII-_Eco_RI fragments with respect to the SAR sequence. _Bam_HI and _Eco_RI sites are a part of the polylinker located at the 5′ end of the SAR element. Different sites were used merely to facilitate the cloning procedure. Retroviral vector plasmid DNAs were cotransfected with a vesicular stomatitis virus G expression plasmid (8) into gp47 cells as described previously (29). At 48 h posttransfection, culture supernatants were used to inoculate amphotropic ProPak-A packaging cells (29). Following transduction, transgene (Lyt-2)-expressing ProPak-A cells were enriched by fluorescence-activated cell sorting (FACS) to generate polyclonal producer cell populations. Retroviral vector supernatants were prepared as described previously (12). The transduction efficiencies of the retroviral vector supernatants were determined on NIH 3T3 cells (12). All producer cells tested negative for replication-competent retrovirus by S+L− assay on PG4 cells.
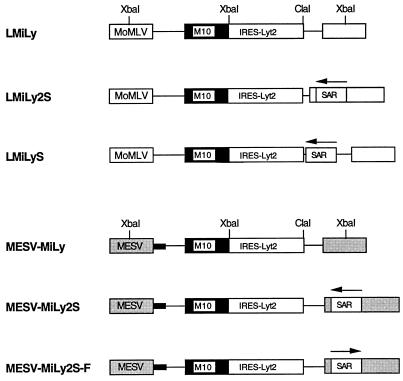
FIG. 1.
Schematic representation of the retroviral vector constructs (not drawn to scale). The arrow above the SAR box indicates the orientation of the element. The small black box in the MESV-MiLy vector series indicates that the primer binding site is derived from the dl587rev retrovirus (9). Abbreviations: M10, RevM10 gene; MoMLV, MoMuLV LTR; MESV, MESV LTR; SAR, IFN-β SAR; IRES-Lyt2, internal ribosomal entry site linked to the Lyt-2 gene.
Transduction of primary T cells and analysis of transgene expression in activated and resting cells.
Primary T cells were isolated from the peripheral blood of healthy donors or from thymus grafts of SCID-hu thymus-liver mice (thymocytes) (27). Both cell populations were enriched for CD4+ cells by depleting CD8+ cells with anti-CD8 biotinylated antibody (Becton Dickinson) and streptavidin-coated magnetic Dynabeads (Dynal). This procedure yielded a 90 to 95% pure CD4+ population. Cells were cultured in TOC medium (RPMI medium supplemented with 1× minimal essential medium vitamin solution) (GIBCO-BRL), insulin-transferrin-sodium selenite supplement (Sigma), 10% fetal bovine serum (Hyclone), phytohemagglutinin (PHA; 2 μg/ml), interleukin-2 (IL-2; 40 U/ml), and allogeneic JY feeder cells for 3 to 4 days (26). Retroviral vector transduction was performed by spinoculation (centrifugation at 2,000 × g) of 5 × 105 cells with 1 ml of retroviral supernatant supplemented with 8 μg of polybrene per ml for 3 h at 34°C. This procedure was repeated on 2 consecutive days. Transduced cells were routinely enriched to >90% Lyt-2+ cells by two rounds of positive selection with biotinylated anti-Lyt-2 antibody (PharMingen) and streptavidin Dynabeads (Dynal). For analysis of retroviral transgene expression, cells were stimulated with PHA plus feeder cells as described above. At various time points poststimulation, aliquots of cells were stained with both anti-Lyt-2 R-phycoerythrin (R-PE; PharMingen)-conjugated and anti-CD25 fluorescein isothiocyanate (FITC; Becton Dickinson)-conjugated antibodies and then analyzed on a FACScan (Becton Dickinson).
HIV infection of primary T cells.
On day 5 following stimulation with PHA and feeder cells, T cells were washed and resuspended in TOC medium containing IL-2 only. Cells (2 × 104 to 3 × 104; 75 μl) were mixed with 75 μl of undiluted JR-CSF HIV type 1 (HIV-1) virus stock (104 to 105 50% tissue culture infective doses/ml) and plated in triplicate in round-bottom 96-well plates. Cells were cultured overnight and on the following day 125 μl of medium was removed and replaced with 135 μl of fresh TOC and containing IL-2. Cell supernatants were harvested on days 3, 5, 7, and 9 postinoculation. Where indicated, 135 μl of TOC–IL-2 containing 2.5 × 105 feeder cells/ml was added to the cells on day 3 to maintain T-cell activation. HIV-1 p24 antigen concentration in the culture supernatants was determined by enzyme-linked immunosorbent assay (Dupont, NEN Research Products).
Isolation of MPB CD34+ cells and retroviral transduction.
Mobilized peripheral blood (MPB) HSPC samples were obtained from healthy donors treated with granulocyte colony-stimulating factor (G-CSF) (0.25 mg/m2) from Baxter Biotech Immunotherapy Division, Irvine, Calif., with informed consent from the donors. Leukapheresis samples were enriched (generally >90%) for CD34+ cells with an immunomagnetic bead selection device (Baxter Healthcare Corp.). CD34+-selected MPB tissues were phenotyped for HLA MA2.1 (FITC-conjugated antibody was prepared at SyStemix from hybridomas obtained from the American Type Culture Collection, Rockville, Md.). Prior to transduction cells were cultured for 2 days at 5 × 105 to 10 × 105 cells/ml in Whitlock-Witte medium (50% Iscove’s modified Dulbecco medium (IMDM), 50% RPMI, 10% fetal calf serum, 4 × 10−5 M 2-mercaptoethanol, 5 mM sodium pyruvate, 10 mM HEPES, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 4 mM glutamine) supplemented with IL-3 (20 ng/ml), IL-6 (20 ng/ml), and stem cell factor (100 ng/ml) and then subjected to two rounds of spinoculation as described above with either LMiLy or LMiLyS supernatants. At 48 h after the second transduction, cells were stained for surface CD34 (SR) and Lyt-2 (PE) and then sorted for the presence of CD34 and Lyt-2 with a FACStar cell sorter (Becton Dickinson Immunocytometry Systems) as described in detail previously (3).
