c-Maf Interacts with c-Myb To Regulate Transcription of an Early Myeloid Gene during Differentiation (original) (raw)
Abstract
The MafB transcriptional activator plays a pivotal role in regulating lineage-specific gene expression during hematopoiesis by repressing Ets-1-mediated transcription of key erythroid-specific genes in myeloid cells. To determine the effects of Maf family proteins on the transactivation of myeloid-specific genes in myeloid cells, we tested the ability of c-Maf to influence Ets-1- and c-Myb-dependent CD13/APN transcription. Expression of c-Maf in human immature myeloblastic cells inhibited _CD13/APN_-driven reporter gene activity (85 to 95% reduction) and required the binding of both c-Myb and Ets, but not Maf, to the promoter fragment. c-Maf’s inhibition of CD13/APN expression correlates with its ability to physically associate with c-Myb. While c-Maf mRNA and protein levels remain constant during myeloid differentiation, formation of inhibitory Myb-Maf complexes was developmentally regulated, with their levels being highest in immature myeloid cell lines and markedly decreased in cell lines representing later developmental stages. This pattern matched that of CD13/APN reporter gene expression, indicating that Maf modulation of c-Myb activity may be an important mechanism for the control of gene transcription during hematopoietic cell development.
The association of the Myb and Ets-1 proteins serves as a paradigm for the functional and cooperative interactions between hematopoietic transcription factors. The E26 virus contains the DNA binding and transactivation domains of both the Myb and Ets-1 proteins fused to form a single oncoprotein (14, 43). While expression of either the single v-Myb or v-Ets proteins is weakly transforming, coexpression of the proteins either as a fusion construct or separately in the same cells results in a much higher transforming activity than the additive effects of the individual proteins, suggesting that their combination potentiates the transformation of avian hematopoietic cells (11, 45). In addition, c-Myb and Ets family members have been shown to cooperate to transactivate myeloid-specific promoters (10, 60). Attempts to demonstrate a physical interaction between Myb and Ets-1 have not been successful (10, 14, 44–46), indicating that accessory proteins may be required to facilitate their cooperative interaction.
While it appears to be the rule that Ets family members interact with accessory proteins, resulting in either positive or negative regulatory effects (52, 59, 65), the isolation of direct protein partners for Myb has proved elusive. Recently, however, two reports have shown that the interaction of Myb with the CREB binding protein transcriptional coactivator results in increased transcriptional ability (6, 55), presumably by linking c-Myb to the basal transcriptional machinery. Similarly, c-Myb interacts with the p100 transcriptional coactivator through the highly conserved EVES motif (8). In addition, a physical association between c-Myb and the C/EBP transcription factor is essential for their cooperative regulation of the avian mim-1 promoter (48) and may account for their combinatorial activation of myeloid genes in heterologous cells (3).
Members of the Maf family of basic region/leucine zipper (bZIP) transcription factors can affect transcription in either a positive or negative fashion, depending on their particular protein partner and the context of the target promoter (21, 26, 29–34, 38, 62). Previous reports (62) have shown that enforced expression of MafB in avian erythroid cells results in its physical interaction with the Ets-1 transcription factor and repression of Ets-1 transcriptional activity. MafB is normally expressed in mature avian myeloid cells but not in erythroid cells, a pattern consistent with MafB repression of the erythroid gene program in myeloid cells. These observations prompted us to investigate the effect of Maf family members on genes regulated by the Ets-1 protein in myeloid cells.
The CD13/APN gene is expressed very early in myeloid cell development and is restricted to cells of the granulocyte/macrophage lineage (15, 16, 23). c-Myb and Ets-1 transcription factors act cooperatively to positively regulate CD13/APN gene transcription in myeloid cell progenitors (60). Here we report that c-Maf inhibits transcription of CD13/APN, and this inhibition profoundly affects the c-Myb–Ets-1 cooperative interaction. Furthermore, c-Maf physically interacts in vitro with the c-Myb DNA binding domain, and the Myb-Maf complex is detected in myeloid cell lines. Finally, although c-Maf mRNA and protein are expressed at equal levels in human myeloid cell lines arrested at progressive stages of differentiation, the formation of Myb-Maf inhibitory complexes appears to be developmentally regulated. These results suggest a crucial role for c-Maf in regulation of cooperative interactions between c-Myb and Ets-1 in early myeloid cell differentiation.
MATERIALS AND METHODS
Cell lines.
Human cell lines included the myeloid leukemia lines HL-60 (ATCC CRL 1593), U937 (ATCC CRL 1593), KG1 (ATCC CCL 246), KG1a, a phenotypically primitive, developmentally arrested revertant of the KG1 cell line that is incapable of further myeloid differentiation (37) (ATCC CCL 246.1), and KCL22 and NB-4 (generous gifts of H. P. Koeffler) as well as the epithelial cell line C33A (ATCC HTB 31). Nonadherent cell lines were grown in RPMI 1640 medium, and adherent lines were maintained in Dulbecco’s modified Eagle medium, each supplemented with 2 mM l-glutamine and 10% fetal calf serum. The HL-60 and U937 cell lines were induced to differentiate by using 12-O-tetradecanoylphorbol diester (TPA; 10−6 M) added to the culture medium; cells were harvested at 6 and 24 h after addition.
Plasmid construction.
The wild-type reporter construct −411luc contains genomic sequences from bp −411 through +65 of the CD13/APN myeloid promoter in the pGL2basic backbone (Promega) (60). The 5′-deletion mutant −291luc was constructed from this parent plasmid by _Dde_I enzyme digestion. The consensus Myb site in the −411luc reporter construct was mutated from TAACGGAC to TCTTGGAC, using a Transformer kit (Clontech) to produce the Mybmutluc plasmid. Etsdelluc, lacking the 30-bp segment between −330 and −360 which includes the three consensus Ets binding sites, was constructed by overlap extension (22) using 5′ primer UP2-PMC (5′-CTGTTGGGAAGGGCGATCGGTGC-3′), 3′ primer DN2-PMC (5′-CCTGGGATGCACCAGGGCTCCTG-3′), and the complementary overlap primer DN(UP)-ESPMC (5′-CACCACCCAGCTGCACGG/GCACAGAGCTCCCTGCGGT-3′). pRc/RSVcMaf was made by inserting the full-length murine c-Maf cDNA (38) into the _Bam_HI/_Xba_I sites of the pRc/RSV plasmid (Invitrogen). The pMT-CB6-cMaf construct used to produce the U937 cell line inducibly expressing c-Maf in response to Zn2+ was made by cloning the full-length c-Maf cDNA (38) into _Hin_dIII/_Xba_I-cut pMT-CB6 (a gift of A. T. Look).
The Myb-LexA fusion protein was constructed in the yeast expression plasmid Y.LexA (a gift from Steven Dalton). In this construct, c-Myb codons 1 to 240 were fused in frame to the lexA operator binding domain (codons 2 to 202) of the LexA protein as an _Nco_I/_Hin_cII restriction fragment. Fusion constructs of the herpes simplex virus protein VP16 transactivation domain with c-Maf (amino acids 123 to 370), c-Fos, and c-Jun (38) as well as USF2 (40) are reported elsewhere. nrl cDNA (34) was a gift from Tom Kerppola. Kreisler cDNA was generated by reverse transcription-PCR using primers based on published sequences (5). The VP16 fusion of Nrl amino acids 118 to 237 and full-length Kreisler were made in pSD.10a (7). In both the Y.LexA and pSD.10a vectors, expression of the hybrid proteins was under the control of a GAL10-CYC1 hybrid yeast promoter. This made bait and prey gene expression glucose repressible and galactose inducible (18).
Transfection of recombinant plasmids and reporter gene assays.
