Neuronal Death Induced by Brain-Derived Human Immunodeficiency Virus Type 1 Envelope Genes Differs between Demented and Nondemented AIDS Patients (original) (raw)
Abstract
Human immunodeficiency virus type 1 (HIV-1) infection of the brain results in viral replication primarily in macrophages and microglia. Despite frequent detection of viral genome and proteins in the brains of AIDS patients with and without HIV dementia, only 20% of AIDS patients become demented. To investigate the role of viral envelope gene variation in the occurrence of dementia, we examined regions of variability in the viral envelope gene isolated from brains of AIDS patients. Brain-derived HIV-1 V1-V2 envelope sequences from seven demented and six nondemented AIDS patients displayed significant sequence differences between clinical groups, and by phylogenetic analysis, sequences from the demented group showed clustering. Infectious recombinant viruses containing brain-derived V3 sequences from both clinical groups were macrophagetropic, and viruses containing brain-derived V1, V2, and V3 sequences from both clinical groups spread efficiently in macrophages. In an indirect in vitro neurotoxicity assay using supernatant fluid from HIV-1-infected macrophages, recombinant viruses from demented patients induced greater neuronal death than viruses from nondemented patients. Thus, the HIV-1 envelope diversity observed in these patient groups appeared to influence the release of neurotoxic molecules from macrophages and might account in part for the variability in occurrence of dementia in AIDS patients.
Human immunodeficiency virus type 1 (HIV-1) is frequently detectable in the brains of HIV-infected individuals at all stages of infection (4), but only 20% of AIDS patients develop HIV dementia (HIV-D) (35). In both HIV-D and HIV nondemented (HIV-ND) AIDS patients, productive HIV-1 infection in the brain is limited to nonneuronal cells, primarily perivascular macrophages and microglia (20, 26, 58) and, to a lesser extent, astrocytes (61, 70). Only certain HIV-1 isolates can infect macrophages (16), and tropism for these cell types is influenced by specific amino acids within and adjacent to the V3 hypervariable region (Fig. 1) of the HIV-1 envelope (10, 41, 54, 69). Similar sequences influence tropism for microglia (23), and not surprisingly, V3 sequences from brain-derived HIV-1 closely resemble those of previously described blood-derived macrophagetropic viruses (48, 51). The extent of HIV-1 infection in macrophages is also affected by other viral envelope regions including the V1 and V2 hypervariable regions (27), which appear to modulate the efficiency of viral spread in macrophages (64). Although altered V3 sequences associated with the syncytium-inducing nonmacrophagetropic phenotype frequently arise during the progression of clinical AIDS (24, 53), there are conflicting results regarding the generation of variant V1-V2 sequences during progression of clinical disease (21, 55, 67).
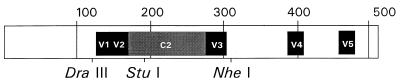
FIG. 1.
The entire HIV-1 gp120 sequence (511 amino acids) showing the regions of interest, including the V1-V2 and C2-V3 fragments and restriction sites used in this study. The V1 and V2 regions lie between the _Dra_III and _Stu_I sites (76 amino acids), and the C2 and V3 regions are within the _Stu_I-to-_Nhe_I fragment (143 amino acids).
Viral envelope gene variation is known to influence incidence of neurological disease in a variety of retroviral and nonretroviral systems (13, 33, 60, 65). In several neurotropic murine retrovirus models, one or more changes in envelope amino acids are sufficient to cause a switch from a virulent to a nonvirulent phenotype (34, 42, 45, 71). Among HIV-1-infected humans, previous studies of HIV-D and HIV-ND AIDS patients indicated that specific brain-derived V3 sequences correlate with the occurrence of dementia (48). Some of these sequences may influence specialized adaptation to growth in microglial cells as opposed to macrophages in general (59), while other sequence differences might determine neurotoxic effects, subsequent to macrophage or microglial cell infection. The induction of neuronal injury or death may proceed from a complex cascade of events involving viral and host molecules to cause the clinical manifestations of HIV-D. Nonetheless, how different viral proteins might contribute to neural damage remains uncertain. It has been proposed that HIV-1 envelope protein might be directly toxic to neurons (14) or might mediate neuronal injury indirectly through induction of toxic cytokines or other host molecules released from infected microglia (32). However, other viral proteins, including Tat (40) and gp41 (2), have also been implicated in HIV-1 neuropathogenesis.
In the present study, brain-derived V1 and V2 envelope sequences from prospectively studied individuals with or without HIV-D were examined (47, 48), and significant differences were detected between sequences of demented and nondemented groups. Recombinant viruses, constructed by using portions of envelope genes obtained from both clinical groups, showed similar patterns of infectivity and spread in macrophages. However, neuronal death induced by supernatants from HIV-infected macrophages was significantly greater among HIV-D-derived viruses than among HIV-ND-derived viruses, suggesting that viral envelope variability might be important in the pathogenesis of HIV-D.
MATERIALS AND METHODS
Sequence amplification and analysis.
V1-V2 envelope sequences were obtained by using previously reported conditions in a nested PCR protocol (48). Briefly, cDNA was synthesized from total RNA, obtained directly from infected brains of patients described previously (68), and was used as a template for the first PCR. Two microliters of the product was added to the second PCR. Oligonucleotides 6575/7330C (9) and 6575/6804C (5′-ACAGGCCTATATAATGACT-3′) were included in the first and second PCRs, respectively. This amplification yielded a fragment of 230 bp that included both the V1 and V2 regions of the HIV-1 envelope gene. Products from multiple amplifications of the same cDNA template were cloned into the pCRII vector (Invitrogen), and the sequence was analyzed by dideoxy sequencing. Sequence fragments were aligned with CLUSTAL (DNAStar) by identity comparison of each residue, and phylogenetic tree construction was performed by neighbor-joining and maximal parsimony analyses. To ensure that contaminant viruses had not been amplified, all sequences were compared to previously reported HIV-1 V1-V2 sequences (38).
Construction of recombinant viruses.