Analysis of thymocytes from SCID-hu mice.
The SCID-hu mice were prepared by surgical transplantation of human fetal thymus and liver fragments into C.B-17 scid/scid mice as previously described (25), and in accordance with the guidelines set forth by the SyStemix Animal Care and Use Committee. All thymus and liver tissues had been identified as negative for expression of the HLA MA2.1 marker. At 3 to 5 months after transplantation of thymus and liver fragments, the mice were given 400 rads of total body irradiation followed by reconstitution with transduced HSPC as described previously in detail (3, 11). Then 105 LMiLy- or LMiLyS-transduced and sorted CD34+ Lyt-2+ HSPC were injected directly into thymus and liver grafts in SCID-hu mice. Control mice were injected with mock-transduced unsorted HSPC. At 8 weeks after reconstitution, thymocytes were recovered from the grafts and analyzed for the level of donor reconstitution (FITC staining for MA2.1) and the expression of the LMiLy- or LMiLyS-encoded Lyt-2 (PE staining for Lyt-2) by flow cytometry (FACScan; Becton Dickinson). Gene marking of thymocytes was examined by depositing donor-derived T cells into 96-well Thermowell PCR plates (Corning, Costar, Cambridge, Mass.) followed by sensitive DNA PCR with the Moloney murine leukemia virus (MoMuLV) LTR U3 region-specific primers lsn7 (5′ dAGACCCCACCTGTAGGTTTG 3′) and lsn346 (5′ dTTGAGCTCGGGGAGCAGAAG 3′). The amplified DNA fragments were denatured with NaOH at 95°C and transferred to nylon membranes with a 96-well dot-blot apparatus (GIBCO-BRL). The immobilized DNA was detected by hybridization with a nested U3 region-specific probe followed by autoradiography. The probe was generated by PCR (with the primers lsn123 [5′ dCTGAATATGGGCCAAACAGG 3′] and lsn320 [5′ dAACAGAAGCGAGAAGCGAAC 3′]) and labelled to ∼108 cpm/μg by the random priming method (Ambion) with [α-32P]dCTP (Amersham).
RESULTS
Production of retroviral vectors.
The MoMuLV-based retroviral vector LMiLy (Fig. 1) expresses two genes from one bicistronic mRNA transcript: the RevM10 gene (21) and the Lyt-2 surface marker (mouse CD8 α′ chain) (31) (kindly provided by G. P. Nolan, Stanford University, Stanford, Calif.). Translation of the Lyt-2 protein is mediated by the internal ribosomal entry site of the human encephalomyocarditis virus (15) and thus is linked to RevM10 protein expression. Double staining of transduced CEMSS and primary T cells for RevM10 and Lyt-2 showed that expression of the two proteins is colinear (3, 30). Flow cytometric analysis of the more easily detected Lyt-2 surface antigen was subsequently used to estimate overall transgene expression. The 800-bp IFN-β–SAR fragment (20) was inserted into the _Cla_I site of the LMiLy, generating the LMiLyS vector (Fig. 1), or into the _Nhe_I site of the 3′ LTR, generating a double-copy type LMiLy2S vector (13). Following transduction with the LMiLy2S, the 3′ LTR SAR sequence is duplicated in the 5′ LTR, generating an integrated provirus that is bordered by two SAR sequences. We have also generated MESV-based vectors (14). The MESV-MiLy2S and MESV-MiLy2S-F vectors were derived from the MESV-MiLy construct (26) (Fig. 1). In the LMiLy2S and MESV-MiLy2S vectors the SAR sequence is in the reverse orientation, and in the MESV-MiLy2S-F vector the SAR sequence is in the forward orientation, as indicated by the arrows in Fig. 1. “Forward” and “reverse” refer to the orientation of the SAR element in its natural human IFN-β gene locus (23). Amphotropic producer cell lines were generated with ProPak-A packaging cells (29). Transduction efficiencies of the retroviral stocks used in this study were determined by measuring the percentage of Lyt-2+ NIH 3T3 cells 2 days postinoculation with 1:3-diluted viral supernatants (12) and were as follows: LMiLy, 53%; MESV-MiLy, 81%; LMiLyS, 74%; LMiLy2S, 21%; MESV-MiLy2S, 14%; and MESV-MiLy2S-F, 7%. Since the vectors do not contain a drug selection marker it was not possible to determine the endpoint viral titers. However, to achieve 50% or greater efficiency of transduction of 3T3 cells with _neo_-containing vector, a viral stock with a titer of at least 0.5 × 106 to 1 × 106 CFU/ml (12) is needed. We can assume therefore that, at least for the LMiLy, MESV-LMiLy, and LMiLyS vectors, the endpoint titers were greater than 106 CFU/ml. All retroviral stocks used in this study were free of replication-competent retrovirus (data not shown).
The SAR sequence improves retroviral vector expression in CD4+ primary T cells.
CD4+ T cells were enriched from PBLs from normal healthy donors or from thymus grafts of SCID-hu thymus-liver mice (thymocytes) (27) by depleting CD8+ cells with immunomagnetic beads, a technique yielding ≈90% enriched cell populations. Cells were stimulated with PHA, IL-2, and irradiated allogeneic JY feeder cells for 3 to 4 days and subsequently transduced with the LMiLy and LMiLy2S retroviral vectors by centrifugation (26). Following this protocol, we detected 4 to 20% Lyt-2+ cells (data not shown) and, after expansion in vitro, positive cells were further enriched to ≥90% purity by immunomagnetic bead selection (Fig. 2B and C). We used the CD25 surface protein (i.e., a low-affinity IL-2 receptor) as a marker for the T-cell activation status. At 3 to 4 days poststimulation, CD25 expression was at a maximum, with >95% CD25+ cells (Fig. 2A). After 4 days the percentage of CD25+ cells started to decline as the T cells became quiescent. By days 9 to 11, cells ceased to proliferate and the CD25 marker was down-regulated (>50% CD25− cells), reflecting the mitotically resting state of the cells (Fig. 2D).