To compare transcriptional activity among cell lines, we electroporated the KG1a, U937, and HL-60 cell lines with 5 μg of the wild-type promoter constructs and 2 μg of the control β-actin–secreted alkaline phosphatase (SEAP) plasmid as described elsewhere (60). The transfection efficiency with each construct was normalized to the control level of SEAP activity (1); the reported values were calculated as relative light units per unit of SEAP activity. To compare results among the cell lines, we expressed transcriptional activity as the fold increase over that produced with the promoterless luciferase vector, pGL2basic, determined in parallel transfections. Each point was determined at least four times. For transactivation assays, C33A epithelial cells (which are negative for CD13/APN and c-Myb expression) were transfected by the calcium phosphate method (60) with 1 μg of each of the −411luc reporter plasmid and β-actin–SEAP control plasmid, plus the indicated amounts of reporter and expression plasmids (Fig. 2): pRc/RSVcMaf, encoding the full-length murine c-Maf protein, pCMV4cMyb, encoding the full-length murine c-Myb protein (60); and pEVRFO-Ets-1, encoding the full-length murine Ets-1 protein (54) (kindly provided by Barbara Graves, University of Utah). Assays to detect luciferase activity were performed as described previously (60); 100-μl aliquots of total cellular protein from lysates representing each transfection condition were tested for luciferase activity, and the resulting values were normalized to SEAP activity, assayed as described elsewhere (1).
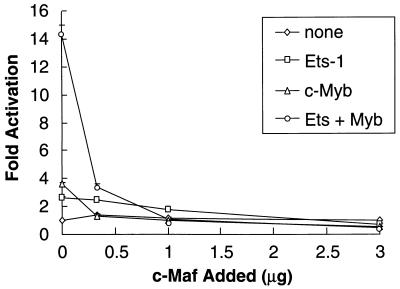
FIG. 2.
c-Maf affects the c-Myb–Ets-1 functional cooperation on the CD13/APN promoter. The C33A human epithelial cell line was cotransfected with 1 μg of the −411luc reporter construct and increasing amounts (0.3 to 3.0 μg) of pRc/RSVcMaf along with either empty vectors only (none), 0.5 μg of pEVRFO-Ets-1 only (Ets-1), 1 μg of pCMV4cMyb only (c-Myb), or 1 μg of pCMV4cMyb cotransfected with 0.5 μg of pEVRFO-Ets-1 (Ets-1 + Myb). All values are normalized to those for a cotransfected control plasmid (MAP1-SEAP) both for transfection efficiency and as a control for the c-Myb, Ets, and Maf effect on other promoters. Results are expressed as fold activation above that obtained with cotransfection of equal amounts of the empty expression plasmid.
Northern and immunoprecipitation/Western blot analyses.
Poly(A)+ RNA was extracted from the various cell lines by using a FastTrack 2.0 kit (Invitrogen). Ten micrograms of poly(A)+ RNA from the indicated cell lines was separated on a 1% agarose-formaldehyde gel, transferred to a nylon membrane, and sequentially probed with the _Bst_EII/_Nco_I fragment containing the 5′ region of murine c-Maf (which excludes the bZIP domain) and the β-actin cDNA probes. Relative expression levels between the cell lines were normalized to β-actin levels by using a Molecular Dynamics PhosphorImager. For immunoprecipitation/Western blot analysis of c-Maf proteins, KG1a or HL-60 cells were immunoprecipitated as specified by the manufacturer (Santa Cruz Biotechnology). Briefly, cells were harvested, washed, resuspended in radioimmunoprecipitation assay buffer, and disrupted by aspiration through a 21-gauge needle. The supernatant was precleared with normal rabbit immunoglobulin G (IgG) and protein A-Sepharose. Rabbit anti-v-Maf antiserum (1 μg; Santa Cruz Biotechnology) was added to 10 μg of total protein for 1 h at 4°C, and antibody complexes were precipitated with protein A-Sepharose. Proteins were separated on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and transferred onto a nitrocellulose membrane. Membranes were blocked with 5% milk in Tris-buffered saline; the primary antibody used was rabbit anti-v-Maf polyclonal antiserum (1 μg/ml), followed by a 1:2,000 dilution of horseradish peroxidase-linked donkey anti-rabbit IgG. Specific horseradish peroxidase-conjugated protein complexes were detected by the enhanced chemiluminescence method (Amersham).
DNA binding assays.
For DNA binding assays, whole-cell lysates from myeloid cell lines were prepared by resuspending and washing pelleted cells in cold phosphate-buffered saline and resuspending the pellet in lysis buffer (20 mM Tris-HCl [pH 7.5], 2 mM dithiothreitol [DTT], 20% glycerol, 50 mM KCl) containing a protease inhibitor cocktail (Boehringer Mannheim Complete). After addition of Triton X-100 to 0.5% (vol/vol) final concentration, the lysate was incubated on ice for 60 min, and the debris was pelleted for 15 min at 14,000 rpm. Cleared lysate was quantitated and stored at −80°C until use. Glutathione _S_-transferase (GST) fusion proteins (1 μg) or cell lysates from myeloid cell lines (20 μg) were preincubated in binding buffer [10 mM HEPES (pH 7.9), 50 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 1 μg of poly(dI-dC)] in 25-μl reactions with or without 0.1 μg of the consensus or mutant double-stranded oligonucleotide competitors mimAmyb (CTAGGACATTATAACGGTTTTTTAGT), mimAmybmut (CTAGGACATTAGCCAGATTTTTTAGT (60), and MARE (TCGAGCTCGGAATTGCTGACTCAGCATTACTC) (31) for 15 min at 4°C before the addition of probe. For supershift experiments, 1 μl of an anti-v-Maf polyclonal antibody (cross-reactive with c-Maf; Santa Cruz Biotechnology) or control antibody was preincubated with lysate for 10 min at 4°C before addition of probe. The 32P-end-labeled genomic fragment probe (_Xba_I/_Dde_I fragment containing bp −426 through −291 of the CD13/APN promoter) was then added, and the mixture was incubated for an additional 15 min at 4°C. Binding reactions were electrophoresed through a 3.5% acrylamide gel containing 10% glycerol in Tris-acetate-EDTA buffer at 4°C. Gels were dried and exposed to X-ray film. For stability comparisons in Fig. 10, binding reaction mixtures containing bacterial protein and probe were incubated for 15 min at 4°C before addition of 50 ng of a 70-bp oligonucleotide (Myb/Ets-70) consisting of sequences corresponding to bp −320 to −390 (including the Myb and Ets consensus sites [60]) of the CD13/APN promoter. Reaction mixtures were loaded on the gel at the times indicated in the figure.
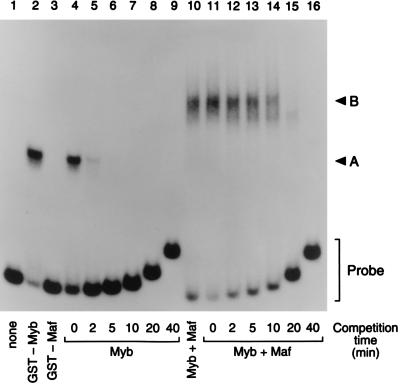
FIG. 10.
Myb-Maf complexes are more stable than Myb alone. Mobility shift assays measuring the off rate of c-Myb alone (lanes 4 to 9) and the Myb-Maf protein complex (lanes 11 to 16) from preformed DNA-protein complexes after incubation for the indicated times with an excess (50 ng) of unlabeled competitor DNA (Myb/Ets-70; see Materials and Methods). An end-labeled 135-bp promoter fragment probe (bp −426 through −291) was incubated with bacterially expressed GST fusion proteins. Lane 1, probe alone; lanes 2 and 4 to 9, GST-Myb-DBD only; lane 3, GST-Maf only; lanes 10 to 16, equal amounts of GST-Myb-DBD and GST-Maf. A, the specific protein complex formed by GST-Myb-DBD; B, the ternary complex containing both Myb and Maf; Probe, free probe.
