CXCR4 and CCR5 Genetic Polymorphisms in Long-Term Nonprogressive Human Immunodeficiency Virus Infection: Lack of Association with Mutations other than CCR5-Δ32 (original) (raw)
Abstract
Polymorphisms in the coding sequences of CCR5 and CXCR4 were studied in a group of human immunodeficiency virus (HIV)-infected long-term nonprogressors. Two different point mutations were found in the CXCR4 coding sequence. One of these CXCR4 mutations was silent, and each was unique to two nonprogressors. The well-described 32-bp deletion within the CCR5 coding sequence (CCR5-Δ32) was found in 4 of 13 nonprogressors, and 12 different point mutations were found scattered over the CCR5 coding sequence from 8 nonprogressors. Most of the mutations created either silent or conservative changes in the predicted amino acid sequence: only one of these mutations was found in more than a single nonprogressor. All nonsilent mutations were tested in an HIV envelope-dependent fusion assay, and all functioned comparably to wild-type controls. Polymorphisms in the CXCR4 and CCR5 coding sequences other than CCR5-Δ32 do not appear to play a dominant mechanistic role in nonprogression among HIV-infected individuals.
The chemokine receptors CCR5 and CXCR4 function as coreceptors for macrophage- and T-cell-line-tropic strains of human immunodeficiency virus type 1 (HIV-1), respectively (1, 5, 9–11, 13). A 32-bp deletion in the CCR5 gene results in a truncated protein product that does not function as a coreceptor (16). The defective CCR5 gene (CCR5-Δ32) affords near-total protection against HIV infection in homozygotes (3, 8, 14, 16, 18, 20, 22, 24, 27) and partial protection against disease progression in HIV-infected CCR5 heterozygotes (6, 8, 12, 14, 18, 27). Other polymorphisms have been identified in the CCR5 coding sequence, although their relevance to HIV coreceptor function and rates of HIV disease progression is unknown. Two different alleles have been identified in Asian populations, including a single base deletion that results in a frameshift and premature stop codon in the region of the gene encoding the C-terminal intracellular domain of CCR5 and a single base substitution causing an amino acid change in the third intracellular loop of CCR5 (2). Another single base substitution resulting in an amino acid change in the C-terminal intracellular domain of CCR5 was identified in African Americans (2). We hypothesized that, similar to the CCR5-Δ32 polymorphism, the frequency in the CXCR4 and CCR5 genes of other polymorphisms that affect rates of HIV disease progression would be enriched among HIV-infected long-term nonprogressors.
Participants in this study included 17 HIV-infected long-term nonprogressors, defined as asymptomatic individuals with documented HIV infection of at least 7 years duration, stable CD4+ T-cell counts of >600 cells/μl, and no history of antiretroviral therapy. Characteristics of the study population are presented in Table 1. Mononuclear cells were obtained from peripheral blood by Ficoll-Hypaque (Organon Teknika, West Chester, Pa.) density centrifugation. Total RNA was extracted from cells with RNAzol (Tel-Test, Friendswood, Tex.), and 2 μg was reverse transcribed with 400 U of Superscript II RNase H− reverse transcriptase (Life Technologies, Baltimore, Md.), 40 μg of random hexamers (Pharmacia Biotech, Piscataway, N.J.)/ml, 0.8 mM (each) deoxynucleotide triphosphate (Pharmacia), and 10 U of RNAsin (Promega Corp., Madison, Wis.) in a volume of 50 μl at 42°C for 45 min. One-fifth of each cDNA (10 μl) was used in each amplification reaction for the coding sequences of CCR5 and CXCR4. The PCR mixture included a final magnesium concentration of 2.5 mM, 200 nM (each) deoxynucleoside triphosphate (Pharmacia), 500 nM each primer, and 5 U of Expand High Fidelity Taq polymerase (Boehringer-Mannheim, Indianapolis, Ind.) per reaction. Fifty cycles of PCR were performed as follows (per cycle): 94°C for 15 s, 55°C for 30 s, and 72°C for 60 s. A final extension step of 72°C for 10 min was added after the 50 PCR cycles. Forward and reverse primers were located in the 5′ and 3′ untranslated regions of the CCR5 and CXCR4 mRNA as follows: CCR5 forward, 5′ GCATTCATGGAGGGCAACTAAA 3′; CCR5 reverse, 5′ GCCCAGGCTGTGTATGAAAACTAA 3′; CXCR4 forward, 5′ AAGTGACGCCGAGGGCCTGAG 3′; and CXCR4 reverse, 5′ CTGTACAATATTGGTCAGTC 3′. Full-length PCR products were purified from 1% agarose gels by using the QIAquick extraction kit (Qiagen, Chatsworth, Calif.). The purified PCR products were then ligated into the pCR 2.1 vector (Invitrogen, San Diego, Calif.) with a TA cloning kit (Invitrogen). Plasmids containing inserts were used to transform Max Efficiency Stable 2 competent cells (Life Technologies). Single colonies of cells resistant to ampicillin were expanded, and DNA was extracted by an alkaline lysis method (Wizard Plasmid Kit; Promega). DNA was sequenced with a dye terminator DNA sequencing kit (Perkin-Elmer, Foster City, Calif.) on an ABI PRISM 377 DNA sequencer (Perkin-Elmer). Sequences were analyzed with Sequencer 3.0 software (Gene Codes, Ann Arbor, Mich.). Mutations were considered relevant if they were present in at least two independent clones from the same patient.
