Virus-Dependent Phosphorylation of the IRF-3 Transcription Factor Regulates Nuclear Translocation, Transactivation Potential, and Proteasome-Mediated Degradation (original) (raw)
Abstract
The interferon regulatory factors (IRF) consist of a growing family of related transcription proteins first identified as regulators of the alpha beta interferon (IFN-α/β) gene promoters, as well as the interferon-stimulated response element (ISRE) of some IFN-stimulated genes. IRF-3 was originally identified as a member of the IRF family based on homology with other IRF family members and on binding to the ISRE of the ISG15 promoter. IRF-3 is expressed constitutively in a variety of tissues, and the relative levels of IRF-3 mRNA do not change in virus-infected or IFN-treated cells. In the present study, we demonstrate that following Sendai virus infection, IRF-3 is posttranslationally modified by protein phosphorylation at multiple serine and threonine residues, which are located in the carboxy terminus of IRF-3. A combination of IRF-3 deletion and point mutations localized the inducible phosphorylation sites to the region -ISNSHPLSLTSDQ- between amino acids 395 and 407; point mutation of residues Ser-396 and Ser-398 eliminated virus-induced phosphorylation of IRF-3 protein, although residues Ser-402, Thr-404, and Ser-405 were also targets. Phosphorylation results in the cytoplasm-to-nucleus translocation of IRF-3, DNA binding, and increased transcriptional activation. Substitution of the Ser-Thr sites with the phosphomimetic Asp generated a constitutively active form of IRF-3 that functioned as a very strong activator of promoters containing PRDI-PRDIII or ISRE regulatory elements. Phosphorylation also appears to represent a signal for virus-mediated degradation, since the virus-induced turnover of IRF-3 was prevented by mutation of the IRF-3 Ser-Thr cluster or by proteasome inhibitors. Interestingly, virus infection resulted in the association of IRF-3 with the CREB binding protein (CBP) coactivator, as detected by coimmunoprecipitation with anti-CBP antibody, an interaction mediated by the C-terminal domains of both proteins. Mutation of residues Ser-396 and Ser-398 in IRF-3 abrogated its binding to CBP. These results are discussed in terms of a model in which virus-inducible, C-terminal phosphorylation of IRF-3 alters protein conformation to permit nuclear translocation, association with transcriptional partners, and primary activation of IFN- and IFN-responsive genes.
Interferons (IFNs) are a large family of multifunctional secreted proteins involved in antiviral defense, cell growth regulation, and immune activation (63). Virus infection induces the transcription and synthesis of multiple IFN genes (33, 52, 63); newly synthesized IFN interacts with neighboring cells through cell surface receptors and the JAK-STAT signalling pathway, resulting in the induction of over 30 new cellular proteins that mediate the diverse functions of the IFNs (17, 35, 39, 58). Among the many virus- and IFN-inducible proteins are the growing family of interferon regulatory factor (IRF) transcription factors, the IRFs. IRF-1 and IRF-2 are the best-characterized members of this family, originally identified by studies of the transcriptional regulation of the human beta IFN (IFN-β) gene (22, 23, 30, 47). Their discovery preceded the recent expansion of this group of IFN-responsive proteins, which now include seven other members, i.e., IRF-3, IRF-4/Pip/ICSAT, IRF-5, IRF-6, IRF-7, ISGF3γ/p48, and ICSBP (48). Structurally, the Myb oncoproteins share homology with the IRF family, although the relationship of this family to the IFN system is unclear (62). Recent evidence also demonstrates the presence of a virally encoded analog of cellular IRFs, i.e., vIRF in the genome of human herpes virus 8 (55).