Bacterial protein preparations and protein-protein interactions.
Recombinant GST fusion protein containing the bacterially expressed Ets-1 DNA binding domain (GST-Ets-1-DBD), Myb DNA binding domain (GST-Myb-DBD), and c-Maf (GST-c-Maf) were purified from bacterial cell lysates by a standard glutathione (GSH)-bead method (58). The GST-Myb-DBD plasmid was the gift of Kevin Ess. GST pull-down assays were performed as described previously (58). Briefly, GSH-beads were preblocked for nonspecific protein interactions using normal rabbit serum, followed by two washes with PC + 100 buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% Nonidet P-40, 20% glycerol, 0.01% bovine serum albumin, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml [58]); 10 μl of in vitro-transcribed and -translated [35S]methionine-labeled protein was added to the GSH-beads and rotated for 30 min. GSH-beads were washed three times in ice-cold PC + 100 buffer and boiled in 20 μl of 2× SDS-gel sample buffer before SDS-polyacrylamide gel electrophoresis (PAGE).
Yeast two-hybrid analysis.
Saccharomyces cerevisiae S260 contained a lexA operator-lacZ (β-galactosidase) reporter fusion gene integrated into its genome and was a gift from Steven Dalton (39). S260 has been cotransformed as described previously (38) with two plasmids, one encoding Myb-LexA (bait) and one encoding a VP16 hybrid (prey). Selection of transformants and assays for β-galactosidase activity were performed as previously described (7, 38). The development of blue color in the yeast colonies was monitored for 6 h.
RESULTS
c-Maf inhibits CD13/APN gene transcription in early myeloid cells by affecting Myb-Ets functional cooperation.
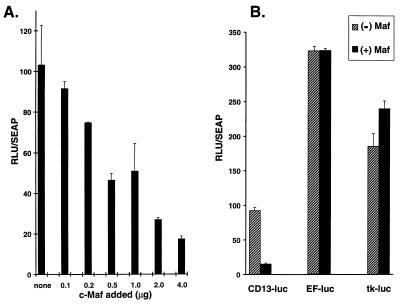
In the hematopoietic compartment, the CD13/APN cell surface glycoprotein is expressed exclusively on cells of the myeloid lineage. To investigate the contribution of Maf family proteins to the transcriptional regulation of this myeloid-specific and Ets-1-regulated gene in human myeloid cells (60), we transiently cotransfected an expression plasmid driving murine c-Maf expression together with a _CD13/APN_-luciferase reporter plasmid containing the minimal promoter necessary to direct CD13/APN transcription in myeloid cells (60). Increasing the amount of c-Maf in KG1a early myeloid cells (as confirmed by Western blot analysis [data not shown]) resulted in a dose-dependent reduction in luciferase activity of the CD13/APN promoter constructs (Fig. 1A). By contrast, there was either no effect or, often, a weak transactivating effect of c-Maf expression on transcription of control luciferase constructs driven by either the human elongation factor-1a promoter (EF-luc [35]) or the thymidine kinase promoter (tk-luc), arguing against a general downregulation of transcription by c-Maf (Fig. 1B). Therefore, inhibition of CD13/APN promoter activity by c-Maf is selective.
FIG. 1.
c-Maf selectively abrogates transcription from CD13/APN promoter constructs in myeloid cells. (A) c-Maf inhibition is dose dependent. Increasing amounts (0.1 to 4.0 μg) of the expression construct pRc/RSVcMaf were cotransfected with 4 μg of the CD13/APN wild-type reporter construct (−411luc) into KG1a immature myeloblastic cells, and luciferase activity was assayed at 24 h. (B) Inhibition by c-Maf is selective. The indicated reporter plasmids (4 μg) were transfected with equal amounts of c-Maf expression plasmids or the empty expression vector. All values are normalized to those for a cotransfected control plasmid (MAP1-SEAP). RLU, relative light units.
To determine whether c-Maf repression of CD13/APN promoter activity was occurring through an effect on c-Myb- and/or Ets-dependent transactivation, we initially assessed effects in C33A human epithelial cells (Fig. 2). These cells do not express CD13/APN or c-myb mRNA (data not shown), allowing us to observe c-Maf’s effect without influence from endogenous proteins. Expression of Ets-1 or c-Myb individually in C33A cells leads to weak activation of the CD13/APN reporter plasmid, while coexpression of these proteins results in higher than additive transactivation (60). However, increasing levels of c-Maf substantially repressed the cooperative transactivation of the CD13/APN promoter by c-Myb and Ets-1 in combination, and transactivation by either c-Myb or Ets-1 alone was also reduced to background levels (Fig. 2). Myb and Ets-1 protein levels were not affected by cotransfection of c-Maf (data not shown). Importantly, c-Maf did not alter CD13/APN basal promoter activity, indicating that c-Maf-mediated repression was specific (Fig. 2).
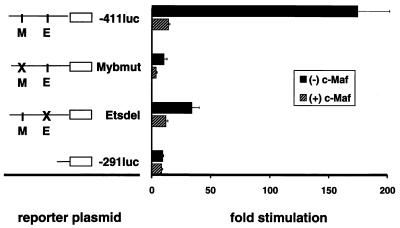
To determine whether c-Maf inhibited Ets-1 or c-Myb transcriptional activity, we tested its ability to inhibit CD13/APN reporter constructs containing either mutations or deletions of the essential c-Myb (Mybmut) or Ets (Etsdel) consensus binding sites (60). The reduced levels of transcription from the single-site-mutated promoters should be unaffected if c-Maf acts exclusively to inhibit the activity of either Myb or Ets-1 alone. Alternatively, if c-Maf suppresses the ability of both c-Myb and Ets-1 to transactivate the CD13/APN promoter, it should retain some or all of its inhibitory capacity when either site is altered. Introduction of c-Maf into _CD13/APN_-expressing KG1a myeloid cells (which contain endogenous c-Myb and Ets-1) inhibited transcription from both the wild-type and mutated promoter constructs (Fig. 3) to near or below baseline levels. By contrast, c-Maf had no effect on a promoter construct lacking both the Myb and Ets binding sites (−291luc), thus localizing the target of c-Maf inhibition to this region.
FIG. 3.
c-Maf inhibits c-Myb and Ets-1 transcriptional activity. Representations of the wild-type reporter plasmid (−411luc) indicating the c-Myb and Ets-1 consensus sites, and those containing deletions from the 5′ end (−291luc), point mutations (Mybmut), or internal deletions (Etsdel) of the CD13/APN myeloid promoter, are depicted on the left. Reporter plasmids (5 μg) were transiently transfected into the immature myeloblastic cell line KG1a along with 5 μg of pRc/RSVcMaf expression construct [(+) c-Maf)] or the empty expression vector [(−) c-Maf] and assayed for luciferase activity. All data are normalized to values for the cotransfected MAP1-SEAP control plasmid both for transfection efficiency and as a control for c-Maf’s effect on other promoters. Results are expressed as fold stimulation above the normalized activity of the empty reporter plasmid pGL2basic.
c-Maf physically interacts with c-Myb both in vitro and in vivo.