TABLE 1.
Characteristics of HIV-infected long-term nonprogressors
| Characteristic | Datum |
|---|---|
| No. of participants | 17 |
| Male (no.) | 14 |
| Female (no.) | 3 |
| Mean duration of HIV infection (yr) | 9.6 ± 0.4 |
| Mean CD4+ T-cell count (cells/μl) | 829 ± 51 |
| Mean plasma viremia (log HIV RNA copies/ml) | 3.30 ± 0.15 |
The abilities of the mutant CXCR4 and CCR5 sequences to function as HIV coreceptors were tested in a fusion assay as previously described (1, 19). CXCR4 and CCR5 variants were subcloned into pcDNA3 (Invitrogen, Carlsbad, Calif.); expression in this system is driven by both cytomegalovirus major immediate-early and T7 promoters. BSC-1 cells, which do not express HIV-1 fusion-entry cofactors (1), were transfected with plasmids carrying CXCR4 or CCR5 variants and infected with recombinant vaccinia viruses vTF7-3 (encoding T7 polymerase) and vCB3 (encoding human CD4). Another set of BSC-1 cells was infected with recombinant vaccinia virus vCB21R (encoding the lacZ gene under the control of the T7 promoter) and either vCB41 (encoding the HIV-1 IIIB envelope gene) or vCB43 (encoding the Ba-L HIV-1 envelope gene). Control experiments were performed both with BSC-1 cells transfected with vector plasmid pcDNA3 and infected with vTF7-3 and vCB3 and with another set of BSC-1 cells infected with vCB21R and vCB16 (encoding a nonfusogenic mutant HIV-1 IIIB envelope gene). Cells were mixed at 37°C for 4 h in the presence of cytosine arabinoside (40 μg/ml), and cell lysates were assayed for β-galactosidase activity by measuring optical density (OD) at 570 nm. The fusion index was calculated as the percentage of the wild-type OD at 570 nm (13).
Seventy-four CXCR4 clones from 11 nonprogressors and 85 CCR5 clones from 13 nonprogressors were sequenced and analyzed. Two point mutations were identified in the CXCR4 coding sequence (Table 2). Neither of these mutations was found in any other nonprogressor analyzed, and one was a silent mutation. The 32-bp deletion at positions 554 to 585 (21) of the CCR5 coding sequence (CCR5-Δ32) was detected in 4 of 13 (31%) patients. These four patients were previously shown to be heterozygous for CCR5-Δ32 (6). Twelve point mutations were detected in the CCR5 coding sequence from eight patients (Table 2). Of the eight patients with point mutations, two were heterozygous for CCR5-Δ32. The mutations were scattered over the CCR5 coding sequence and were all unique except for one point mutation which occurred in two patients (c→t mutation at position 746). Most of the point mutations created predicted amino acid changes which were either silent or conservative. With the exception of the CCR5-Δ32 mutation, no insertions, deletions, or nonsense mutations were found.
TABLE 2.