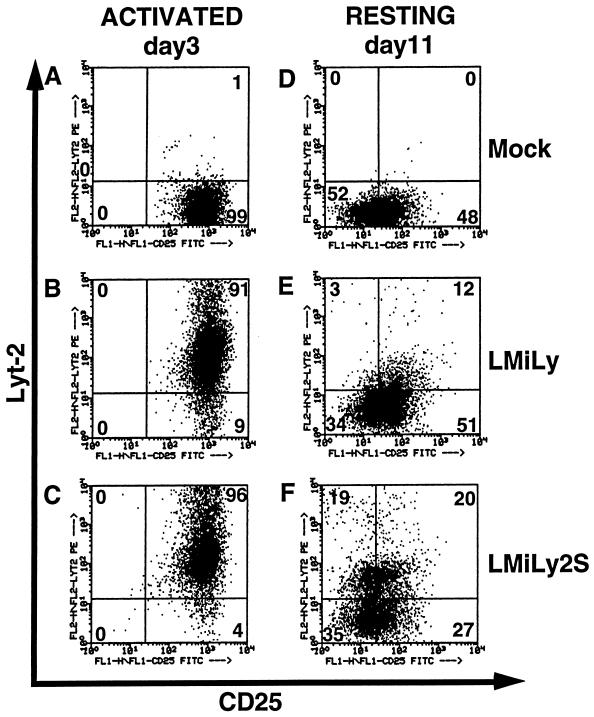
FIG. 2.
The LMiLy2S vector is efficiently expressed in resting T cells. Lyt-2-enriched LMiLy- and LMiLy2S-transduced CD4+ primary T cells were activated with PHA, IL-2, and irradiated allogenic feeder cells. On days 3 and 11 poststimulation, cell aliquots were stained with anti-CD25 FITC-conjugated and anti-Lyt-2 PE-conjugated antibodies and analyzed by FACscan. The numbers indicate the percentage of Lyt-2+ cells in the respective quadrants. Gates for background fluorescence were set based on control isotype antibodies. Mock, untransduced control cells.
We analyzed Lyt-2 transgene expression in activated (day 3 poststimulation) and resting (day 11 poststimulation) cells. The results obtained with one representative tissue sample are shown in Fig. 2. There was no marked difference in the percentage of the Lyt-2+ activated T cells between the control vector LMiLy (91%) and the SAR-containing vector LMiLy2S (96%) (Fig. 2B and C). In the resting LMiLy-transduced cells, Lyt-2 expression was low (15%) (Fig. 2E) and the decrease in transgene expression correlated with the decrease of the CD25 marker (data not shown) as previously reported (26). With the SAR-containing LMiLy2S vector, however, we observed 2.6-fold-higher levels of Lyt-2+ resting T cells (39%; Fig. 2F). Also, the mean Lyt-2 fluorescence intensity of LMiLy2S-transduced resting cells was 3- to 4-fold higher than that of the LMiLy-transduced cells (data not shown). Upon restimulation, both LMiLy- and LMiLy2S-transduced cells expressed comparable high levels of Lyt-2 (87 and 95%, respectively), demonstrating that the observed loss of expression was not caused by loss of integrated vector. Similar expression patterns were observed irrespective of the source of the primary T cells. The data obtained with four independent tissues (two PBL and two thymocyte sources) are summarized in Table 1. Although the absolute percentage of Lyt-2+ resting T cells varied from tissue to tissue, the LMiLy2S vector consistently yielded higher values (on average 2.4 ± 0.9-fold more Lyt-2+ cells; P < 0.1) than the LMiLy vector (Table 1).
TABLE 1.
Lyt-2 surface marker expression in activated and resting primary T cells transduced with the LMiLy and LMiLy2S vectorsa
| Tissueb | Activated | Resting | ||||
|---|---|---|---|---|---|---|
| % Lyt-2+ cells | LMiLy2S/LMiLy ratio | % Lyt-2+ cells | LMiLy2S/ LMiLy ratio | |||
| LMiLy | LMiLy2S | LMiLy | LMiLy2S | |||
| 1 | 91 | 96 | 1.05 | 15 | 39 | 2.6 |
| 2 | 65 | 64 | 0.98 | 7 | 26 | 3.7 |
| 3 | 73 | 86 | 1.18 | 33 | 48 | 1.5 |
| 4 | 95 | 96 | 1.01 | 35 | 55 | 1.6 |
| Avg | 1.06 ± 0.08 | 2.4 ± 0.9 (P < 0.1c) |
Detailed analysis of the FACS data revealed that the effect of the SAR sequence on transgene expression was most significant in the CD25− compartment of resting T cells (Table 2). On average, there were 5.7 ± 3.4-fold (P < 0.01) more Lyt-2+ cells in the CD25− fraction of the LMiLy2S-transduced populations than in that of the LMiLy-transduced populations, whereas in the CD25+ fraction the difference was only 1.7 ± 0.5-fold (P < 0.3) (Table 2).
TABLE 2.
Lyt-2 surface marker detection in the CD25− and CD25+ subpopulations of resting primary T cells transduced with the LMiLy and LMiLy2S vectorsa
| Tissueb | CD25− fraction | CD25+ fraction | ||||
|---|---|---|---|---|---|---|
| % Lyt-2+ cells | LMiLy2S/LMiLy ratio | % Lyt-2+ cells | LMiLy2S/LMiLy ratio | |||
| LMiLy | LMiLy2S | LMiLy | LMiLy2S | |||
| 1 | 3 | 19 | 6.3 | 12 | 20 | 1.7 |
| 2 | 1 | 11 | 11 | 6 | 15 | 2.5 |
| 3 | 7 | 20 | 2.9 | 26 | 28 | 1.1 |
| 4 | 9 | 22 | 2.4 | 26 | 33 | 1.3 |
| Avg | 5.7 ± 3.4 (P < 0.01c) | 1.7 ± 0.5 (P < 0.3) |
Analysis of individual T-cell clones.