The observation that c-Maf showed a functional effect on Ets-1/c-Myb transactivation suggested that these proteins might physically interact. In a GST pull-down assay, bacterially expressed Myb or Ets-1 DNA binding domain fusion proteins were incubated with [35S]methionine-labeled, in vitro-translated murine c-Maf (Fig. 4A). As shown in lanes 3 and 4, c-Maf was specifically retained by the Myb DNA binding domain (lane 3), as well as by the Ets-1 DNA binding domain that interacts with MafB (lane 4) (62). By contrast, GST alone (lane 2) failed to retain c-Maf, despite loading of equivalent amounts of protein in these lanes (Fig. 4B).
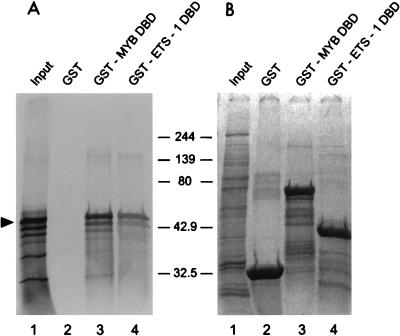
FIG. 4.
Full-length c-Maf binds to the c-Myb and Ets-1 DNA binding domains. (A) [35S]methionine-labeled c-Maf produced in vitro was incubated with GST alone (lanes 2) or with GST-Myb-DBD (amino acids 1 to 240) (lane 3) or GST-Ets-1-DBD (amino acids 322 to 440) (lane 4) fusion protein. The interacting proteins were purified on GSH-beads and analyzed by SDS-PAGE and autoradiography. The arrowhead indicates full-length c-Maf; molecular size markers are indicated in kilodaltons. Input (lane 1) shows 10% of the c-Maf used in each pull-down assay. GST acts as a negative control for c-Maf binding. (B) Coomassie blue staining of the same gel indicates that comparable amounts of GST fusion proteins were used in each assay.
The in vivo interaction between c-Myb and c-Maf was demonstrated by a yeast two-hybrid assay using the amino-terminal third of c-Myb (amino acids 1 to 240) fused to the yeast lexA DNA binding domain as bait. Prey constructs contained hybrids between different leucine zipper-containing proteins and the VP16 transcriptional activation domain. Development of blue color in bait-prey-cotransformed yeast colonies in a β-galactosidase activity assay indicated an association between the bait and prey proteins. Colonies of c-Myb-LexA–c-Maf-VP16-cotransformed yeast became blue within 60 min, indicating a strong interaction between c-Myb and c-Maf (Table 1). The result depended on the presence of both c-Myb and c-Maf (data not shown). Two additional Maf family members with bZIP domains closely related to that of c-Maf, Nrl (63) and the putative murine MafB homolog Kreisler (5), also interacted with c-Myb in this system, albeit with apparently weaker affinity (blue colonies within 120 min [Table 1]). Finally, three c-Maf-interacting proteins, including c-Fos and c-Jun (34), as well as the bHLH-ZIP protein USF2 (40), failed to interact with c-Myb in this sensitive assay.
TABLE 1.
Protein-protein interactions of Myb-LexA in the yeast two-hybrid system
| Prey construct | Interaction with Myb (amino acids 1–240)-LexAa |
|---|---|
| c-Maf (amino acids 123–370)-VP16 | ++ |
| Nrl (amino acids 118–237)-VP16 | + |
| Kreisler-VP16 (MafB) | + |
| c-Fos-VP16 | − |
| c-Jun-VP16 | − |
| USF2-VP16 | − |
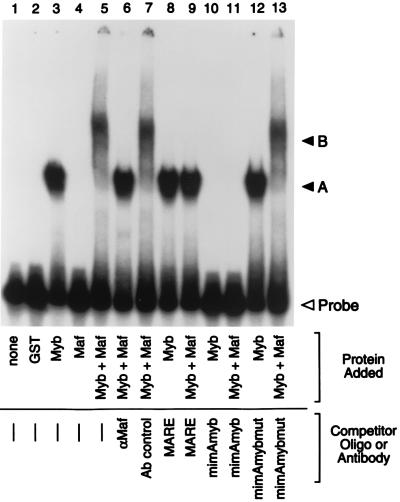
Mutation of the Myb binding site diminished c-Maf’s ability to inhibit CD13/APN transcription, indicating that c-Maf-mediated repression requires Myb binding to its cognate recognition site on the promoter. To address whether the Myb-Maf interactions can occur on the relevant CD13/APN promoter fragment, we performed electrophoretic mobility shift assays (EMSAs) (Fig. 5). Bacterially expressed GST fusion proteins incorporating either the c-Myb DNA binding domain (GST-Myb) or full-length c-Maf fused to GST (GST-Maf) were incubated with a 135-bp _Xba_I/_Dde_I genomic DNA fragment containing the region necessary for c-Maf inhibition of CD13/APN transcription (bp −426 through −291 [Fig. 3]). Incubation of both Myb and Maf proteins with this probe resulted in the formation of a complex of lower mobility (lane 5, complex B) compared with that seen with GST-Myb alone (lane 3, complex A). Importantly, GST-Maf alone (lane 4) did not retard the probe, showing that c-Maf acts not by binding to the DNA but through its physical association with Myb. Complex B contained c-Maf, since incubation with c-Maf antibody abrogated complex formation, presumably by binding to c-Maf and inhibiting Myb-Maf interactions (lane 6). Both of the shifted complexes (GST-Myb and GST-Myb plus GST-Maf) were abolished by the addition of excess unlabeled oligomer containing the Myb consensus site from the mim-1 promoter (mimAmyb [lanes 10 and 11]) but not by a competitor oligomer containing point mutations in the Myb consensus site (mimAmybmut [lanes 12 and 13]), indicating that complex B also contains Myb protein and that Myb binding to the CD13/APN Myb consensus site is necessary for secondary complex formation. Finally, addition of an excess of unlabeled oligomer containing the Maf consensus binding site (MARE [31]) also eliminated formation of the more slowly migrating complex B, suggesting that Maf is unable to interact with Myb when Maf is bound to DNA. Importantly, no higher-order complexes are formed in experiments combining probe with GST and GST-Myb or GST and GST-Maf, indicating that Myb and Maf do not interact via dimerization of the GST portions of the fusion proteins (data not shown).
FIG. 5.
c-Myb and Maf proteins form higher-order complexes on the CD13/APN promoter in vitro. An end-labeled 135-bp promoter fragment probe (bp −426 through −291) containing the functionally defined Maf target sequences was incubated with bacterially expressed GST fusion proteins. Lane 1, probe alone; lane 2, GST control; lanes 3, 8, 10, and 12, GST-Myb-DBD only; lane 4, GST-Maf only; lanes 5 to 7, 9, 11, and 13, equal amounts of GST-Myb-DBD and GST-Maf. Antibodies recognizing the c-Maf protein (lane 6) or an isotype-matched control antibody (Ab) (lane 7) were added in equal amounts to binding reactions. Unlabeled competitor oligonucleotides containing either the consensus Maf binding site (MARE; lanes 8 and 9), the consensus Myb site from the mim-1 promoter (mimAmyb; lanes 10 and 11), or a mutated Myb site (mimAmybmut; lanes 12 and 13) were added to the assays in 100-fold molar excess. A, the specific DNA-protein complex formed by GST-Myb-DBD; B, the ternary complex containing DNA, c-Myb, and c-Maf; Probe, free probe.
Myb-Maf complex formation correlates with CD13/APN transcription rates at different stages of myeloid differentiation.