The presence of IRF-like binding sites in the promoter region of the IFN-α and -β genes implicated the IRFs as essential mediators of the induction of IFN genes. The original results of Harada et al. (30, 32) indicated that IFN gene induction was activated by IRF-1, while the related IRF-2 factor suppressed IFN expression. However, the essential role of IRF-1 and IRF-2 in the regulation of IFN-α and -β gene expression has become controversial with the observation that mice containing a homozygous deletion of IRF-1 or IRF-2 or fibroblasts derived from these mice induced IFN-α and -β gene expression after virus infection to the same level as that for the wild-type mice or cells (44). On the other hand, IRF-1 was shown to have an important role in the antiviral effects of IFNs (44, 54). IRF-1 binds to the interferon-stimulated response element (ISRE) present in many IFN-inducible gene promoters and activates expression of some of these genes (54). However, activation of ISG genes by IFN-α and -β was shown to be mediated generally by the multiprotein ISGF3 complex (31, 36, 38). The binding of this complex to DNA is mediated by another member of the IRF family, ISGF3γ/p48, which in IFN-treated cells interacts with phosphorylated STAT1 and STAT2 transcription factors, forming the heterotrimeric complex ISGF3 (8, 39, 62). The homozygous deletion of p48 in mice abolished the sensitivity of these mice to the antiviral effects of IFNs, and virus-infected macrophages from p48−/− mice showed an impaired induction of IFN-α and -β genes (31).
Several other members of the IRF family have been identified. The ICSBP gene is expressed exclusively in the cells of the immune system (18, 64), and its expression can be enhanced by IFN-γ. ICSBP was shown to form a complex with IRF-1 and to inhibit the transactivating activity of IRF-1 (9, 59). The homozygous deletion of ICSBP in mice leads to defects in myeloid cell lineage development and chronic myelogenous leukemia (34). Another lymphoid cell-specific IRF, Pip/LSIRF/IRF-4, that interacts with phosphorylated PU.1, a member of the Ets family of transcription factors (15), was identified (19, 43, 66). The Pip/PU.1 heterodimer can bind to the immunoglobulin light-chain enhancer and function as a B-cell-specific transcriptional activator. Expression of Pip/LSIRF was induced by antigenic stimulation but not by IFN, and Pip/LSIRF/IRF-4−/− mice failed to develop mature T and B cells (46). A novel member of the IRF family was recently identified by its ability to bind to an ISRE-like element in the promoter region of the Qp gene of Epstein-Barr virus (69).
Another unique member of the human IRF family, IRF-3, was recently characterized (2). The IRF-3 gene encodes a 55-kDa protein which is expressed constitutively in all tissues. Recombinant IRF-3 binds to the ISRE element of the IFN-induced gene ISG-15 and stimulates this promoter in transient expression assays. In contrast to IRF-1, which contains both DNA binding and transactivating domains, IRF-3—like IRF-2 and ICSBP—does not contain a well-defined transactivation domain. Viral induction or IFN treatment does not stimulate expression of the IRF-3 gene. In previous studies, we showed that IRF-3 binds to the IE and PRDIII regions of the IFN-α and -β promoters, respectively, but has different effects on their transcriptional activity (56). While the induction of the IFN-α4 promoter activated by IRF-1 or virus infection was inhibited in the presence of IRF-3, the fusion protein containing the IRF-3 DNA binding domain and the RelA(p65) transactivation domain effectively activated both IFN-α and -β promoters. In contrast, coexpression of IRF-3 and RelA plasmids transactivated the IFN-β gene promoter, but not the promoter of the IFN-α4 gene (56).
In the present study, we examined the virus-induced, posttranslational modulation of IRF-3 protein following Sendai virus infection. Our studies demonstrate that phosphorylation represents an important posttranslational modification of IRF-3, leading to cytoplasm-to-nucleus translocation of phosphorylated IRF-3, stimulation of DNA binding and transcriptional activity, association of IRF-3 with the transcriptional coactivator CBP/p300, and, ultimately, proteasome-mediated degradation.
MATERIALS AND METHODS
Plasmid constructions and mutagenesis.