Recombinant HIV-1 clones containing V1, V2, and V3 regions from each patient were generated with a two-step construction process. First, brain-derived V1-V2 sequences from patients were excised from the pCRII cloning sites, _Dra_III and _Stu_I (Fig. 1). These fragments were ligated into the _Dra_III and _Stu_I sites of a plasmid which contained the envelope sequence from the _Eco_RI (5743) to _Dra_III (6591) sites of the HIV-1 clone NL4-3, with an adjacent polylinker encoding _Dra_III-_Stu_I-_Xba_I-_Mlu_I-_Nhe_I-_Bsu_I-_Bam_HI sites. This cassette was in a vector, p4-8b, derived from pBluescript KS(+) but lacking the _Dra_III site at position 230, thus making the _Dra_III site in the HIV-1 envelope unique. Next, HIV-1 sequences from _Eco_RI to _Stu_I were excised from clones generated from each patient’s cDNA and were ligated into plasmids described previously (47), containing C2-V3 sequences from _Stu_I and _Nhe_I sites from each patient in the NL4-3 background. To facilitate this cloning step, each of these vectors was previously modified by replacement of the original NL4-3 sequence from _Eco_RI to _Stu_I by a synthetic oligonucleotide. The resulting vectors were transfected into CD4-expressing HeLa cells that were then cocultivated for 24 h with uninfected phytohemagglutinin-stimulated human peripheral blood mononuclear cells (PBMC). PBMC were then maintained in interleukin-2-containing medium to obtain infectious virus stocks as previously described (6).
In vitro infectivity assays.
All viruses derived from transfection studies were titrated in PBMC as previously reported (6). HIV-1 infectivity studies were performed in CD4-positive HeLa (HeLa-CD4) cells (clone 1022) (8), CD4-positive, CKR5-positive HeLa (HeLa-CD4/CKR5) cells (clone JC37) (44), and primary human macrophages by using established methods (6). Viral infection and replication were measured as supernatant p24 levels or by staining with anti-p24 antibody in a focal immunoassay (7). Controls included uninfected cells or cells infected with viruses of known cell tropism, including NL4-3 and JR-FL.
Neurotoxicity assay.
Neuronal cultures were prepared from 12- to 15-week gestational fetuses with approval of the Human Ethics Committee at the University of Manitoba as previously reported (40). Briefly, the meninges and blood vessels were removed, the tissue was mechanically dissociated, cells were resuspended in Opti-MEM (GIBCO) with 1% heat-inactivated fetal bovine serum, 1% N2 supplement (GIBCO), and 1% antibiotic solution (104 U of penicillin G per ml and 10 mg of streptomycin B per ml in 0.9% NaCl), seeded in 96-well microtiter plates at 105 per well, and maintained for a minimum of 4 weeks prior to use. Sample wells were immunostained for the neuronal marker microtubule-associated protein 2, and only cultures in which >70% of the cells stained positive for the marker were used for experiments. The remaining cells were principally astrocytes, as indicated by glial acidic fibrillary protein immunopositivity with rare microglia (<1%) which immunostained with EBM-11 (46).
Primary human macrophages were infected with 102 to 103 50% tissue culture infective doses (TCID50) of different HIV recombinants containing brain-derived C2-V3 or V1-V3 sequences or of JR-FL. Culture supernatant was harvested as conditioned medium (CM) at days 3, 7, and 10 postinfection and stored at −80°C. Prior to application to neuronal cultures, CM was centrifuged at 13,000 rpm for 10 min to clear cellular debris, mixed at several dilutions with Opti-MEM containing 0.1% fetal bovine serum, and applied to the cultured neurons for 3 to 24 h. The neuronal cultures were stained subsequently with trypan blue to identify dead cells and fixed in 4% paraformaldehyde, and the number of neurons with trypan blue-positive nuclei per unit area was determined over five randomly chosen fields by an examiner unaware of the specific treatment. Neuronal death was expressed as the percentage of trypan blue-stained neurons to total number of neurons counted. Background neuronal death in untreated fetal neuronal cultures varied 4 to 8%, depending on the age of the fetus, and thus was subtracted from total neuronal death for each experiment. To assess the role of the glutamate receptors in the neurotoxicity observed, neuronal cultures were pretreated with the _N_-methyl-d-aspartate (NMDA) receptor antagonist AP5 and the amino-3-hydroxy-5-methyl-4-isoazole propionate (AMPA) receptor antagonist CNQX (RBI, Natick, Mass.) before application of the CM. Individual experiments were conducted in triplicate wells and repeated at least twice; means and standard errors of the means (SEM) were determined.
Statistical tests.
Statistical analyses comparing groups were made by using nonparametric (Mann-Whitney U), parametric (Student’s t), or Fisher’s exact test.
RESULTS
HIV-1 envelope sequence analysis.
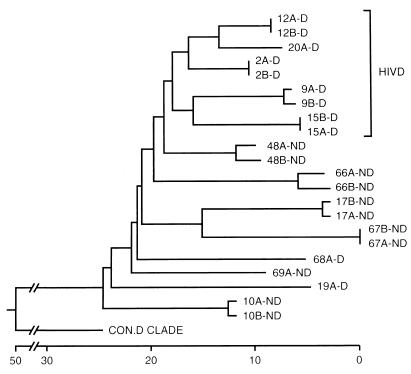
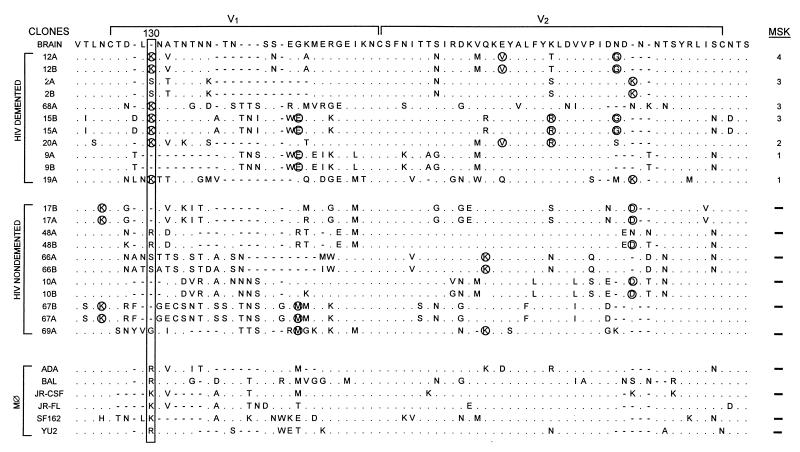
We compared the V1-V2 sequences (Fig. 1) derived from brains of HIV-D and HIV-ND patients in our prospectively studied population (2, 20, 48, 68) (Table 1). Phylogenetic analysis of V1-V2 sequences from HIV-D and HIV-ND patients showed that clones from the same patient were closely related, but individual patients displayed marked sequence divergence with low bootstrap values (Fig. 2). V1-V2 sequences in five of the seven HIV-D patients whose severity of dementia was measured by the Memorial Sloan-Kettering (MSK) scale (49) were clustered together, while no clusters of three or more patients within the HIV-ND group were observed. When specific residues at each position of all brain-derived sequences were compared (Fig. 3), a lysine at position 130 predominated in the HIV-D group in 7 of 11 clones and was never present at the same position in the ND group. There were numerous positions at which two or more patients in one group shared an identical residue that was not present at the same position in the other clinical group. These were termed unique amino acids (Fig. 3, circled) and were more frequently observed among clones from HIV-D clones. Thus, the above findings resembled those previously observed in brain-derived C2-V3 envelope sequences in which HIV-D and HIV-ND sequences differed significantly at specific positions and the number of unique amino acids was greater in the HIV-D group (48). Together with the phylogenetic analysis indicating the clustering of five of seven HIV-D sequences, these observations suggest that specific V1 and V2 sequences were also closely associated with the development of HIV-D.