If Maf is present in myeloid cells and inhibits transcription of CD13/APN, why is the gene expressed in such cells? One possibility is that c-Maf expression levels and CD13/APN transcription shift coordinately during myeloid cell differentiation. To demonstrate the activity of CD13/APN reporter constructs during myeloid cell development, we transiently transfected the CD13/APN reporter construct into human cell lines representing three progressive stages of myeloid differentiation: KG1a (immature myeloblastic), HL-60 (promyelocytic), and U937 (monoblastic) (50). The CD13/APN reporter gene transcription levels were lower in cell lines arrested at later stages of myeloid cell development (Fig. 6), suggesting that c-Maf levels may correlate with decreases in CD13/APN transcription. However, there were no significant differences in the low-level c-Maf mRNA (Fig. 7A) or protein (Fig. 7B) levels between the KG1a and HL-60 cell lines. Therefore, differences in the functional properties of the c-Myb and c-Maf proteins in these cell types could be responsible for the downregulation of CD13/APN transcription seen during myeloid cell differentiation.
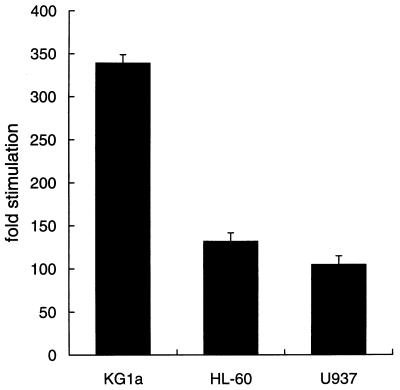
FIG. 6.
The activity of CD13/APN promoter constructs is diminished in later-stage myeloid cells. The −411luc reporter construct containing sequences sufficient for wild-type level, tissue-appropriate expression from the CD13/APN myeloid promoter (60) was transiently transfected into the immature myeloblastic cell line KG1a, the promyelocytic cell line HL-60, or the monoblastic cell line U937 and assayed for luciferase activity at 6 h. Luciferase activities were normalized for differences in transfection efficiency between cell lines with SEAP activity produced by the cotransfected plasmid MAP1-SEAP. Results are expressed as fold stimulation above the normalized activity of the empty reporter plasmid pGL2basic in each cell line.
FIG. 7.
Expression of c-Maf and c-Myb in myeloid cell lines. (A) c-Maf mRNA expression levels in human myeloid cell lines as determined by Northern blot analysis of poly(A)+ RNA from the KG1a immature myeloblastic cell line or the HL-60 promyelocytic cell line. A single blot was sequentially probed, stripped, and reprobed with the β-actin probe as a control for RNA loading and integrity. Exposure times for optimal detection of c-Maf were routinely significantly longer than for β-actin (1 week for c-Maf versus 1 day for β-actin). (B) c-Maf protein levels in human myeloid cell lines as determined by immunoprecipitation followed by Western blot analysis. Cell lysates from the KG1a and HL-60 cell lines were immunoprecipitated with either an antiserum recognizing the c-Maf protein (lanes 3 and 4) or an isotype-matched control (lanes 5 and 6). Immunoprecipitates were analyzed beside control lysates from an induced (lane 7) or uninduced (lane 8) cell line engineered to inducibly express a full-length c-Maf expression construct or IgG protein alone (lane 9). The gel was transferred and probed for c-Maf protein with the anti-Maf antiserum. The arrowhead marks the specific c-Maf band; asterisks indicate IgG bands recognized by the secondary donkey anti-rabbit IgG antiserum.
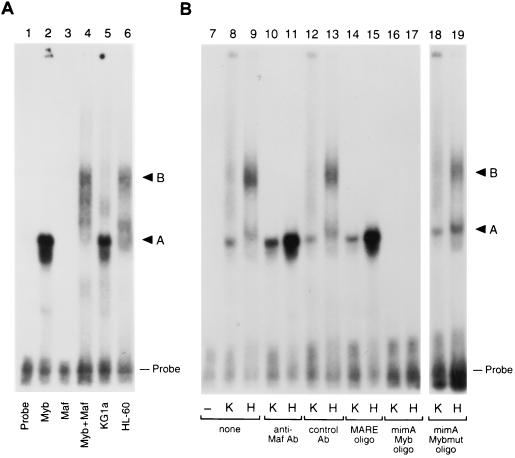
To assess the functional properties of the endogenous Myb and Maf proteins in different myeloid cell lines, we determined the amounts of Myb-Maf complex in lysates prepared from KG1a immature myeloblasts and the later-stage HL-60 promyelocytes. EMSA analysis using the 135-bp genomic promoter fragment as a probe detected markedly increased amounts of a slowly migrating complex (complex B) in the HL-60 cell lysate compared to lysates from the developmentally more primitive KG1a cell line (Fig. 8A), and this slower complex comigrated with the Myb-Maf complex formed with bacterially expressed GST fusion proteins (lane 4). This complex contained both Myb and Maf, since complex formation was eliminated by the addition of antiserum against the Maf protein, unlabeled Myb consensus oligomers (mimAmyb), or oligomers containing the Maf consensus binding site (MARE), while competitor oligomers with a mutated Myb site (mimAmybmut) had no effect (Fig. 8B). While it is possible that disparate Myb levels were responsible for discrepant complex formation (HL-60 cells express approximately twofold-higher Myb message levels than KG1a cells [61]), addition of GST-Myb-DBD to KG1a lysates did not result in higher-order complex formation (data not shown). These observations suggest that CD13/APN transcriptional downregulation in HL-60 cells versus KG1a cells results from higher levels of Myb-Maf inhibitory complexes binding to DNA in these cells.
FIG. 8.
Myb-Maf complex formation is regulated differently in early- and later-stage myeloid cell lines. An end-labeled 135-bp promoter fragment probe containing the functionally defined Maf target sequences was incubated with bacterially expressed GST fusion proteins or whole-cell lysates from KG1a immature myeloblasts or HL-60 promyelocytes. (A) HL-60 lysates contain a complex that comigrates with the Myb-Maf complex (GST-Myb-DBD–GST-Maf complex, 57 kDa + 69 kDa = 126 kDa; native Myb-Maf complex, 80 kDa + 42 kDa = 122 kDa). Lane 1, probe alone; lane 2, GST-Myb-DBD only; lane 3, GST-Maf only; lane 4, equal amounts of GST-Myb-DBD and GST-Maf; lane 5, KG1a lysate; lane 6, HL-60 lysate. (B) HL-60 lysates contain a Myb-Maf complex that binds to the Myb site. Lane 7, probe alone; lanes 8, 10, 12, 14, 16, and 18, KG1a lysate (K); lanes 9, 11, 13, 15, 17, and 19, HL-60 lysate (H). Antibodies (Ab) recognizing the c-Maf protein (lanes 10 and 11) or an isotype-matched control antibody (lanes 12 and 13) were added in equal amounts to binding reactions. Unlabeled competitor oligonucleotides containing either the consensus Maf binding site (MARE; lanes 14 and 15), the consensus Myb site from the mim-1 promoter (mimAmyb; lanes 16 and 17), or a mutated Myb site (mimAmybmut; lanes 18 and 19) were added to the assays in 100-fold molar excess. A, the DNA-protein complex containing Myb; B, the ternary complex containing both Myb and Maf; Probe, uncomplexed probe.