Mutations detected in HIV coreceptor coding sequences
| Coreceptor | Positiona | Mutation | Amino acid change | Patient(s) |
|---|---|---|---|---|
| CXCR4 | 204 | a→g | K→K | 7 |
| 278 | t→c | F→S | 6 | |
| CCR5 | 140 | g→t | G→V | 9 |
| 164 | t→a | L→Q | 11 | |
| 268 | g→a | A→T | 11 | |
| 315 | a→t | T→T | 12 | |
| 496 | t→c | F→L | 3 | |
| 554–585 | Deletion | 1, 2, 6, 9 | ||
| 744 | g→t | W→C | 13 | |
| 746 | c→t | A→V | 5, 8 | |
| 792 | t→c | F→F | 11 | |
| 823 | t→g | L→V | 8 | |
| 928 | g→a | V→I | 13 | |
| 990 | g→a | E→E | 1 | |
| 1037 | a→g | Q→R | 5 |
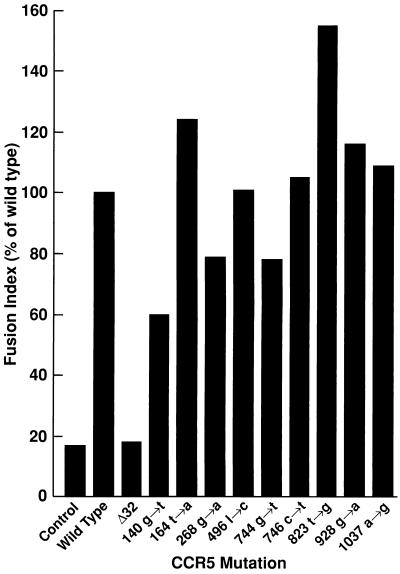
Each of the nonsilent mutations in the CXCR4 and CCR5 coding sequences was tested for the ability to support HIV envelope-dependent cell membrane fusion. The CXCR4 variant with a t→c mutation at position 278 supported T-cell-tropic HIV-1-IIIB envelope-dependent fusion to a degree (84%) comparable to that of the wild-type CXCR4. Similarly, all of the CCR5 variants with nonsilent single point mutations supported macrophage-tropic HIV-1-Ba-L envelope-dependent fusion to a degree (60 to 155%) comparable to that of the wild-type CCR5 (Fig. 1). In contrast, CCR5-Δ32, similar to the negative control, failed to support fusion (Fig. 1).
FIG. 1.
CCR5 variants with point mutations at the indicated positions function comparably to wild-type CCR5 in an HIV envelope-dependent fusion assay. BSC-1 cells were transfected with plasmids carrying CCR5 variants isolated from HIV-infected long-term nonprogressors and infected with vTF7-3 (encoding T7 polymerase) and vCB3 (encoding human CD4). Another set of BSC-1 cells was infected with vCB21R (encoding the lacZ gene under the control of the T7 promoter) and vCB43 (encoding the Ba-L HIV-1 envelope gene). Cells were mixed at 37°C for 4 h in the presence of cytosine arabinoside (40 μg/ml), and cell lysates were assayed for β-galactosidase activity by measuring OD at 570 nm. The fusion index was calculated as the percentage of the wild-type OD at 570 nm.
The CCR5-Δ32 allele affords partial protection against disease progression in HIV-infected CCR5-Δ32 heterozygotes, as indicated by the increased frequency of this allele among HIV-infected long-term nonprogressors (6, 8, 12, 14, 18, 27). However, the CCR5-Δ32 allele is clearly not the sole or even dominant factor responsible for nonprogression of HIV infection (6). We therefore searched for other mutations in the CXCR4 and CCR5 coding sequences that might be found at high frequencies in long-term nonprogressors. Only one nonsilent mutation was detected in the CXCR4 coding sequence. Although point mutations were identified scattered throughout the CCR5 coding sequence, only one of these was found in more than a single patient. Furthermore, all of the nonsilent CXCR4 and CCR5 mutations that were identified had no substantial impact on HIV coreceptor function measured in a fusion assay. The possibility that some of the putative mutations that were identified represent PCR artifacts must also be considered. In this regard, both of the mutations identified in the CXCR4 coding sequence, and 7 of the 12 mutations in the CCR5 coding sequence were identified in only two clones each. Mutations identified in 3 or more clones each included CCR5-140 (3 of 10 clones from patient 9), CCR5-496 (5 of 10 clones from patient 3), CCR5- 744 (4 of 8 clones from patient 13), CCR5-746 (7 of 7 clones from patient 5), and CCR5-928 (6 of 8 clones from patient 13). These data indicate that HIV coreceptor polymorphisms other than CCR5-Δ32 do not play a dominant role in nonprogression among HIV-infected individuals. Other possible mechanisms of nonprogression related to HIV coreceptors are under investigation. In this regard, several polymorphisms in genes encoding HIV coreceptor molecules or their ligands have recently been shown to influence rates of HIV disease progression (23, 25); such genetic polymorphisms and/or the immunoregulation of these genes in certain hosts may account for constitutive low-level expression of coreceptors (15, 23), downregulation of coreceptor expression (4, 26), or secretion of high levels of coreceptor ligands capable of inhibiting cellular entry of HIV (7).