The IRF-3 expression plasmid was prepared by cloning the _Eco_RI-_Xho_I fragment containing the IRF-3 cDNA from the pSKIRF-3 plasmid downstream of the cytomegalovirus (CMV) promoter of the CMVBL vector. CMVt-IRF-3 was constructed by cloning of IRF-3 cDNA downstream of the doxycycline-responsive promoter CMVt at the _Bam_HI site of the neoCMVtBL vector (49). cDNAs encoding IRF-3 carboxyl-terminal deletion mutations were generated by 28 cycles of PCR amplification with Vent DNA polymerase. DNA oligonucleotide primers were synthesized with an Applied Biosystems DNA/RNA synthesizer. The amino-terminal primer was synthesized with an _Eco_RI restriction enzyme site, and the carboxyl-terminal primers were synthesized with _Xba_I restriction enzyme sites at their ends. The PCR products were purified by phenol-chloroform extraction and ethanol precipitation, digested with _Eco_RI and _Xba_I, and inserted into _Eco_RI/_Xba_I sites of the CMVBL vector. The point mutations of IRF-3 were generated by overlap PCR mutagenesis with Vent DNA polymerase. Mutations were confirmed by sequencing. The N-terminal deletion mutations (ΔN, ΔN2A, ΔN3A, and ΔN5A) of IRF-3 were generated by digestion of the related IRF-3/CMVBL plasmid with _Bam_HI (filled in with Klenow enzyme), partial digestion with _Sca_I, and religation. Green fluorescence protein (GFP)–IRF-3 expression plasmids were generated by cloning of cDNAs encoding wild-type or mutated forms of IRF-3 into the downstream region of EGFP in the pEGFP-C1 vector (Clontech). For construction of plasmids encoding myc-tagged CREB-binding protein (CBP)-truncated proteins, the cDNAs coding for CBP were generated from the pRC-RSV/mCBP plasmid (provided by Dimitris Thanos) by PCR amplification. The cDNA fragments were cloned in the downstream of region myc tag in the 5′ myc-PCDNA3 vector (provided by Stephane Richard).
Generation of IRF-3 cell lines.
Plasmid CMVt-rtTA (49) was introduced into 293 cells by a calcium phosphate-based method. Cells were selected beginning at 48 h after transfection for about 1 week in alpha modified Eagle medium (αMEM; GIBCO-BRL) containing 10% heat-inactivated calf serum, glutamine, antibiotics, and 2.5 ng of puromycin (Sigma) per μl. Resistant cells carrying the CMVt-rtTA plasmid (rtTA-293 cells) were then transfected with the CMVt-IRF-3 plasmid. Cells were selected beginning at 48 h for a period of approximately 2 weeks in αMEM containing 10% heat-inactivated calf serum, glutamine, antibiotics, 2.5 ng of puromycin per μl, and 400 μg of G418 (Life Technologies, Inc.) per ml.
Cell culture and transfections.
All transfections for chloramphenicol acetyltransferase (CAT) assaying were carried out with human embryonic kidney 293 cells or NIH 3T3 cells grown in αMEM (293 cells) or Dulbecco’s MEM (NIH 3T3 cells; GIBCO-BRL) supplemented with 10% calf serum, glutamine, and antibiotics. Subconfluent cells were transfected with 5 μg of CsCl-purified CAT reporter and expression plasmids by the calcium phosphate coprecipitation method (293 cells) or Lipofectamine (NIH 3T3 cells). The reporter plasmids were the SVoβ CAT and ISG15 CAT reporter genes (56); the transfection procedures were also previously described (41, 56). For individual transfections, 100 μg (SVoβ CAT) or 10 μg (ISG15 CAT) of total protein extract was assayed for 1 to 2 h at 37°C. CAT activities were normalized with the β-galactosidase (β-Gal) assay. All transfections were performed three to six times.
Western blot analysis of IRF-3 modification and degradation.
To characterize the posttranslational regulation of the IRF-3 protein, stable or transiently transfected IRF-3-expressing cells were infected with Sendai virus (80 hemagglutinating units [HAU]/ml) or were treated with 5 ng of tumor necrosis factor alpha per ml, either with or without the addition of 50 μg of cycloheximide per ml. In some experiments, the cells were treated with either 100 μM calpain inhibitor I (ICN), 40 μM MG132 proteasome inhibitor, or an equivalent volume of their respective solvents (ethanol) as a control. The cells were washed with phosphate-buffered saline (PBS) and lysed in 10 mM Tris-Cl (pH 8.0)–200 mM NaCl–1 mM EDTA–1 mM dithiothreitol (DTT)–0.5% Nonidet P-40 (NP-40)–0.5 mM phenylmethylsulfonyl fluoride (PMSF)–5-μg/ml leupeptin, 5-μg/ml pepstatin–5-μg/ml aprotinin. Equivalent amounts of whole-cell extract (20 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% polyacrylamide gel. After electrophoresis, the proteins were transferred to a Hybond transfer membrane (Amersham) in a buffer containing 30 mM Tris, 200 mM glycine, and 20% methanol for 1 h. The membrane was blocked by incubation in PBS containing 5% dried milk for 1 h and then was probed with IRF-3 antibody in 5% milk–PBS at a dilution of 1:3,000. These incubations were done at 4°C overnight or at room temperature for 1 to 3 h. After four 10-min washes with PBS, the membranes were reacted with a peroxidase-conjugated secondary goat anti-rabbit antibody (Amersham) at a dilution of 1:2,500. The reaction was then visualized with the enhanced chemiluminescence detection system (ECL) as recommended by the manufacturer (Amersham Corp.).