TABLE 1.
Clinical features of HIV-D and HIV-ND patients
| Clinical groupa | Mean age (yr) ± SD | Mean CD4+ cells/mm3 ± SD | No. with HIV encephalitisb | No. receiving antiretroviral drug therapy |
|---|---|---|---|---|
| HIV-D (n = 7) | 33 ± 3.8 | 56 ± 75 | 3 | 6 |
| HIV-ND (n = 6) | 38 ± 9.4 | 38 ± 42 | 0 | 4 |
FIG. 2.
Phylogenetic comparison of brain-derived V1-V2 sequences obtained from HIV-D (D) and HIV-ND (ND) individuals with AIDS and the consensus sequence from clade D (con.D clade) (38), using neighbor-joining analysis (30). Numbers refer to individual patients, and letters (A and B) refer to clones from the same patient. Two clones were analyzed for each patient. Clones from five of seven HIV-D patients clustered together, while none of the HIV-ND patients showed close associations. The horizontal axis represents the number of substitution events. Analysis of the V1 and V2 sequences separately revealed no clustering of sequences from the same clinical group. Similar topologies were obtained with different phylogenetic methods, but bootstrap values were low (<70) between individuals.
FIG. 3.
Brain-derived V1-V2 envelope sequences from HIV-D (D) and HIV-ND (ND) patients aligned with the brain consensus sequence and corresponding sequences from established macrophagetropic viruses. HIV-D sequences are presented in order of severity of dementia according to MSK score shown at the right. Two clones were sequenced from each patient except for patients 68, 20, 19, and 69. Position 130 (boxed) shows significant difference between HIV-D and HIV-ND groups (Fisher’s exact, P < 0.01), with a lysine predominating in the HIV-D group and a variable amino acid in the HIV-ND group. Comparison of the frequency of the Lys130 in established databases (38) revealed that it was detected significantly more frequently in the HIV-D sequences than in the database (Fisher’s exact, P < 0.01). Circled amino acids identified in two or more patients in one group, but not present in the other group at a specific position, were termed unique. The mean number of unique amino acids per clone ± SEM was significantly greater in the HIV-D group (3.0 ± 0.44) than in the HIV-ND group (1.6 ± 0.20) (Student’s t test, P < 0.05). In patients with two or more clones, the difference between clones varied from zero to three amino acids. The length of the V2 region, the number of positively charged amino acids, and number of potential glycosylation sites did not differ between HIV-D and HIV-ND groups.
Tropism of brain-derived recombinant HIV clones.
Previous studies of recombinant viruses containing brain-derived C2-V3 envelope sequences indicate that these sequences influence the ability of HIV-1 to infect macrophages and mixed glial cells (47). The V1-V2 envelope regions of HIV-1 appear to enhance the efficiency of replication in macrophages in vitro, at least in part, by increasing viral spread (64). To further examine the effect of brain-derived V1-V2 sequences, V1-V2 sequences from two HIV-D patients and three HIV-ND patients were inserted into recombinant viruses already containing C2-V3 sequences from these same patients. These clones were compared to the recombinant viruses containing only the C2-V3 envelope sequences from the same patients. All recombinant viruses with either C2-V3 or V1-V3 brain-derived envelope sequences infected HeLa-CD4/CKR5 cells efficiently but failed to infect HeLa-CD4 cells lacking CKR5 expression (Table 2). Thus, by these criteria, all recombinant viruses utilized the CKR5 coreceptor, similar to what occurs with other macrophagetropic viruses (5, 56), but no difference between recombinant viruses containing brain-derived C2-V3 versus V1-V3 sequences was seen.
TABLE 2.
Comparison of infectivities of recombinant HIV-1 clones containing brain-derived V1-V3 and C2-V3 sequences in different cell types
| Patient | Status | Clonea | Envelope insertb | HIV titerc | ||
|---|---|---|---|---|---|---|
| PBMCd | HeLa-CD4/CKR5 | HeLa-CD4 | ||||
| 48 | ND | 48-1C | C2-V3 | 9.8 × 103 | 5.8 × 104 | <5 |
| 234-11 | V1-V3 | 9.3 × 103 | 8.2 × 104 | <5 | ||
| 10 | ND | 10-1A | C2-V3 | 2.8 × 104 | 3.7 × 104 | <5 |
| 215-7 | V1-V3 | 2.0 × 104 | 5.0 × 104 | <5 | ||
| 17 | ND | 17-2B | C2-V3 | 1.1 × 103 | 5.4 × 104 | <5 |
| 214-4 | V1-V3 | 2.3 × 104 | 3.4 × 104 | <5 | ||
| 2 | D | 2-1B | C2-V3 | 2.5 × 103 | 1.3 × 104 | <5 |
| 218-14 | V1-V3 | 5.4 × 103 | 9.2 × 104 | <5 | ||
| 15 | D | 15-2B | C2-V3 | 1.0 × 103 | 3.4 × 104 | <5 |
| 237-5 | V1-V3 | 6.3 × 103 | 2.0 × 104 | <5 | ||
| JR | D | JR-FL | 3.1 × 103 | 3.3 × 103 | <5 | |
| NL4-3 | 4.4 × 103 | 4.9 × 103 | 3.9 × 103 |
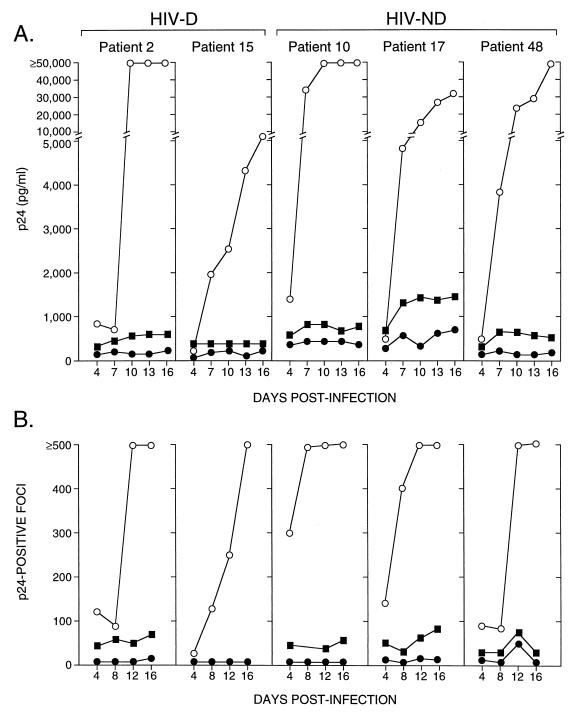
In experiments where infectivity of these clones was also analyzed in human macrophages, V1-V3 recombinant viruses showed a large increase in p24 levels and the number of positive cells with time postinfection. In contrast, the recombinant viruses containing only C2-V3 sequences from patients produced p24 levels and numbers of infected macrophages that were lower and failed to increase with time after infection even when 10-fold-higher amounts of virus were used to infect cells (Fig. 4). Thus, the addition of the V1 and V2 regions of brain-derived HIV-1 to the C2-V3 containing clones from each patient enhanced viral replication and spread in macrophages. However, infectivity of the V1-V3 clones did not differ significantly between HIV-D- and HIV-ND-derived viruses. These results indicated that the ability to replicate efficiently and spread in macrophages might be a property common to all brain-derived HIV-1 envelope sequences but did not correlate with the development of HIV-D.