Acknowledgments
O.J.C. and S.P. contributed equally to this work.
We acknowledge the kind gift of recombinant vaccinia viruses encoding HIV envelopes from E. Berger and C. Broder; the technical support of M. Gonda, J. Greenwood, and L. Rasmussen; and the editorial assistance of M. Rust and P. Walsh.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C, Feng Y, Kennedy P, Murphy P, Berger E. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Ansari-Lari M A, Liu X-M, Metzker M L, Rut A R, Gibbs R A. The extent of genetic variation in the CCR5 gene. Nat Genet. 1997;16:221–222. doi: 10.1038/ng0797-221. [DOI] [PubMed] [Google Scholar]
- 3.Biti R, French R, Young J, Bennetts B, Stewart G. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 4.Bleul C, Wu L, Hoxie J, Springer T, Mackay C. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P, Wu L, Mackay C, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1138. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Cohen O J, Vaccarezza M, Arthos J, Lam G, Baird B, Wildt K, Murphy P, Zimmerman P, Nutman T, Fox C, Hoover S, Adelsberger J, Baseler M, Davey R, Dewar R, Metcalf J, Schwartzentruber D, Orenstein J, Buchbinder S, Saah A, Detels R, Phair J, Rinaldo C, Margolick J, Pantaleo G, Fauci A. Heterozygosity for a defective gene for CC chemokine receptor 5 is not the sole determinant for the immunologic and virologic phenotype of HIV-infected long term non-progressors. J Clin Invest. 1997;100:1581–1589. doi: 10.1172/JCI119682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen O J, Kinter A, Fauci A S. Host factors in the pathogenesis of HIV disease. Immunol Rev. 1997;159:31–48. doi: 10.1111/j.1600-065x.1997.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, Huttley G, Smith M, Allikmets R, Goedert J, Buchbinder S, Vitinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; San Francisco City Clinic Cohort; ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R, Hill C, Davis C, Peiper S, Schall T, Littman D, Landau N. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.Doranz B, Rucker J, Yi Y, Smyth R, Samson M, Peiper S, Parmentier M, Collman R, Doms R. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Dragic T, Litwin V, Allaway G, Martin S, Huang Y, Nagashima K, Cayanan C, Maddon P, Koup R, Moore J, Paxton W. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 12.Eugen-Olsen J, Iversen A K N, Garred P, Koppelhus U, Pedersen C, Benfield T L, Sorensen A M. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS free survival and slower CD4 T cell fall in a cohort of HIV seropositive individuals. AIDS. 1997;11:305–310. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Broder C, Kennedy P, Berger E. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane domain, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Paxton W, Wolinsky S, Neumann A, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N, Phair J, Ho D, Koup R. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 15.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Paxton W, Choe S, Ceradini D, Martin S, Horuk R, MacDonald M, Stuhlmann H, Koup R, Landau N. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 17.Loetscher M, Geiser T, O’Reilly T, Zwahlen R, Baggiolini M, Moser B. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J Biol Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 18.Michael N, Chang G, Louie L, Mascola J, Dondero D, Birx D, Sheppard H. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 19.Moriuchi H, Moriuchi M, Arthos J, Hoxie J, Fauci A S. Promonocytic U937 subclones expressing CD4 and CXCR4 are resistant to infection with and cell-to-cell fusion by T-cell-tropic human immunodeficiency virus type 1. J Virol. 1997;71:9664–9671. doi: 10.1128/jvi.71.12.9664-9671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Brien T, Winkler C, Dean M, Nelson J, Carrington M, Michael N, White G. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. [DOI] [PubMed] [Google Scholar]
- 21.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new CC-chemokine receptor gene, CC-CKR5. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 22.Samson M, Libert F, Doranz B, Rucker J, Liesnard C, Farber C, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R, Collman R, Doms R, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 23.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science. 1997;267:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 24.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. HIV-1 infection in an individual homozygous for CCR5 delta 32. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- 25.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J ALIVE Study; Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman R A K W, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1692. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmerman P, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P, Kumraraswami V, Giorgi J, Detels R, Hunter J, Chopek M, Berger E, Fauci A, Nutman T, Murphy P. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial backgrounds and quantified risks. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]