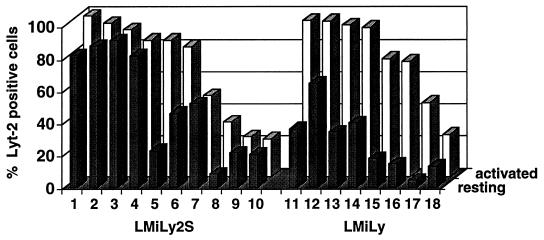
Reproducibly, we observed two types of resting LMiLy2S-transduced T cells: 30 to 40% of the cells were Lyt-2+, and the rest were Lyt-2− (Fig. 2F). To further characterize these populations, we generated individual LMiLy2S- and LMiLy-transduced cell clones and analyzed Lyt-2 expression in activated and resting cells (Fig. 3). Both vectors had comparable distributions of high- and low-expressing activated T-cell clones. All 8 LMiLy clones showed a 2- to 10-fold decrease in Lyt-2 expression in the resting state (Fig. 3, clones 11 to 18). In sharp contrast, 7 of 10 LMiLy2S clones (clones 1, 2, 3, 4, 7, 9, and 10) showed no marked decrease in Lyt-2 expression, and for 3 clones (clones 5, 6, and 8) expression decreased to the same extent (two- to eightfold) as that observed for the LMiLy vector. The protective effect of the SAR sequence did not correlate with the level of Lyt-2 expression in activated cells (for example, compare clones 5 and 7). Since double-copy-type vectors can be unstable (18), we carefully analyzed the structure of the LMiLy2S proviral DNA by DNA PCR with primers that span the 5′ and 3′ LTRs and the Lyt-2 gene (data not shown). Although we cannot rule out small deletions of less than 50 bp, SAR and LTR sequences in all 10 LMiLy2S clones appeared to be intact. Overall these data show that, in at least some clones, the SAR sequence can prevent the attenuation of retroviral vector expression in resting cells. The finding that in approximately 30% of resting LMiLy2S clones expression was nevertheless decreased explains why we observed Lyt-2+ and Lyt-2− fractions in populations of resting LMiLy2S-transduced T cells.
FIG. 3.
Analysis of LMiLy2S vector expression in individual primary T-cell clones. Ten LMiLy2S (no. 1 through 10) and eight LMiLy (no. 11 through 18) T-cell clones were analyzed for Lyt-2 expression on days 3 (activated) and 11 (resting) after stimulation with PHA, IL-2, and irradiated allogenic feeder cells.
The SAR element functions in an orientation-dependent manner.
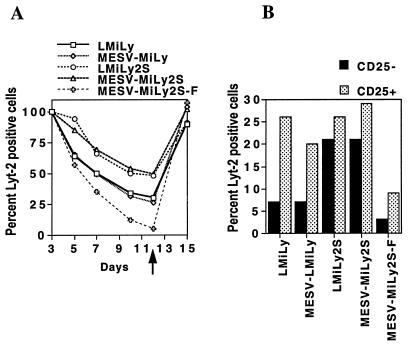
The SAR sequence was able to rescue expression of the MESV-based retroviral vector MESV-MiLy (Fig. 1), which is also down-regulated in resting primary T cells (26). Kinetic analysis of Lyt-2 expression in transduced T-cell cultures demonstrated that the MESV-MiLy2S vector behaves like the LMiLy2S vector (Fig. 4A). Also, when resting cells were analyzed for the Lyt-2+ CD25− phenotype, the results for the LMiLy2S and MESV-MiLy2S vectors were comparable (Fig. 4B). The enhancing effect was observed only when the SAR sequence was present in the reverse orientation (compare the MESV-MiLy2S and MESV-MiLy2S-F vectors in Fig. 4). Interestingly, when the SAR element was in the forward orientation (e.g., vector MESV-MiLy2S-F) transgene expression was even lower than with the parental MESV-MiLy vector. A similar lower transgene expression was also seen with the LMiLy2S-F vector, which carries the SAR sequence in the forward orientation. There was no difference in the magnitude of expression (as shown by the mean fluorescence intensity of the Lyt-2 staining) between the LMiLy2S and the MESV-MiLy2S vectors (data not shown).
FIG. 4.
The SAR effect is orientation dependent. (A) Lyt-2-enriched CD4+ primary T cells transduced with the MESV-MiLy, MESV-MiLy2S, MESV-MiLy2S-F, LMiLy, and LMiLy2S vectors were stimulated with PHA, IL-2, and feeder cells. Transgene expression was analyzed on days 3, 5, 7, 10, and 12 poststimulation as described in the legend to Fig. 2. On day 12, the cells were restimulated (indicated by arrow) and analyzed 3 days later (day 15 on the graph). (B) The percentage of Lyt-2+ cells in the CD25+ and CD25− fractions of resting T cells was determined on day 10 poststimulation. Results of a representative experiment are shown. Results were reproduced with two separate tissues.
Reconstitution of SCID-hu thymus-liver grafts with retrovirally transduced HSPCs and analysis of Lyt-2 expression on donor-derived T cells.