FIG. 4.
Infectivity of brain-derived C2-V3- and V1-V3-containing recombinant viruses measured by p24 levels in macrophage culture supernatant (A) or p24-positive foci (B) over time. At an input titer of 102 TCID50/0.1 ml, V1-V3 clones (open circles) replicated to a greater extent in macrophages, reflected by higher p24 levels in supernatant (A) and progressively increasing numbers of p24-positive cells (B), compared to the matched C2-V3 clones (closed circles) for both HIV-D- and HIV-ND-derived viruses. When a 10-fold-higher input of C2-V3 viruses was used (closed squares), higher p24 values and focus counts were observed for all viruses, confirming that these C2-V3 clones were macrophagetropic. The increasing p24 level in patient 17 did not reflect an increase in focus number and probably was due to accumulation of p24 in the medium, since all medium was not removed when cells were fed every third day.
Virus-induced neuronal toxicity.
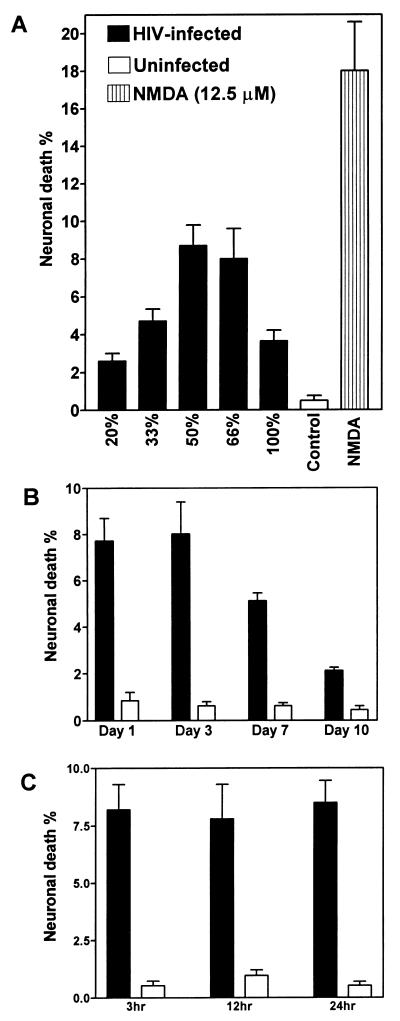
Since neurons are rarely, if ever, infected by HIV-1, the mechanism of neuronal injury and/or death resulting from HIV-1 infection of the brain is assumed to be indirect, possibly due to the release of neurotoxic molecules by HIV-1-infected macrophages or microglia (17, 50). Therefore, we examined human fetal neuronal cultures after treatment with CM from macrophage cultures infected with different HIV-1 clones. CM from macrophages infected with JR-FL, a brain-derived isolate from a patient with HIV-D (29), produced a maximum neuronal death when diluted to 50 to 66% with fresh medium, but 100% CM produced a reduced neuronal death rate (Fig. 5A). Comparison of neurotoxicity induced by CM harvested at days 1, 3, 7, and 10 after infection of macrophages with JR-FL revealed that maximal neuronal death was induced by CM from days 1 and 3 (Fig. 5B). Consistent with previous experiments, viral reverse transcriptase assays indicated that background values were detected on days 1 and 3 and significantly higher virus release occurred on days 7 and 10 (data not shown). Thus, there was no correlation between neurotoxicity and the presence of virus and viral proteins in supernatant fluids. To determine if the duration of exposure to CM influenced the extent of neuronal death induced by CM, neuronal death rates after treatment of neuronal cultures for 3, 12, and 24 h were compared. Neuronal death did not differ significantly at the three time points tested (Fig. 5C), suggesting that neuronal death occurred rapidly and that only a subpopulation of neurons were susceptible to killing in this assay.
FIG. 5.
Comparison of mean percentage (±SEM) of neuronal death induced by CM from macrophages infected with HIV-1 JR-FL, uninfected macrophages (control), or NMDA treatment (A), CM harvested from different time points after infection (B), and different times of exposure of neuronal cultures to CM (C). JR-FL induced neurotoxicity at various dilutions of CM (A), with maximum neuronal death at a 50 to 66% dilution. Neuronal cultures treated with 50% CM from uninfected macrophages showed significantly less neuronal death than CM at the same concentration from JR-FL-infected macrophages (Student’s t test, P < 0.001). A decline in neuronal killing was observed with 50% CM harvested at later time points postinfection (B). The extent of neuronal death did not change with different times of exposure to CM, harvested at 3 days postinfection (C). Macrophages were infected at an input titer of 103 TCID50/0.1 ml.