Phosphatase treatment.
Twenty to sixty micrograms of whole-cell extract was treated with 0.3 U of potato acidic phosphatase (PPA; Sigma) in a final volume of 30 μl of piperazine-N,_N_′-bis(2-ethanesulfonic acid) (PIPES) buffer (10 mM PIPES [pH 6.0], 0.5 mM PMSF, 5-μg/ml aprotinin, 1-μg/ml leupeptin, 1-μg/ml pepstatin) or 5 U of calf intestine alkaline phosphatase (CIP; Pharmacia) in 30 μl of CIP buffer. The phosphatase inhibitor mix contained 10 mM NaF, 1.5 mM Na2MoO4, 1 mM β-glycerophosphate, 0.4 mM Na3VO4, and 0.1 μg of okadaic acid per ml.
Subcellular localization of GFP–IRF-3 proteins.
To analyze the subcellular localization of wild-type and mutated forms of IRF-3 proteins in uninfected and virus-infected cells, the GFP–IRF-3 expression plasmids (5 μg) were transiently transfected into COS-7 cells by the calcium phosphate coprecipitation method. For virus infection, transfected cells were infected with Sendai virus (80 HAU/ml for 2 h) at 24 h post transfection. GFP fluorescence in living cells was analyzed with a Leica fluorescence microscope with a ×40 objective.
EMSA.
Nuclear extracts were prepared from 293 cells at different times after infection with Sendai virus (80 HAU/ml). In some experiments, extracts were prepared from cells transfected with different IRF-3 expression plasmids, as indicated in individual experiments. The cells were washed in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF) and were resuspended in buffer A containing 0.1% NP-40. The cells were then chilled on ice for 10 min before centrifugation at 10,000 × g. The pellets were then resuspended in buffer B (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 5-μg/ml leupeptin, 5-μg/ml pepstatin, 0.5 mM spermidine, 0.15 mM spermine, 5-μg/ml aprotinin). Samples were incubated on ice for 15 min before being centrifuged at 10,000 × g. Nuclear extract supernatants were diluted with buffer C (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 50 mM KCl, 0.5 mM DTT, 0.5 mM PMSF). Nuclear extracts were subjected to electromobility shift assaying (EMSA) by using a 32P-labeled probe corresponding to the PRDIII region of the IFN-β promoter (5′-GGAAAACTGAAAGGG-3′) or the ISRE region of the ISG-15 promoter (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′). The resulting protein-DNA complexes were resolved by 5% PAGE and exposed to X-ray film. To demonstrate the specificity of protein-DNA complex formation, 125-fold molar excess of unlabeled oligonucleotide was added to the nuclear extract before adding labeled probe.
Immunoprecipitation and Western analysis of CBP-associated proteins.
Whole-cell extracts (300 μg) were prepared from either transfected or untransfected cells and precleared with 5 μl of preimmune rabbit serum and 20 μl of protein A-Sepharose beads (Pharmacia) for 1 h at 4°C. The extract was incubated with 10 μl of anti-CBP antibody A-22 (Santa Cruz) or 2 μl of anti-myc antibody 9E10 (21) and 30 μl of protein A-Sepharose beads for 2 to 3 h at 4°C. Precipitates were washed five times with lysis buffer, which was eluted by boiling the beads for 3 min in 1× SDS sample buffer. Eluted proteins were separated by SDS-PAGE and transferred to Hybond transfer membranes. The membranes were incubated with anti-IRF-3 (1:3,000) or anti-myc antibody 9E10 (1:1,000). Immunocomplexes were detected by using a chemiluminescence-based system.
RESULTS
Virus-induced phosphorylation of IRF-3 protein.