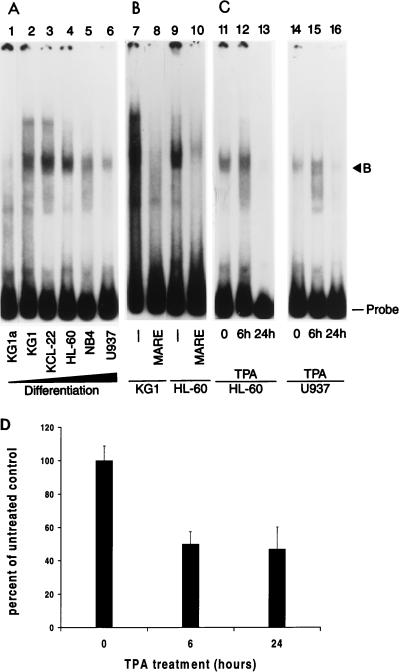
Gel shift assays using lysates from an expanded panel of cell lines arrested at progressively later stages of myeloid differentiation (KG1a, phenotypically primitive, developmentally arrested revertant of the KG1 cell line that is incapable of further myeloid differentiation [37]); KG1, early myeloblastic; KCL22, late myeloblastic; HL-60 and NB4, promyelocytic; U937, myelomonoblastic [36a, 51]) indicate that the level of inhibitory Myb-Maf complexes appears to peak near the late myeloblast stage and decreases as the cells mature (Fig. 9A and B). To confirm that the complexes are regulated in response to developmental signals, HL-60 or U937 cells were induced to differentiate toward the monocytic lineage by treatment with TPA. Lysates from both cell lines show a slight increase in Myb-Maf complex levels in response to TPA before near complete ablation by 24 h of treatment. Therefore, Myb-Maf complex formation is regulated in response to differentiating signals and is not simply due to inherent differences between cell lines. Finally, CD13/APN promoter constructs show a correspondent decrease in luciferase activity in HL-60 cells after 6 and 24 h of TPA treatment (Fig. 9D), possibly indicating an irreversible downregulation of transcription.
FIG. 9.
Levels of Myb-Maf complexes change with differentiation stage of myeloid cell lines and in response to monocytic differentiation signals and correlate with CD13/APN transcriptional activity. An end-labeled 135-bp promoter fragment probe containing the functionally defined Maf target sequences was incubated with whole-cell lysates from myeloid cell lines arrested at progressively more differentiated stages (37, 51) or from cell lines treated with TPA. (A) Myb-Maf complex formation appears to peak near the late myeloblast stage. Lane 1, KG1a (phenotypically primitive, developmentally arrested revertant of the KG1 cell line); lane 2, KG1 (early myeloblastic); lane 3, KCL22 (late myeloblastic); lane 4, HL-60 (promyelocytic); lane 5, NB4 (promyelocytic); lane 6, U937 (myelomonoblastic). (B) The differentially regulated complex contains Maf. Unlabeled competitor oligonucleotides containing the consensus Maf binding site (MARE; lanes 8 and 10) were added to binding reactions containing KG1 or HL-60 lysates in 100-fold molar excess. (C) Myb-Maf complexes are regulated during monocytic differentiation. HL-60 (lanes 11 to 13) or U937 (lanes 14 to 16) cells were treated with 10−6 M TPA and harvested at the indicated times. B, the secondary complex containing both Myb and Maf; Probe, uncomplexed probe. (D) Reporter gene levels correlate with complex formation. HL-60 cells were transiently transfected with 4 μg of the CD13/APN wild-type reporter construct (−411luc). Cultures were treated with 10−6 M TPA, and luciferase activity was assayed at 6 and 24 h.
If Myb-Maf complexes bind to the CD13/APN Myb site to inhibit transcription, binding of these negative regulatory complexes might be preferred over the positive regulatory binding of Myb alone. To address this issue, we determined the relative stability of the Myb-Maf-DNA ternary complexes and the Myb-DNA binary complexes by measuring the dissociation of preformed complexes in the presence of vast excess of cold competitor DNA (Fig. 10). Results of EMSAs using the CD13/APN promoter probe, GST fusion proteins, and a fixed amount of a 70-bp competitor oligonucleotide encompassing bases −320 through −390 of the CD13/APN promoter (which includes the Myb and Ets consensus sites) indicate that the half-life of the Myb-Maf-DNA complexes is between 10 and 20 min, compared to less than 2 min for the Myb-DNA complexes. Therefore, binding of Myb-Maf inhibitory complexes to the CD13/APN promoter would be preferred over the binding of uncomplexed c-Myb, which is required for maximal cooperative transactivation (60).
DISCUSSION
Myeloid-specific expression of MafB inhibits Ets-1-dependent expression of avian erythroid-restricted genes, and inhibition of Ets-1 activity has been postulated to play a pivotal role in myeloid/erythroid lineage specification (62). Ets-1 and c-Maf are also expressed in human myeloid cells, leading us to investigate the role of Maf family proteins in modulating the transcription of a prototypical Ets-1-dependent myeloid gene, CD13/APN. Here we have shown that murine c-Maf inhibits the capacity of both Ets-1 and c-Myb to transactivate CD13/APN gene transcription in early myeloid cells. This inhibition depends on binding of Myb and Ets-1 to their cognate promoter sites and is effective at disrupting the cooperativity between c-Myb and Ets-1. Dissection of the mechanism of Maf’s inhibitory effect illustrated that Maf does not bind to the CD13/APN promoter itself but rather interacts with c-Myb to form higher-order complexes that appear to bind preferentially to the Myb site of the CD13/APN promoter. Finally, although c-Maf mRNA and protein are expressed at essentially equivalent levels at different stages of myeloid development, formation of the Myb-Maf complex on the CD13/APN promoter appears to be developmentally regulated, suggesting a model of transcription inhibition by preferential binding of regulated inhibitory complexes.
c-Maf protein is capable of inhibiting CD13/APN transcription in early myeloid cells. Because the cooperative transactivation of CD13/APN by c-Myb and Ets-1 is implicated in this inhibition, two different mechanisms can be invoked. First, Maf could inhibit transactivation by forming a ternary complex with Myb and Ets-1, thereby interfering with their activity. Even though c-Maf can interact with Ets-1 in GST pull-down assays (Fig. 4), and the probe used in our EMSAs contains functional Ets consensus sites, the Myb-Maf-containing protein complexes bound to the CD13/APN promoter in later-stage myeloid cells do not contain Ets since they are unaffected by the addition of Ets competitor oligomers or antibodies against Ets-1 (20). Additionally, we were unable to demonstrate the formation of tertiary complexes by using bacterially expressed Myb, Maf, and Ets-1 proteins in EMSA under conditions supporting Myb-Maf complex formation (20), arguing against an association of these three proteins on the CD13/APN promoter.
Alternatively, the presence of detectable Myb-Maf complexes in later-stage myeloid cells suggests that Myb may be the primary target of c-Maf and that its association with Myb is sufficient to negate Myb-Ets cooperation in CD13/APN transcription. An extension of this model predicts that inhibitory Myb-Maf complexes would preferentially bind to the CD13/APN promoter, interfering with the positive consequences of c-Myb and Ets-1 binding. Indeed, our data indicate that the Myb-Maf-DNA complex is more stable than the binding of uncomplexed c-Myb required for full transactivation (Fig. 10). Finally, while Ets-1 interacts with c-Maf in vitro and it appears that Ets-1 is functionally affected by c-Maf in our system, our inability to demonstrate Maf-Ets complexes in our human myeloid cells suggests that Maf inhibits Ets-1 by mechanisms other than stable complex formation.