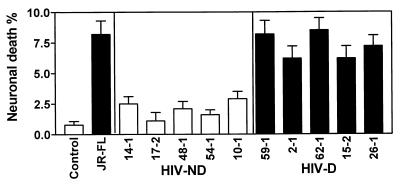
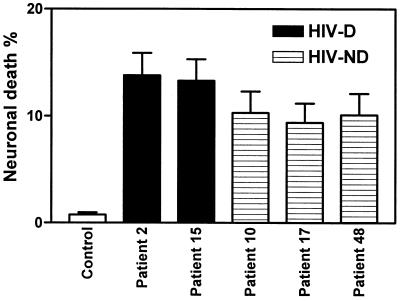
With respect to neuronal death induced by CM from macrophage cultures infected with recombinant HIV-1 clones containing brain-derived envelope sequences, the viruses containing the brain-derived C2-V3 regions from the HIV-D group induced significantly higher levels of neuronal death than the HIV-ND group (Fig. 6). Comparison of individual HIV-D-derived and individual HIV-ND-derived viruses showed that each HIV-D-derived virus induced significantly higher levels of neuronal death than each HIV-ND-derived virus. Comparing the five clones containing brain-derived V1-V3 sequences in the neurotoxicity assay, we found a trend toward increased neuronal death induced by the two HIV-D clones compared to the HIV-ND clones, but too few clones were tested to give this observation statistical significance (Fig. 7). Nevertheless, the percentage of neuronal death induced by all the V1-V3-containing clones (9 to 14%) was higher than that induced by the C2-V3-containing clones (1 to 8.5%), implying that neuronal death may be influenced by more than one region of the HIV-1 envelope. This latter effect may be due to the fact that more macrophages were infected on day 3 by the brain-derived V1-V3-containing clones than by the C2-V3-containing clones (Fig. 4).
FIG. 6.
Mean percentage of neuronal death (±SEM) caused by CM from control (uninfected) or JR-FL-infected macrophage cultures or macrophage cultures infected with recombinant viruses containing brain-derived C2-V3 sequences from HIV-D and HIV-ND individuals. Mean neuronal death caused by all recombinant HIV-D viruses was significantly greater than neuronal death caused by all HIV-ND viruses (Student’s t test, P < 0.0001). Each HIV-D-derived virus caused significantly more neuronal death than each HIV-ND-derived virus when the isolates were compared individually (P < 0.05). Macrophages were infected at an input titer of 103 TCID50/0.1 ml.
FIG. 7.
Comparison of mean percentages (±SEM) of neuronal death induced by CM for brain-derived V1-V3-containing recombinant viruses from two HIV-D and three HIV-ND patients compared to CM from control uninfected CM. For all five patients, the recombinant viruses caused greater neuronal death than the uninfected CM (Student’s t test, P < 0.001). HIV-D clones caused greater neurotoxicity than corresponding HIV-ND clones. Macrophages were infected at an input titer of 102 TCID50/0.1 ml.
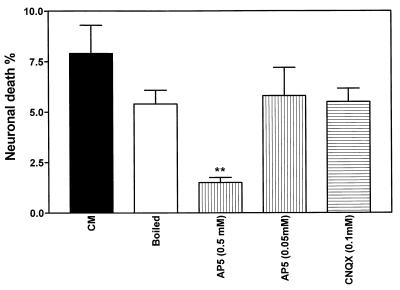
Several neurotoxic molecules that vary in stability and mechanism of action have been implicated in different assays of HIV-induced neuronal death (19, 40). To characterize the neurotoxin released by macrophages in the present assay, we determined the stability of the CM from macrophages infected by a recombinant virus from an HIV-D patient (clone 62-1). CM was boiled for 10 min, brought to 37°C, and applied to neuronal cultures. The percentage of neurotoxicity of the boiled CM did not differ significantly from that of the untreated supernatant, indicating that the neurotoxic molecule(s) was heat stable (Fig. 8). Since glutamate receptor-mediated mechanisms have been suggested to participate in HIV-induced neurotoxicity (3, 14), we pretreated the neuronal cultures with an NMDA (AP5) or AMPA (CNQX) receptor antagonist. Application of CM from macrophages infected with the virus clone 62-1 revealed that after the NMDA receptor was blocked with 0.5 mM AP5, neuronal death was significantly reduced (Fig. 8). In contrast, blocking the AMPA receptor with CNQX caused no inhibition of neurotoxicity.
FIG. 8.
Comparison of mean neuronal death rates induced by CM from macrophages infected by the recombinant virus 62-1, containing the brain-derived C2-V3 sequences from HIV-D patient 62, after boiling for 10 min or pretreatment with AP5 or CNQX. AP5 (0.5 mM) significantly reduced neuronal killing by CM (Student’s t test, P < 0.01). AP5 or CNQX at identical concentrations without CM did not influence neuronal death rates (data not shown). Macrophages were infected at an input titer of 103 TCID50/0.1 ml.
DISCUSSION
The results presented above indicate that HIV-1 envelope sequence diversity influenced induction of neuronal death. In addition, the ability of a recombinant virus to cause neuronal death was associated with the clinical status of the individual patient from which the envelope sequence was derived. These findings suggest that envelope protein itself may be important in causing neurological disease and that envelope sequence variation may account in part for the variation in occurrence of clinical HIV-D. The current studies also indicate that in both HIV-D and HIV-ND patients, brain infection appears to select for viruses that express a V3 envelope region capable of mediating infection of macrophages and microglia and using CCR5 as a coreceptor. Furthermore, the patients in both groups also selected viruses with V1 and V2 regions that could cause a spreading infection in macrophage cultures in vitro. Thus, a spreading infection in macrophages and microglia may be a necessary feature of HIV infection in brain, although the spreading phenotype by itself is not sufficient to cause clinical HIV-D. In addition, induction of dementia may require the presence of specific viral sequences such as the dementia-associated sequences detected in the V1 and V3 regions (Fig. 3) (48).
The phylogenetic clustering observed in the V1-V2 sequences of many of the demented patients in this study was unexpected. The HIV-1 strains which appear early after seroconversion resemble macrophagetropic viruses (36), suggesting that such viruses are selected during transmission between individuals. However, no similar selection for common V1 or V2 sequences has yet been observed. HIV replication in patients is known to give rise to a wide diversity of mutant or variant viruses which would be subjected to many selective pressures during the course of the infection. The finding of a cluster among V1-V2 sequences from HIV-D patients might suggest either that these patients were initially infected with a similar and somewhat unique virus or that these patients have exerted similar selective pressures on their viruses during infection. This could be due to similar immune response genes leading to selection for or against certain viral variants, or it could be due to other host genetic or nongenetic factors which these patients have in common. Whatever viral or host factors account for this cluster may also have the capability of influencing the occurrence of clinical dementia.