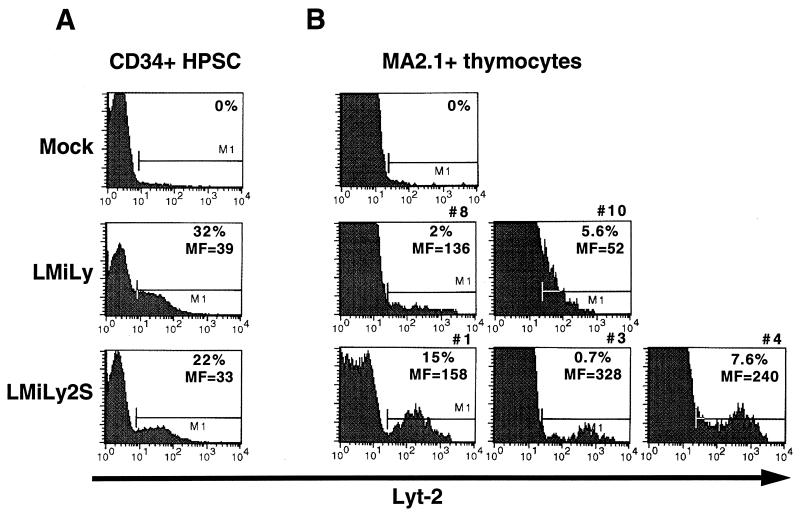
To evaluate the influence of the SAR sequence on vector expression in T cells derived from retrovirally transduced HSPC, we used the SCID-hu thymus-liver mouse model. Reconstitution of SCID-hu thymus-liver grafts with transduced HSPC was as described in detail previously (3). In brief, mobilized peripheral blood CD34+ HSPC from G-CSF-mobilized healthy donors were enriched to >90% by positive selection with immobilized antibodies directed against surface CD34, cultured in vitro for 2 days in the presence of IL-3, IL-6, and stem cell factor, and then subjected to two rounds of transduction by spinoculation with either LMiLy or LMiLyS vectors. Single-copy LMiLyS and double-copy LMiLy2S vectors (Fig. 1) are comparably expressed in resting primary T cells (data not shown); however, the LMiLyS vector gives a three- to fivefold-higher titer and was therefore chosen for transduction of HSPC. At 48 h after the final round of transduction we detected 32 and 22% CD34+ Lyt-2+ cells with the LMiLy and LMiLyS vectors, respectively (Fig. 5). Cells were sorted to 84 to 86% purity for CD34 and Lyt-2 (data not shown) and then injected directly into the conjoint organ of irradiated SCID-hu thymus-liver mice.
FIG. 5.
Analysis of Lyt-2 transgene expression in thymocytes from HSPC-reconstituted SCID-hu thymus-liver grafts. (A) Mobilized peripheral blood CD34+ HSPCs were transduced with the LMiLy and LMiLyS vectors and analyzed for Lyt-2 expression 2 days posttransduction. (B) At 8 weeks after transplantation of SCID-hu thymus-liver mice with Lyt-2-enriched HSPC, thymus-liver grafts were harvested and freshly isolated thymocytes were examined for surface Lyt-2 (transgene) expression on donor HLA-positive cells (MA2.1+ thymocytes in a MA2.1− host background). Percentages of Lyt-2+ cells were determined after subtraction of the background noise observed in mock-transduced reconstituted controls. Numbers (#) above FACS plots are as in Table 3 and indicate the individual SCID-hu animals. MF, relative Lyt-2 mean fluorescence intensity.
At 8 weeks post-HSPC transplantation, thymus-liver grafts were removed and examined for donor cell content by staining for HLA marker MA2.1. We detected significant levels (>2%) of MA2.1-positive cells in seven of nine mice reconstituted with the LMiLyS- and in five of seven mice reconstituted with the LMiLy-transduced HSPC. Overall, the percent MA2.1-positive cells ranged from 11 to 82%, and there was no detectable difference between the LMiLyS- and the LMiLy-transduced HSPC regarding their ability to reconstitute thymus-liver grafts (data not shown). Lyt-2 surface marker expression was detected in three of seven mice reconstituted with the LMiLyS-transduced HSPC and in two of five mice reconstituted with the LMiLyS-transduced HSPC. Data obtained with Lyt-2-expressing animals are summarized in Table 3. When corrected for the percentage of donor cells in the grafts the levels of Lyt-2+ cells were somewhat higher for the LMiLyS vector (15, 0.73, and 7.6%) than for the LMiLy vector (2 and 5.6%) (Table 3). However, the most significant difference between the two vectors was in the relative expression level per cell, which was measured as Lyt-2 mean fluorescence intensity (Fig. 5 and Table 3). Cells harboring the SAR-containing LMiLyS vector showed distinct Lyt-2+ populations that were three to six times brighter than the LMiLy-marked cells (Fig. 5 and Table 3). The frequencies of gene-marked cells were determined by depositing individual MA2.1-positive cells into 96-well format PCR plates and performing sensitive DNA PCR. Overall, 27 to 55% of thymocytes were transgene positive and there was no marked difference between the LMiLyS and the LMiLy vectors (Table 3), indicating that irrespective of the vector used only a fraction of marked cells expressed levels of Lyt-2 transgene high enough to be detected by FACS analysis (3). Most (>90%) of the cells recovered from the SCID-hu thymus-liver grafts were immature CD4+ CD8+ thymocytes (data not shown). We have expanded in vitro the CD4+ thymocytes from LMiLy- and LMiLyS-marked grafts and observed similar transgene expression patterns in activated and resting cells as with directly transduced T cells (data not shown). Overall, these data show that the SAR element can enhance retroviral vector expression levels in the T cells derived from transduced HSPC.
TABLE 3.
Summary of SCID-hu thymus-liver reconstitution experiments for LMiLy- and LMiLyS-transduced HSPC
| Mouse no. | Vector | % MA2.1+ donor cells in engrafted animals | % DNA-PCR+ donor-derived cells | % Lyt-2+ donor-derived cells | Lyt-2 mean fluorescence |
|---|---|---|---|---|---|
| 1 | LMiLyS | 20 | NDa | 15 | 158 |
| 3 | LMiLyS | 68 | 46 | 0.73 | 328 |
| 4 | LMiLyS | 37 | 35 | 7.6 | 240 |
| 8 | LMiLy | 43 | 27 | 2 | 136 |
| 10 | LMiLy | 42 | 55 | 5.6 | 52 |
Improved anti-HIV efficacy of the LMiLy2S vector.