The ability of Maf to inhibit c-Myb activity appears, on the surface, to conflict with recent reports illustrating that MafB is unable to inhibit Myb’s transactivation of a reporter construct containing multimerized Myb consensus sites in quail fibroblast cells (62). This discrepancy, however, is likely due to the context of both the promoter and cell type in transcriptional regulation. Indeed, while CD13/APN reporter constructs are inhibited by c-Maf in C33A human epithelial cells, c-Maf has no effect on a reporter construct containing five Myb consensus sites fused to the thymidine kinase promoter in the same cell line (61). In addition, Myb and Maf complexes were not found in EMSA when we used shorter (23- to 26-bp) oligonucleotide probes consisting of the isolated CD13/APN Myb consensus site or the consensus Myb site from the mim-1 promoter (20). In both cases, while uncomplexed c-Myb binds to DNA, no higher-order aggregates are formed upon addition of c-Maf protein. This observation suggests that c-Myb binding to its cognate site is not by itself sufficient for Myb-Maf complex formation and that the promoter context contributes to the ability of the inhibitory complex to bind to DNA and, consequently, to inhibit transcription. Similar results have been obtained in studies in which complexes containing the Oct-1 transcription factor and its coactivator Bob-1 bound only a small subset of the sequence elements that bound Oct-1 alone, leading to the differential regulation of promoters containing octamer sequences (17). Additionally, the functional consequences of transcription factor binding can differ greatly depending on the context of the promoter, presumably via the binding of distinct proteins to contiguous regulatory elements (9, 13, 19, 24, 42, 53, 57). In like manner, DNA signals can specifically alter the conformation of bound proteins, and these specific conformations may be selectively recognized by interacting regulatory proteins (49), thereby influencing their transregulatory capabilities. Each of these is a plausible model for the promoter context dependence of the Myb-Maf interaction.
Cell context is obviously another key determinant of either the formation or binding of Myb-Maf complexes, as illustrated by disparate complex levels between myeloid cell lines arrested at progressively later stages of differentiation (Fig. 9A). Similarly, cells induced to differentiate show an initial increase in Myb-Maf complexes (Fig. 9C). Such results suggest the presence of an inhibitory protein modification or molecule in the early stage lysates that is lost as cells progress toward more differentiated stages, perhaps in response to specific differentiation signals that can also be triggered by treatment with TPA. Indeed, distinct patterns of transcription factor binding between different cell types or developmental stages have been described as the result of cell-type-specific sequestration of a transcription factor by an inhibitory molecule (12, 28), interference with DNA binding (36, 47), or protein modification (4, 41, 64). One of us has recently found that the USF2/FIP member of the bHLH-ZIP protein family can also associate with c-Maf and subsequently inhibit c-Maf DNA binding to its consensus site (40), supporting the concept of an interacting inhibitory molecule present in early myeloid cells that blocks Myb-Maf inhibitory function. Similarly, small Mafs have been shown to form heterocomplexes with two classes of proteins via their leucine zippers to form either repressive or transactivating complexes, depending on the cell context (27, 56). It is therefore likely that similar interactions are responsible for the developmentally regulated interaction of c-Maf and c-Myb observed among the myeloid cell lines tested, at least in the early stages of myeloid differentiation.
By contrast, later-stage myeloid cell lines and cells induced to differentiate for a prolonged period show a disappearance of Myb-Maf complexes (Fig. 9). While this reduction may attributable to decreased Myb levels in cells induced to differentiate (2, 61), high Myb expression in uninduced U937 cells (equivalent to that seen in HL-60 cells [61]) does not result in high levels of Myb-Maf complexes (Fig. 9A), arguing against Myb expression levels as the sole determinant of complex formation. It is possible, therefore, that inhibition of Myb activity by Myb-Maf complex formation is no longer a functionally active mechanism in later stages of myelopoiesis. The fact that _CD13/APN_-driven luciferase levels remain low at 24 h in the absence of Myb-Maf complexes may merely reflect an irreversible downregulation of target gene activity due to other developmentally dependent factors.
The ability of the Maf family of proteins to inhibit the activity of two hematopoietically important proto-oncogenes is a potentially exciting mechanism for guiding gene transcription. Together with data implicating Maf proteins as both positive and negative regulators of tissue-specific gene expression in multiple hematopoietic lineages (12, 21, 25, 62), our observations suggest an important role for Maf proteins in the commitment and lineage determination of hematopoietic cells.
ACKNOWLEDGMENTS
We thank Elizabeth Mann for expert technical assistance, Rick Bram, David Shapiro, John Cleveland, and Paul Brindle for helpful comments, H. P. Koeffler for his gift of myeloid cell lines, Barbara Graves for her gift of ETS-1 plasmids, and John Gilbert for editorial assistance.
This study was supported by NIH grant CA-70909 to L.H.S., by National Cancer Institute Cancer Center Support (CORE) grant CA-21765, and by the American Lebanese Syrian Associated Charities, St. Jude Children’s Research Hospital.
REFERENCES
- 1.Bram R J, Hung D T, Martin P K, Schreiber S L, Crabtree G R. Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: roles of calcineurin binding and cellular location. Mol Cell Biol. 1993;13:4760–4769. doi: 10.1128/mcb.13.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brelvi Z S, Studzinski G. Coordinate expression of c-myc, c-myb, and histone H4 genes in reversibly differentiating HL-60 cells. J Cell Physiol. 1987;131:43–49. doi: 10.1002/jcp.1041310108. [DOI] [PubMed] [Google Scholar]
- 3.Burk O, Mink S, Ringwald M, Klempnauer K H. Synergistic activation of the chicken mim-1 gene by v-myb and C/EBP transcription factors. EMBO J. 1993;12:2027–2038. doi: 10.1002/j.1460-2075.1993.tb05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkhoven C F, Ab G. Multiple steps in the regulation of transcription-factor level and activity. Biochem J. 1996;317:329–342. doi: 10.1042/bj3170329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordes S P, Barsh G S. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 6.Dai P, Akimaru H, Tanaka Y, Hou D-X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 7.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 8.Dash A B, Orrico F C, Ness S A. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 9.Diagana T T, North D L, Jabet C, Fiszman M Y, Takeda S, Whalen R G. The transcriptional activity of a muscle-specific promoter depends critically on the structure of the TATA element and its binding protein. J Mol Biol. 1997;265:480–493. doi: 10.1006/jmbi.1996.0752. [DOI] [PubMed] [Google Scholar]
- 10.Dudek H, Tantravahi R V, Rao V N, Reddy E S, Reddy E P. Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc Natl Acad Sci USA. 1992;89:1291–1295. doi: 10.1073/pnas.89.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton J, Kouzarides T, Doderlein G, Graf T, Weston K. Influence of the v-Myb transactivation domain on the oncoprotein’s transformation specificity. EMBO J. 1993;12:1333–1341. doi: 10.1002/j.1460-2075.1993.tb05778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francastel C, Augery-Bourget Y, Prenant M, Walters M, Martin D I, Robert-Lezenes J. c-Jun inhibits NF-E2 transcriptional activity in association with p18/maf in Friend erythroleukemia cells. Oncogene. 1997;14:873–877. doi: 10.1038/sj.onc.1200902. [DOI] [PubMed] [Google Scholar]
- 13.Galvin K M, Shi Y. Multiple mechanisms of transcriptional repression by YYI. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf T. Myb: a transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr Opin Genet Dev. 1992;2:249–255. doi: 10.1016/s0959-437x(05)80281-3. . (Erratum, 2:504.) [DOI] [PubMed] [Google Scholar]
- 15.Griffin J D, Ritz J, Beveridge R P, Lipton J M, Daley J F, Schlossman S F. Expression of MY7 antigen on myeloid precursor cells. Int J Cell Cloning. 1983;1:33–48. doi: 10.1002/stem.5530010106. [DOI] [PubMed] [Google Scholar]
- 16.Griffin J D, Ritz J, Nadler L M, Schlossman S F. Expression of myeloid differentiation antigens on normal and malignant myeloid cells. J Clin Invest. 1981;68:932–941. doi: 10.1172/JCI110348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- 18.Guarente L, Yocum R R, Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA. 1982;79:7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:4057–4066. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 20.Hegde, S. P., and L. H. Shapiro. 1997. Unpublished data.