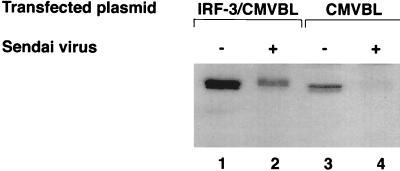
IRF-3 is expressed constitutively in various cells, and its expression is not enhanced by viral infection or by IFN treatment. To investigate whether the IRF-3 protein is regulated by posttranslational modification after virus infection, 293 cells were transiently transfected with an IRF-3 expression plasmid and subsequently infected with Sendai virus 24 h later. In cells transfected with the CMVBL vector alone, endogenous IRF-3 protein was easily detected with a polyclonal IRF-3 antibody, and in cells transfected with the IRF-3 expression plasmid, IRF-3 protein levels were significantly increased (Fig. 1, lanes 1 and 3). Interestingly, Sendai virus infection resulted in two alterations in the expression of IRF-3: an overall decrease in the amount of IRF-3 in transfected and control cells (Fig. 1, lanes 2 and 4), and the generation of a more slowly migrating form of IRF-3 (Fig. 1; compare lanes 1 and 2). In all experiments, the turnover of IRF-3 after virus infection was more pronounced with the endogenous protein than with the transfected proteins (Fig. 1, as well as other figures). Because the transfected proteins were driven by the CMV promoter, ongoing synthesis of transfected IRF-3 may partially obscure the turnover of IRF-3.
FIG. 1.
Sendai virus infection induces IRF-3 degradation. IRF-3 expression plasmid CMVBL-IRF3 (lanes 1 and 2) or CMVBL vector alone (lanes 3 and 4), both at 5 μg, was transiently transfected into 293 cells by the calcium phosphate method. At 24 h posttransfection, the cells were infected with Sendai virus for 16 h (lanes 2 and 4) or were left uninfected (lanes 1 and 3). Whole-cell extracts (20 μg) were prepared and analyzed by immunoblotting with anti-IRF-3 antibody.
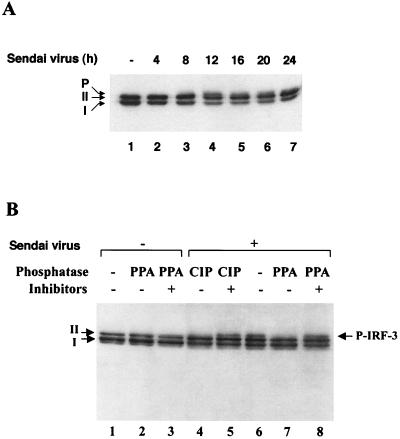
The kinetics of virus-induced modification of IRF-3 in a 293 cell line that expressed IRF-3 inducibly under the control of the tetracycline-responsive promoter CMVt were characterized (26, 27). Infection of this cell line (designated rtTA-IRF-3) with Sendai virus resulted in a decrease in the amount of IRF-3 between 12 and 24 h after infection (Fig. 2A). Two forms of IRF-3 protein (designated I and II) were detected in uninfected cells (Fig. 2A, lane 1), and, following virus infection, a third slowly migrating form of IRF-3 was also detected (Fig. 2A, lanes 4 to 7). To determine whether the slowest form of IRF-3 was due to virus-induced phosphorylation (P-IRF-3), the different forms of IRF-3 were subjected to treatment in vitro with PPA or CIP and/or phosphatase inhibitors (Fig. 2B). These treatments did not affect the mobilities of forms I and II in uninfected cells (Fig. 2B, lanes 1 to 3). However, in rtTA-IRF-3-expressing 293 cells infected with Sendai virus for 12 h, an additional slowly migrating, presumably phosphorylated form of IRF-3 was also detected (Fig. 2B, lane 6); this form of IRF-3 completely disappeared following CIP or PPA treatment (Fig. 2B, lanes 6 and 7) but was maintained in the presence of CIP or PPA when phosphatase inhibitors were also added to the reaction (Fig. 2B, lanes 5 and 8).
FIG. 2.