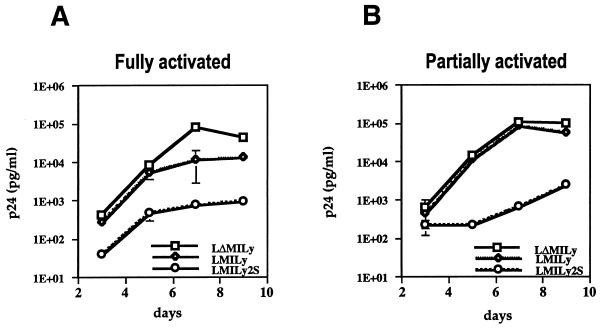
HIV-1 efficiently infects and replicates in fully activated (inoculated on days 2 to 3 postactivation) and partially activated CD4+ T cells (inoculated on days 4 to 5 postactivation), but it does not replicate in resting cells (inoculated on day 8 or later postactivation) (26, 36). We have previously reported that the inhibitory effect of the LMiLy-encoded RevM10 gene on HIV-1 replication is drastically reduced in partially activated T cells, probably because of a rapid decrease in vector expression during the infection experiment (26). To analyze SAR-mediated improved transgene expression with respect to anti-HIV efficacy, primary CD4+ T cells transduced with the LMiLy and LMiLy2S vectors were inoculated with the HIV-1 JR-CSF strain, and viral replication was monitored for 9 days (Fig. 6). The LΔMiLy vector, which does not encode the RevM10 protein, was used as a negative control (26). Cells were harvested on day 5 after stimulation with PHA, IL-2, and feeder cells and then inoculated with HIV-1. One-half of the cultures were maintained in medium with IL-2 only (“partially activated” samples), and the other half were supplemented with fresh PHA and feeder cells 3 days postinoculation to maintain activation of T cells (“fully activated” samples). As shown in Fig. 6, the LMiLy2S vector was over 1 order of magnitude more potent in inhibiting HIV replication in fully activated cells (Fig. 6A), and it maintained its efficacy even in partially activated cells in which the LMiLy vector lost its antiviral effect (Fig. 6B). The antiviral effect of the LMiLy2S vector was solely due to the RevM10 protein expression, since a control SAR vector (LΔMiLy2S) which does not encode RevM10 protein had no effect on HIV-1 replication (data not shown).
FIG. 6.
HIV-1 infection experiment. Primary T cells were harvested on day 5 after stimulation with PHA, IL-2, and feeder cells and then inoculated with the HIV-1 JR-CSF virus. Viral replication was monitored for 9 days by measuring the p24 antigen concentration in cell supernatants. “Fully activated” samples (A) were supplemented with fresh PHA, IL-2, and feeder cells on day 3 after inoculation with HIV-1 to maintain activation of T cells, whereas “partially activated” samples (B) were maintained in medium with IL-2 only. All values are averages from triplicate samples; vertical bars indicate the standard errors (where not shown the error value was below the resolution of the graphics program). Results from a representative experiment are shown. Results were reproduced with two separate tissues.
DISCUSSION
The expression of retroviral-vector-delivered transgenes depends on the activation status of primary T cells: expression is high in activated cells and low in quiescent cells. In this study, we report improved transgene expression in quiescent T cells by incorporation of the human IFN-β–SAR sequence into a retroviral vector. The mechanisms through which SAR elements influence transgene expression are not well understood. It has been proposed that SARs can facilitate the generation of “open” chromatin, allowing access of transcription factors to neighboring enhancer-promoter elements (16). The SAR-containing retroviral vectors used in the present study yielded higher steady-state RNA levels than did the control vector, indicating that the expression is regulated at a transcriptional level (unpublished results), but the finding that the effect is orientation dependent and that the IFN-β–SAR actually suppresses vector expression in the opposite orientation (Fig. 4) argues against the possibility that it may act as a classical transcriptional enhancer element. Analysis of activated T-cell clones, as well as analysis of gene marking and expression in SCID-hu-derived thymocytes, shows that the IFN-β–SAR did not confer position-independent expression to a transgene. Nevertheless, the SAR sequence prevented the attenuation of vector expression in a majority (7 of 10) of resting T-cell clones, and it also improved the level of transgene expression in Lyt-2+ SCID-hu-derived thymocytes. The reason why the SAR sequence failed to prevent attenuation of vector expression in 3 of 10 LMiLy2S clones is not clear, but this could be due to epigenetic factors such as the influence of the integration site or clonal differences in regulatory proteins that interact with the SAR or the LTR enhancer-promoter elements. Considering all our findings and the proposed mechanism of action for SAR elements (2, 16), we speculate that the IFN-β–SAR sequence can influence chromatin at the proviral integration site to promote binding of transcription factors to the LTR enhancer-promoter and in this way enhance transgene expression. On its own, however, it cannot induce formation of a transcriptionally active domain, a feature that would be required to confer position-independent expression on a linked transgene. This hypothesis is a subject of our current investigations.
We observed the positive effect of the SAR element on vector expression in mature CD4+ and CD8+ T cells isolated from PBL samples (data not shown), as well as in immature CD4+ CD8+ thymocytes derived from transduced HSPC in the SCID-hu thymus-liver mice (Fig. 5B), but there was no marked influence on vector expression in transduced HSPC (Fig. 5A). The reason for the T-cell-specific effect is not clear, particularly because expression of the human IFN-β gene is not restricted to T cells, but we can offer two possible hypotheses. First, the effect of the SAR on gene expression was detected mainly in resting T cells (Table 2). At the time of analysis, transduced HSPCs were actively replicating, resembling active T cells more than resting T cells. At present, there is no procedure available to arrest transduced HSPC in vitro, but the effect of SARs on expression in resting HSPC could be tested in vivo in mouse bone marrow transplantation model. Second, T cells but not HSPC may express a regulatory protein(s) that interacts with the SAR sequence. At least one such protein (SATB1) that binds to SAR and is expressed predominantly in thymocytes has been identified (10).