Since HIV does not directly infect neurons, the pathogenesis of HIV-D is likely a complex indirect multistep process extending from virus entry into the brain to infection of brain microglial cells and production of molecules capable of damaging neurons (32). Many different viral and host factors could simultaneously influence the extent and/or tempo of the disease. In fact, even a single viral gene such as env could have several different effects that might map to similar or different regions of the envelope protein. For example, certain V3 amino acid residues affect macrophage tropism (10), V1 and V2 sequences affect virus spread in macrophages (64), and certain V1 and V3 sequences correlate with dementia (48). In the present study, the addition of the brain-derived V1-V2 fragment increased replication in macrophages, resulting in greater induction of neurotoxicity by viruses from both nondemented and demented patients. Hence, replication level and neurotoxicity potential are interrelated viral features that may influence the occurrence of neurological disease. Similar phenomena have been observed in brain disease induced by other retroviruses. Tropism of simian immunodeficiency virus (SIV) for brain microglial cells is correlated with the selection of particular sequences in several different envelope regions (25, 31); however, additional specific envelope sequences in the transmembrane region may also be required for induction of clinical brain disease (33). Similarly, the murine polytropic retrovirus Fr98 has two separate regions of the envelope gene which contain determinants of neurovirulence that may act by different mechanisms to facilitate the same clinical neurological disease (22, 45, 52).
These studies used an indirect neurotoxicity assay to determine the cytotoxic effects of viral envelope sequences derived from brain tissue of AIDS patients. Although viruses from the demented patients induced significantly more neuronal killing than those from the nondemented patients, this analysis may underestimate the extent of true neuronal damage, as some cells might be injured, but not killed, by exposure to supernatant fluid of infected macrophages. The percentage of neuronal death in the present assay was maximal at 50 to 66% macrophage CM. The reduced toxicity observed with 100% CM might be due to a possible neuroprotective effect of higher serum concentrations (15). In addition, the extent of neuronal killing was greater for supernatants derived from macrophage cultures infected with recombinant viruses containing brain-derived V1-V3 sequences that also displayed higher levels of viral spreading, indicating that an increased number of infected cells results in enhanced neurotoxicity. However, the levels of neurotoxicity in the present study were lower than those in other assays of neurotoxicity (12, 18). Our findings were based on the use of human neuronal cultures in which a subpopulation of neurons may be susceptible to injury and/or death, and rates of neuronal death are low with several different neurotoxins (40). In our assay of neurotoxicity, the rate of neuronal death declined with supernatants harvested from later time points postinfection, indicating that perhaps the macrophage cultures change in their capacity to release the neurotoxic molecule(s) with time. It is not yet clear which molecules released by macrophages are responsible for the observed neuronal death, although many reports indicate that host molecules released by cells infected by HIV-1 (18, 50), feline immunodeficiency virus (37), and SIV (1, 39) contribute to neuronal damage. The present study showed that the neurotoxic molecule(s) released by infected macrophages was heat stable, and in agreement with earlier studies (3, 12), our results implicate the NMDA receptor in the mechanism of neurotoxicity. Several candidate molecules that are released from macrophages are heat stable and influence NMDA receptor-mediated neuronal death. These include quinolinic acid (43), Ntox (19), and nitric oxide and its metabolites (12). Individual HIV-1 strains with differing envelope sequences have been shown to vary in the ability to induce quinolinic acid (11) and NO (28) production. Thus, it is conceivable that brain-derived HIV-1 strains vary in pathogenic potential, and some of this variability could be due to variation in envelope protein sequences as documented in the present and previous studies (48).
Our findings of an association between clinical status, viral envelope sequence, and in vitro biological effects conferred by the viral sequences suggest a possible relationship between viral protein variation and occurrence of clinical dementia. However, the actual in vivo mechanisms influencing the development of dementia in AIDS patients remain unclear. To determine the true pathogenic effects of different HIV-1 envelope sequences in the brain, in vivo assays will be essential. Potential models include SCID mice inoculated with HIV-infected macrophages in the brain (66), transgenic animals (62, 63), or SIV/HIV recombinant viruses constructed for testing in primates (57).
ACKNOWLEDGMENTS
We thank John Portis, Jonathan Geiger, and Kevin Coombs for helpful discussions, Jonathan Glass for assistance with collection of human tissues, and Carol Martin and Mark Bernier for preparation of the neuronal cultures.
C.P. is an NHRDP/MRC Scholar. This study was supported by the MHRC, NHRDP/MRC, and grants NS26643, AI35042, and RR00722.
REFERENCES
- 1.Adamson D C, Dawson T M, Zink M C, Clements J E, Dawson V L. Neurovirulent simian immunodeficiency virus infection induces neuronal, endothelial, and glial apoptosis. Mol Med. 1996;2:417–428. [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson D C, Wildemann B, Sasaki M, Glass J D, McArthur J C, Christov V I, Dawson T M, Dawson V L. Immunologic NO synthase: elevation in severe AIDS dementia and induction by HIV-1 gp41. Science. 1996;274:1917–1921. doi: 10.1126/science.274.5294.1917. [DOI] [PubMed] [Google Scholar]
- 3.Barks J D E, Liu X-H, Sun R, Silverstein F S. gp120, a human immunodeficiency virus-1 coat protein, augments excitotoxic hippocampal injury in perinatal rats. Neuroscience. 1996;76:397–409. doi: 10.1016/s0306-4522(96)00373-9. [DOI] [PubMed] [Google Scholar]
- 4.Bell J E, Busuttil A, Ironside J W, Rebus S, Donaldson Y K, Simmonds P, Peutherer J F. Human immunodeficiency virus and the brain: investigations of virus load and neuropathologic changes in pre-AIDS subjects. J Infect Dis. 1993;168:818–824. doi: 10.1093/infdis/168.4.818. [DOI] [PubMed] [Google Scholar]