- 21.Ho I-C, Hodge M R, Rooney J W, Glimcher L H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Hogg N, Horton M J. Myeloid antigens: new and previously defined clusters. In: McMichael A J, editor. Leukocyte typing III. Proceedings of the Third International Workshop on Human Leukocyte Differentiation Antigens. New York, N.Y: Oxford University Press; 1987. pp. 576–621. [Google Scholar]
- 24.Huang W, Bateman E. Transcription of the Acanthamoeba TATA-binding protein gene. A single transcription factor acts both as an activator and a repressor. J Biol Chem. 1997;272:3852–3859. doi: 10.1074/jbc.272.6.3852. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Conditional expression of the ubiquitous transcription factor MafK induces erythroleukemia cell differentiation. Proc Natl Acad Sci USA. 1995;92:7445–7449. doi: 10.1073/pnas.92.16.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 27.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jupin I, Chua N H. Activation of the CaMV as-1 cis-element by salicylic acid: differential DNA-binding of a factor related to TGA1a. EMBO J. 1996;15:5679–5689. [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka K, Fujiwara K T, Noda M, Nishizawa M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol Cell Biol. 1994;14:7581–7591. doi: 10.1128/mcb.14.11.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kataoka K, Igarashi K, Itoh K, Fujiwara K T, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataoka K, Noda M, Nishizawa M. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos, and small Maf proteins. Oncogene. 1996;12:53–62. [PubMed] [Google Scholar]
- 33.Kerppola T K, Curran T. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene. 1994;9:3149–3158. [PubMed] [Google Scholar]
- 34.Kerppola T K, Curran T. Maf and Nrl can bind to AP-1 sites and from heterodimers with Fos and Jun. Oncogene. 1994;9:675–684. [PubMed] [Google Scholar]
- 35.Kim D W, Uetsuki T, Kaziro Y, Yamaguchi N, Sugano S. Use of the human elongation factor 1 alpha promoter as a versatile and efficient expression system. J Biol Chem. 1990;91:217–223. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 36.Kirov N C, Lieberman P M, Rushlow C. The transcriptional corepressor DSP1 inhibits activated transcription by disrupting TFIIA-TBP complex formation. EMBO J. 1996;15:7079–7087. [PMC free article] [PubMed] [Google Scholar]
- 36a.Koeffler, H. P. Personal communication.
- 37.Koeffler H P, Billing R, Lusis A J, Sparkes R, Golde D W. An undifferentiated variant derived from the human acute myelogenous leukemia cell line (KG-1) Blood. 1980;56:265–273. [PubMed] [Google Scholar]
- 38.Kurschner C, Morgan J I. The maf proto-oncogene stimulates transcription from multiple sites in a promoter that directs Purkinje neuron-specific gene expression. Mol Cell Biol. 1995;15:246–254. doi: 10.1128/mcb.15.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurschner C, Morgan J I. Analysis of interaction sites in homo- and heteromeric complexes containing Bcl-2 family members and the cellular prion protein. Mol Brain Res. 1996;37:249–258. doi: 10.1016/0169-328x(95)00323-k. [DOI] [PubMed] [Google Scholar]
- 40.Kurschner C, Morgan J I. USF2/FIP associates with the b-Zip transcription factor, c-Maf, via its bHLH domain and inhibits c-Maf DNA binding activity. Biochem Biophys Res Commun. 1997;231:333–339. doi: 10.1006/bbrc.1997.6097. [DOI] [PubMed] [Google Scholar]
- 41.Leggett R W, Armstrong S A, Barry D, Mueller C R. Sp1 is phosphorylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J Biol Chem. 1995;270:25879–25884. doi: 10.1074/jbc.270.43.25879. [DOI] [PubMed] [Google Scholar]
- 42.Luo X, Sawadogo M. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luscher B, Eisenman R N. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990;4:2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- 44.Melotti P, Calabretta B. Ets-2 and c-Myb act independently in regulating expression of the hematopoietic stem cell antigen CD34. J Biol Chem. 1994;269:25303–25309. [PubMed] [Google Scholar]
- 45.Metz T, Graf T. Fusion of the nuclear oncoproteins v-Myb and v-Ets is required for the leukemogenicity of E26 virus. Cell. 1991;66:95–105. doi: 10.1016/0092-8674(91)90142-l. [DOI] [PubMed] [Google Scholar]
- 46.Metz T, Graf T. v-myb and v-ets transform chicken erythroid cells and cooperate both in trans and in cis to induce distinct differentiation phenotypes. Genes Dev. 1991;5:369–380. doi: 10.1101/gad.5.3.369. [DOI] [PubMed] [Google Scholar]
- 47.Miller S D, Moses K, Jayaraman L, Prives C. Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single-stranded DNA. Mol Cell Biol. 1997;17:2194–2201. doi: 10.1128/mcb.17.4.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mink S, Kerber U, Klempnauer K H. Interaction of C/EBPβ and v-Myb is required for synergistic activation of the mim-1 gene. Mol Cell Biol. 1996;16:1316–1325. doi: 10.1128/mcb.16.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misra V, Walter S, Yang P, Hayes S, O’Hare P. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Mol Cell Biol. 1996;16:4404–4413. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morosetti R, Grignani F, Liberatore C, Pelicci P G, Schiller G J, Kizaki M, Bartram C R, Miller C W, Koeffler H P. Infrequent alterations of the RAR alpha gene in acute myelogenous leukemias, retinoic acid-resistant acute promyelocytic leukemias, myelodysplastic syndromes, and cell lines. Blood. 1996;87:4399–4403. [PubMed] [Google Scholar]
- 51.Morosetti R, Park D J, Chumakov A M, Grillier I, Shiohara M, Gombart A F, Nakamaki T, Weinberg K, Koeffler H P. A novel, myeloid transcription factor, C/EBPe, is upregulated during granulocytic, but not monocytic, differentiation. Blood. 1997;90:2591–2600. [PubMed] [Google Scholar]
- 52.Moulton K S, Semple K, Wu H, Glass C K. Cell-specific expression of the macrophage scavenger receptor gene is dependent on PU.1 and a composite AP-1/ets motif. Mol Cell Biol. 1994;14:4408–4418. doi: 10.1128/mcb.14.7.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol Cell Biol. 1995;15:5975–5982. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelsen B, Tian B, Erman B, Gregoire J, Maki R A, Graves B J, Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 55.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luchser B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 56.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palvimo J J, Parnanen M, Janne O A. Characterization of cell-specific modulatory element in the murine ornithine decarboxylase promoter. Biochem J. 1996;316:993–998. doi: 10.1042/bj3160993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reddy M A, Yang B S, Yue X, Barnett C J, Ross I L, Sweet M J, Hume D A, Ostrowski M C. Opposing actions of c-ets/PU.1 and c-myb protooncogene products in regulating the macrophage-specific promoters of the human and mouse colony-stimulating factor-1 receptor (c-fms) genes. J Exp Med. 1994;180:2309–2319. doi: 10.1084/jem.180.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shapiro L H. MYB and ETS proteins cooperate to transactivate an early myeloid gene. J Biol Chem. 1995;270:8763–8771. doi: 10.1074/jbc.270.15.8763. [DOI] [PubMed] [Google Scholar]
- 61.Shapiro, L. H. 1997. Unpublished data.
- 62.Sieweke M H, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of ETS-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 63.Swaroop A, Xu J, Pawer H, Jackson A, Scolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper protein. Proc Natl Acad Sci USA. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner S, Green M R. DNA-binding domains: targets for viral and cellular regulators. Curr Opin Cell Biol. 1994;6:410–414. doi: 10.1016/0955-0674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 65.Wu H, Moulton K, Horvai A, Parik S, Glass C K. Combinatorial interactions between AP-1 and ets domain proteins contribute to the developmental regulation of the macrophage scavenger receptor gene. Mol Cell Biol. 1994;14:2129–2139. doi: 10.1128/mcb.14.3.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]