The SAR element had a somewhat different effect on transgene expression in T cells cultured in vitro (i.e., a higher percentage and a higher magnitude of expression in resting cells) from that in thymocytes produced in the SCID-hu thymus-liver model (i.e., only a higher magnitude of expression). This could be due to the different cell types that were analyzed. While in vitro-cultured cells were single-positive mature CD4+ T cells, the majority (>90%) of the thymocytes recovered from the SCID-hu thymus-liver grafts were immature CD4+ CD8+ cells which cannot be cultured in vitro. When we expanded in vitro the CD4+ thymocytes from the LMiLy- and the LMiLyS-marked grafts we observed transgene expression patterns in activated and resting cells similar to that seen with directly transduced T cells (unpublished results). It is possible therefore that the difference in expression profiles between CD4+ and CD4+ CD8+ cells is due to the cell-specific differences in the type or quantity of the regulatory proteins that interact with the SAR or the LTR enhancer-promoter elements.
We have used an in vitro culture system to study transgene expression at various stages of T-cell activation. Recently, Bunnell et al. have reported in vivo analysis of transgene expression in T cells in rhesus macaques that have been infused with retrovirally transduced CD4+ T cells (7). The expression was analyzed 20 to 90 days postinfusion. While freshly isolated T cells expressed virtually undetectable levels of transgene RNA, presumably because of their resting state, the expression was readily detected after in vitro culture and activation of T cells, a finding that is in agreement with our results. We suggest therefore that although the in vitro culture system may not be an entirely accurate model for the in vivo resting T cell, it can be used to analyze the transgene expression in activated and resting cells.
The SAR-mediated increase in vector expression was observed with the double-copy-type LMiLy2S and the single-copy-type LMiLyS vectors, demonstrating that the phenomenon was not specific to a particular type of construct. For practical use, however, the single-copy LMiLyS vector is more suitable because it yields higher virus titers. While it was not possible to obtain definitive data regarding the endpoint viral titer for the LMiLyS vector, we can predict based on previous experience with the _neo_-containing vectors (12) that the endpoint titers of the LMiLyS viral stocks were probably greater than 106 CFU/ml, results that compare favorably to those obtained with the existing retroviral vectors used in clinical trials (reviewed in reference 17).
A number of laboratories including ours have demonstrated inhibition of HIV-1 replication in primary T cells by using retroviral vectors carrying the RevM10 gene (3, 26, 33, 35). However, we have also shown that relatively high levels of RevM10 protein are required for the anti-HIV effect and that there is a window in time between fully activated and fully quiescent primary T cells (partially activated cells) during which HIV-1 can still replicate but retroviral vector expression is decreased to a level where the antiviral efficacy was measurably reduced (26). With the SAR-containing vector we have achieved significant improvement in RevM10-mediated anti-HIV efficacy (Fig. 6). Most importantly, the antiviral activity was maintained under conditions in which the standard retroviral vector failed to inhibit HIV-1 replication. The activation status of HIV-infected T cells in vivo is being investigated in many laboratories. Nevertheless, our data indicate that sustained transgene expression in T cells at all stages of activation, which can be achieved with SAR-containing vectors, will help to develop more effective RevM10-based HIV gene therapies and can also be useful for other gene therapies that rely on persistent transgene expression levels in the T-cell compartment.
ACKNOWLEDGMENTS
We thank Jennifer Auten, Kathy Moss, Creton Kalfoglou, and Michele Pineda for technical assistance, Jürgen Bode for the IFN-β–SAR element, and Mike Cooke for critical reading of the manuscript. CEMSS cells and JR-CSF HIV-1 virus were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
REFERENCES
- 1.Blaese R M, Culver K W, Miller A D, Carter C S, Fleisher T, Clerici M, Shearer G, Chang L, Chiang Y, Tolstoshev P, Greenblatt J J, Rosenberg S A, Klein H, Berger M, Mullen C A, Ramsey W J, Muul L, Morgan R A, Anderson W F. T lymphocyte-directed gene therapy for ADA-SCID: initial trial results after 4 years. Science. 1995;270:475–480. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 2.Bode J, Schlake T, Ríos-Ramírez M, Mielke C, Stengert M, Kay V, Klehr W D. Scaffold/matrix-attachment regions (S/MAR): structural properties creating transcriptionally active loci. Orlando, Fla: Academic Press, Inc.; 1995. [DOI] [PubMed] [Google Scholar]
- 3.Bonyhadi M, Moss K, Voytovitch A, Auten J, Kalfoglou C, Plavec I, Forestell S, Su L, Böhnlein E, Kaneshima H. RevM10-expressing T cells derived in vivo from transduced human hematopoietic stem-progenitor cells inhibit human immunodeficiency virus replication. J Virol. 1997;71:4707–4716. doi: 10.1128/jvi.71.6.4707-4716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordignon C, Notarangelo L D, Nobili N, Ferrari G, Casorati G, Panina P, Mazzolari E, Maggioni D, Rossi C, Servida P, Ugazio A G, Mavilio F. Gene therapy in peripheral blood lymphocytes and bone marrow for ADA-immundeficient patients. Science. 1995;270:470–475. doi: 10.1126/science.270.5235.470. [DOI] [PubMed] [Google Scholar]
- 5.Boulikas T. Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J Cell Biochem. 1993;52:14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- 6.Bunnell B A, Mesler Muul L, Donahue R E, Blease R M, Morgan R A. High-efficiency retroviral-mediated gene transfer into human and nonhuman primate peripheral blood lymphocytes. Proc Natl Acad Sci USA. 1995;92:7739–7743. doi: 10.1073/pnas.92.17.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunnell B A, Metzger M, Byrne E, Morgan R A, Donahue R E. Efficient in vivo marking of primary CD4+ T lymphocytes in nonhuman primates using a gibbon ape leukemia virus-derived retroviral vector. Blood. 1997;89:1987–1995. [PubMed] [Google Scholar]