- 5.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism and human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesebro B, Nishio J, Perryman S, Cann A, O’Brien W, Chen I S Y, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesebro B, Wehrly K, Metcalf J, Griffin D E. Use of a new CD4-positive HeLa cell clone for direct quantification of infectious human immunodeficiency virus from blood cells of AIDS patients. J Infect Dis. 1991;163:64–70. doi: 10.1093/infdis/163.1.64. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham A L, Naif H, Saksena N, Lynch G, Chang J, Li S, Jozwiak R, Alali M, Wang B, Fear W, Sloane A, Pemberton L, Brew B. HIV-1 infection of macrophages and pathogenesis of AIDS dementia complex: interaction of the host cell and viral genotype. J Leukoc Biol. 1996;62:117–125. doi: 10.1002/jlb.62.1.117. [DOI] [PubMed] [Google Scholar]
- 12.Dawson V L, Dawson T M, Uhl G R, Snyder S H. Human immunodeficiency virus type 1 coat protein neurotoxicity mediated by nitric oxide in primary cortical cultures. Proc Natl Acad Sci USA. 1993;90:3256–3259. doi: 10.1073/pnas.90.8.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietzschold B, Wunner W H, Wiktor T J, Lopes A D, Lafon M, Smith C L, Koprowski H. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc Natl Acad Sci USA. 1983;80:70–74. doi: 10.1073/pnas.80.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreyer E B, Kaiser P K, Offermann J T, Lipton S A. HIV-1 coat protein neurotoxicity prevented by calcium channel antagonists. Science. 1990;248:364–367. doi: 10.1126/science.2326646. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari G, Batistatou A, Greene L A. Ganglosides rescue neuronal cells from death after factor deprivation. J Neurosci. 1993;13:1879–1887. doi: 10.1523/JNEUROSCI.13-05-01879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 17.Giulian D, Vaga K, Noonan C A. Secretion of neurotoxins by mononuclear phagocytes infected with HIV-1. Science. 1990;250:1593–1596. doi: 10.1126/science.2148832. [DOI] [PubMed] [Google Scholar]
- 18.Giulian D, Wendt E, Vaca K, Noonan C A. The envelope glycoprotein of human immunodeficiency virus type 1 stimulates release of neurotoxins from monocytes. Proc Natl Acad Sci USA. 1993;90:2769–2773. doi: 10.1073/pnas.90.7.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giulian D, Yu J, Li X, Tom D, Li J, Wendt E, Lin S N, Schwarcz R, Noonan C. Study of receptor-mediated neurotoxins released by HIV-1-infected mononuclear phagocytes found in human brain. J Neurosci. 1996;16:3139–3153. doi: 10.1523/JNEUROSCI.16-10-03139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass J D, Fedor H, Wesselingh S L, McArthur J C. Immunocytochemical quantitation of human immunodeficiency virus in the brain: correlations with dementia. Ann Neurol. 1995;38:755–762. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- 21.Groenink M, Fouchier R A M, Broersen S, Baker C H, Koot M, van’t Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 22.Hasenkrug K J, Robertson S J, Porti J, McAtee F, Nishio J, Chesebro B. Two separate envelope regions influence induction of brain disease by a polytropic murine retrovirus (FMCF98) J Virol. 1996;70:4825–4828. doi: 10.1128/jvi.70.7.4825-4828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan C A, Watkins B A, Kufta C, Dubois-Dalcq M. Infection of brain microglial cells by human immunodeficiency virus type 1 is CD4 dependent. J Virol. 1991;65:736–742. doi: 10.1128/jvi.65.2.736-742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson A K, Parsmyr E, Sandstrom E, Fenyo E M, Albert J. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol. 1994;32:364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama T, Mori K, Kawahara T, Ringler D J, Desrosiers R C. Analysis of simian immunodeficiency virus sequence variation in tissues of rhesus macaques with simian AIDS. J Virol. 1993;67:6522–6534. doi: 10.1128/jvi.67.11.6522-6534.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig S, Gendelman H E, Orenstein J M, Dal Canto M C, Pezeshpour G H, Yungbluth M, Janotta F, Akasimit A, Martin M A, Fauci A S. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 27.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong L-Y, Wilson B C, McMillian M K, Bing G, Hudson P M, Hong J-S. The effects of the HIV-1 envelope protein gp120 on the production of nitric oxide and proinflammatory cytokines in mixed glial cell cultures. Cell Immunol. 1996;172:77–83. doi: 10.1006/cimm.1996.0217. [DOI] [PubMed] [Google Scholar]
- 29.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S Y. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 31.Lane T E, Buchmeier M J, Watry D D, Jakubowski D B, Fox H S. Serial passage of microglial SIV results in selection of homogeneous env quasispecies in the brain. Virology. 1995;212:458–465. doi: 10.1006/viro.1995.1503. [DOI] [PubMed] [Google Scholar]
- 32.Lipton S D, Gendelman H E. Dementia associated with the acquired immunodeficiency syndrome. N Engl J Med. 1995;332:934–940. doi: 10.1056/NEJM199504063321407. [DOI] [PubMed] [Google Scholar]
- 33.Mankowski J L, Flaherty M T, Spelman J P, Hauer D A, Didier P J, Amadee A M, Murphey-Corb M, Kirstein L M, Munoz A, Clements J E, Zink M C. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055–6060. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda M, Hanson C A, Alvord W G, Hoffman P M, Ruscetti S K, Masuda M. Effects of subtle changes in the SU protein of ecotropic murine leukemia virus on its capillary endothelial cell tropism and interference proteins. Virology. 1996;215:142–151. doi: 10.1006/viro.1996.0017. [DOI] [PubMed] [Google Scholar]
- 35.McArthur J C, Hoover D R, Bacellar H, Miller E N, Cohen B A, Becker J T, Graham N M H, McArthur J H, Selnes O A, Jacobson L P, Visscher B R, Concha M, Saah A. Dementia in AIDS patients: incidence and risk factors. Neurology. 1993;43:2245–2252. doi: 10.1212/wnl.43.11.2245. [DOI] [PubMed] [Google Scholar]
- 36.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meeker R, English R, Tompkins M. Enhanced excitoxicity in primary feline neural cultures exposed to feline immunodeficiency virus (FIV) J NeuroAIDS. 1996;1:1–27. doi: 10.1300/j128v01n03_01. [DOI] [PubMed] [Google Scholar]
- 38.Myers G. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group T-10. Los Alamos, N.Mex: Los Alamos National Library; 1995. [Google Scholar]
- 39.Namboodiri A M, Venkateshan C N, Narayanan R, Blinder K, Moffett J R, Gajdusek D C, Gravell M, Gibbs C J., Jr Increased quinolinate immunoreactivity in the peripheral blood monocytes/macrophages from SIV-infected monkeys. J Neurovirol. 1996;2:433–438. doi: 10.3109/13550289609146910. [DOI] [PubMed] [Google Scholar]