- 8.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colicelli J, Goff S P. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1986;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 11.DiGiusto D L, Lee R, Moon J, Moss K, O’Toole T, Voytovich A, Webster D, Mulé J J. Hematopoietic potential of cryopreserved and ex vivo manipulated cord blood progenitors evaluated in vitro and in vivo. Blood. 1996;87:1261–1271. [PubMed] [Google Scholar]
- 12.Forestell S P, Böhnlein E, Rigg R J. Retroviral end-point titer is not predictive of gene transfer efficiency: implications for vector production. Gene Ther. 1995;2:723–730. [PubMed] [Google Scholar]
- 13.Hantzopoulos P A, Sullenger B A, Ungers G, Gilboa E. Improved gene expression upon transfer of the adenosine deaminase minigene outside the transcriptional unit of a retroviral vector. Proc Natl Acad Sci USA. 1989;86:3519–3523. doi: 10.1073/pnas.86.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley R G, Fong A Z C, Burns B F, Hawley T S. Transplantable myeloproliferative disease induced in mice by an interleukin 6 retrovirus. J Exp Med. 1992;176:1149–1164. doi: 10.1084/jem.176.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenuwein T, Forrester W C, Fernández-Herrero L A, Laibe G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 17.Jolly D. Viral vector system for gene therapy. Cancer Gene Ther. 1994;1:51–64. [PubMed] [Google Scholar]
- 18.Junker U, Böhnlein E, Veres G. Genetic instability of a MoMLV-based antisense double-copy retroviral vector designed for HIV-1 gene therapy. Gene Ther. 1995;2:639–646. [PubMed] [Google Scholar]
- 19.Kalos M, Fournier R E K. Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol Cell Biol. 1995;15:198–207. doi: 10.1128/mcb.15.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klehr D, Maass K, Bode J. Scaffold-attached regions from the human interferon β domain can be used to enhance stable expression of genes under the control of various promoters. Biochemistry. 1991;30:1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- 21.Malim M H, Böhnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 22.McKnight R A, Shamay A, Sankaran L, Wall R J, Hennighausen L. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mielke C, Kohwi Y, Kohwi-Shigematsu T, Bode J. Hierarchical binding of DNA fragments derived from scaffold-attached regions: correlation of properties in vitro and function in vivo. Biochemistry. 1990;29:7475–7485. doi: 10.1021/bi00484a017. [DOI] [PubMed] [Google Scholar]
- 24.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 25.Namikawa R, Weilbaecher K N, Kaneshima H, Yee E J, McCune J M. Long-term human hematopoiesis in the SCID-hu mouse. J Exp Med. 1990;172:1055–1063. doi: 10.1084/jem.172.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plavec I, Agarwal M, Ho K E, Pineda M, Auten J, Baker J, Matsuzaki H, Escaich S, Bonyhadi M, Böhnlein E. High trans-dominant RevM10 protein levels are required to inhibit HIV-1 replication in cell lines and primary T cells: implication for gene therapy of AIDS. Gene Ther. 1997;7:128–139. doi: 10.1038/sj.gt.3300369. [DOI] [PubMed] [Google Scholar]
- 27.Plavec I, Voytovich A, Moss K, Webster D, Hanley M B, Escaich S, Ho K E, Böhnlein E, DiGiusto D L. Sustained retroviral gene marking and expression in lymphoid and myeloid cells derived from transduced hematopoietic progenitor cells. Gene Ther. 1996;3:717–724. [PubMed] [Google Scholar]
- 28.Poljak L, Seum C, Mattioni T, Laemmli U K. SARs stimulate but do not confer position independent gene expression. Nucleic Acids Res. 1994;22:4386–4394. doi: 10.1093/nar/22.21.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigg J R, Chen J, Dando J S, Forestell S P, Plavec I, Böhnlein E. A novel human amphotropic packaging cell line: high titer, complement resistance, and improved safety. Virology. 1996;218:290–295. doi: 10.1006/viro.1996.0194. [DOI] [PubMed] [Google Scholar]
- 30.Su L, Lee R, Bonyhadi M, Matsuzaki H, Forestell S, Escaich S, Böhnlein E, Kaneshima H. Hematopoietic stem cell-based gene therapy for acquired immunodeficiency syndrome: efficient transduction and expression of RevM10 in myeloid cells in vivo and in vitro. Blood. 1997;89:2283–2290. [PubMed] [Google Scholar]
- 31.Tagawa M, Nakauchi L, Herzenberg L A, Nolan G P. Formal proof that different-size Lyt-2 polypeptides arise from differential splicing and post-transcriptional regulation. Proc Natl Acad Sci USA. 1986;83:3422–3426. doi: 10.1073/pnas.83.10.3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson E M, Christians E, Stinnakre M-G, Renard J-P. Scaffold attachment region stimulate HSP70.1 expression in mouse preimplantation embryos but not in differentiated tissues. Mol Cell Biol. 1994;14:4694–4703. doi: 10.1128/mcb.14.7.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandendriesche T, Chuah M K L, Chiang L, Chang H-K, Ensoli B, Morgan R A. Inhibition of clinical human immunodeficiency virus (HIV) type 1 isolates in primary CD4+ T lymphocytes by retroviral vectors expressing anti-HIV genes. J Virol. 1995;69:4045–4052. doi: 10.1128/jvi.69.7.4045-4052.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woffendin C, Ranga U, Yang Z-Y, Xu L, Nabel G J. Expression of a protective gene prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc Natl Acad Sci USA. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woffendin C, Yang Z Y, Udaykumar R, Xu L, Yang N S, Sheehy M J, Nabel G J. Nonviral and viral delivery of a human immunodeficiency virus protective gene into primary human T cells. Proc Natl Acad Sci USA. 1994;91:11581–11585. doi: 10.1073/pnas.91.24.11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]