- 40.Nath A, Psooy K, Martin C, Knudsen B, Magnuson D S, Haughey N, Geiger J D. Identification of a human immunodeficiency virus type 1 Tat epitope that is neuroexcitatory and neurotoxic. J Virol. 1996;70:1475–1480. doi: 10.1128/jvi.70.3.1475-1480.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 42.Paquette Y, Hanna Z, Savard P, Brousseau R, Robitaille Y, Jolicoeur P. Retrovirus-induced murine motor neuron disease: mapping the determinant of spongiform degeneration within the envelope gene. Proc Natl Acad Sci USA. 1989;86:3896–3900. doi: 10.1073/pnas.86.10.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawley A C, Flesher S, Beninger R J, Jhamamanda K H. Different actions of NMDA antagonists on cholinergic neurotoxicity produced by N-methyl-d-aspartate and quiniolinc acid. Br J Pharmacol. 1996;117:1059–1064. doi: 10.1111/j.1476-5381.1996.tb16697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulsen D J, Robertson S J, Favara C A, Portis J L, Chesebro B. Mapping of a neurovirulence determinant within the envelope protein of a polytropic murine retrovirus: induction of CNS disease by low levels of virus. Virology. 1998;248:199–207. doi: 10.1006/viro.1998.9258. [DOI] [PubMed] [Google Scholar]
- 46.Power C, Kong P A, Crawford T O, Wesselingh S, Glass J D, McArthur J C, Trapp B D. Cerebral white matter changes in acquired immunodeficiency syndrome dementia: alterations in the blood-brain barrier. Ann Neurol. 1993;34:339–350. doi: 10.1002/ana.410340307. [DOI] [PubMed] [Google Scholar]
- 47.Power C, McArthur J C, Johnson R T, Griffin D E, Glass J D, Dewey R, Chesebro B. Distinct HIV-1 env sequences are associated with neurotropism and neurovirulence. Curr Top Microbiol Immunol. 1995;202:89–104. doi: 10.1007/978-3-642-79657-9_7. [DOI] [PubMed] [Google Scholar]
- 48.Power C, McArthur J C, Johnson R T, Griffin D E, Glass J D, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68:4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price R W, Brew B J. The AIDS dementia complex. J Infect Dis. 1988;158:1079–1083. doi: 10.1093/infdis/158.5.1079. [DOI] [PubMed] [Google Scholar]
- 50.Pulliam L, Herndier B G, Tang N M, McGrath M S. Human immunodeficiency virus-infected macrophages produce soluble factors that cause histological and neurochemical alterations in cultured human brains. J Clin Investig. 1991;87:503–512. doi: 10.1172/JCI115024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy R T, Achim C L, Sirko D A, Tehranchi S, Kraus F G, Wong Staal F, Wiley C A. Sequence analysis of the V3 loop in brain and spleen of patients with HIV encephalitis. AIDS Res Hum Retroviruses. 1996;12:477–482. doi: 10.1089/aid.1996.12.477. [DOI] [PubMed] [Google Scholar]
- 52.Robertson S, Hasenkrug K, Chesebro B, Portis J. Neurologic disease induced by polytropic murine retroviruses: neurovirulence determined by efficiency of spread to microglial cells. J Virol. 1997;71:5287–5294. doi: 10.1128/jvi.71.7.5287-5294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuitemaker H M, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schttenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 variants detectable at all stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan M K, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71:4871–4881. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speck R, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephens E B, Mukherjee S, Sahni M, Zhuge W, Raghavan R, Singh D K, Leung K, Atkinson B, Li Z, Joag S V, Liu Z Q, Narayan O. A cell-free stock of simian-human immunodeficiency virus that causes AIDS in pig-tailed macaques has a limited number of amino acid substitutions in both SIVmac and HIV-1 regions of the genome and has altered cytotropism. Virology. 1997;231:313–321. doi: 10.1006/viro.1997.8534. [DOI] [PubMed] [Google Scholar]
- 58.Stoler M H, Eskin T A, Benn S, Angerer R C, Angerer L M. Human T-cell lymphotropic virus type III infection of the central nervous system. A preliminary in situ analysis. JAMA. 1986;256:2360–2364. [PubMed] [Google Scholar]
- 59.Strizki J M, Albright A V, Sheng H, O’Connor M, Perrin L, Gonzalez-Scarano F. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J Virol. 1996;70:7654–7662. doi: 10.1128/jvi.70.11.7654-7662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szurek P F, Floyd E, Yuen P H, Wong P K. Site-directed mutagenesis of the codon for Ile-25 in gPr80env alters the neurovirulence of ts1, a mutant of Moloney murine leukemia virus TB. J Virol. 1990;64:5241–5249. doi: 10.1128/jvi.64.11.5241-5249.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi K, Wesselingh S L, Griffin D E, McArthur J C, Johnson R T, Glass J D. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39:705–711. doi: 10.1002/ana.410390606. [DOI] [PubMed] [Google Scholar]
- 62.Thomas F P, Chalk C, Lalonde R, Robitaille Y, Jolicoeur P. Expression of human immunodeficiency virus type 1 in the nervous system of transgenic mice leads to neurological disease. J Virol. 1994;68:7099–7107. doi: 10.1128/jvi.68.11.7099-7107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toggas S M, Masliah E, Rockenstein E M, Rall G F, Abraham C R, Mucke L. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature. 1994;367:188–193. doi: 10.1038/367188a0. [DOI] [PubMed] [Google Scholar]
- 64.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 65.Tucker P C, Strauss E G, Kuhn R J, Strauss J H, Griffin D E. Viral determinants of age-dependent virulence of Sindbis virus for mice. J Virol. 1993;67:4605–4610. doi: 10.1128/jvi.67.8.4605-4610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tyor W R, Power C, Gendelman H E, Markham R B. A model of human immunodeficiency virus encephalitis in SCID mice. Proc Natl Acad Sci USA. 1993;90:8658–8662. doi: 10.1073/pnas.90.18.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W K, Essex M, Lee T H. The highly conserved aspartic acid residue between hypervariable regions 1 and 2 of human immunodeficiency virus type 1 gp120 is important for early stages of virus replication. J Virol. 1995;69:538–542. doi: 10.1128/jvi.69.1.538-542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wesselingh S L, Power C, Glass J D, Tyor W R, McArthur J C, Farber J M, Griffin J W, Griffin D E. Intracerebral cytokine messenger RNA expression in acquired immunodeficiency syndrome dementia. Ann Neurol. 1993;33:576–582. doi: 10.1002/ana.410330604. [DOI] [PubMed] [Google Scholar]
- 69.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiley C A, Schrier R D, Nelsen J A, Lampert P W, Oldstone M B. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong P K, Floyd E, Szurek High susceptibility of FV/N mice to the paralytic disease induced by ts1, a mutant of Moloney murine leukemia virus TB. Virology. 1991;180:365–371. doi: 10.1016/0042-6822(91)90041-9. [DOI] [PubMed] [Google Scholar]