Sendai virus-induced phosphorylation and degradation of IRF-3 protein. (A) rtTA-IRF-3 cells, which were selected as described in Materials and Methods, were induced to express IRF-3 by doxycycline treatment for 24 h. At 24 h after doxycycline addition, the cells were infected with Sendai virus for 4, 8, 12, 16, 20, or 24 h (lanes 2 to 7) or were left uninfected (lane 1). IRF-3 protein was detected in whole-cell extracts (10 μg) by immunoblotting. Two forms of IRF-3, which were designated form I and form II, were detected. (B) At 24 h post-Dox induction, rtTA-IRF-3 cells were infected with Sendai virus for 16 h (lanes 4 to 8) or were left uninfected (lanes 1 to 3). Whole-cell extracts from untreated cells (20 μg) or Sendai virus-infected cells (60 μg) were incubated with 0.3 units of PPA (lanes 2, 3, 7, and 8) or 5 U of CIP (lanes 4 and 5) in the absence (lanes 1, 2, 4, 6, and 7) or presence (lanes 3, 5, and 8) of phosphatase inhibitors. Phosphorylated IRF-3 protein appears as a distinct band in immunoblots, migrating more slowly than IRF-3 forms I and II.
Mapping the IRF-3 phosphorylation sites.
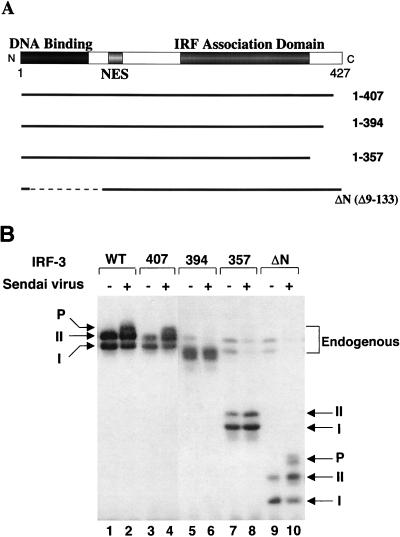
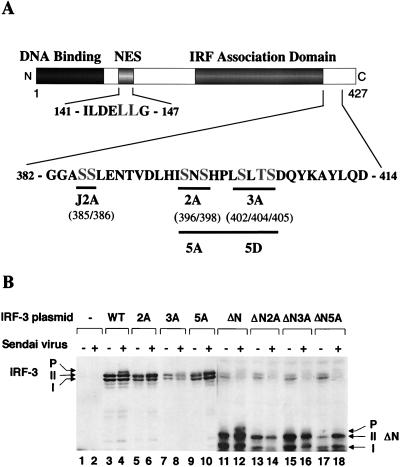
A series of deletions of IRF-3 were generated to identify the virus-induced phosphorylation site(s) of IRF-3 (Fig. 3A). 293 cells were transiently transfected with IRF-3 deletion mutants, and virus-mediated phosphorylation was measured by immunoblotting (Fig. 3B). The results indicated that virus-induced phosphorylation of IRF-3 occurs at the C-terminal end of IRF-3, since the mutations that contained only the N-terminal part of IRF-3 protein (133, 240, 328, 357, or 394 amino acids [aa]) were not phosphorylated (Fig. 3B and data not shown). Full-length and 407-aa forms of IRF-3 were phosphorylated as a consequence of virus infection (Fig. 3B, lanes 1 to 4). C-terminal truncation of IRF-3 to a protein of 394 or 357 aa removed the site(s) of inducible phosphorylation (Fig. 3B, lanes 5 to 8), although the shortened versions of forms I and II were still observed. Also, in the IRF-3 Δ9-133 mutation (ΔN) from which the DNA binding, N-terminal amino acids (aa 9 to 133) were removed, both virus-induced phosphorylation of IRF-3 and the differential migration of the shortened forms I and II were easily detected (Fig. 3B, lanes 9 and 10). Degradation of the endogenous forms of IRF-3 by virus infection was also detected in this experiment (Fig. 3B; compare lanes 7 and 9 with lanes 8 and 10). Thus, by deletion analysis, a phosphorylation domain of IRF-3 protein was localized to the region -ISNSHPLSLTSDQ- between aa 395 and 407. Point mutations in the several putative Ser and Thr phosphorylation residues within this region were generated in the full-length protein and the Δ9-133 (ΔN) protein (Fig. 4A). In the IRF-3 cDNA encoding these proteins, the Ser-396, Ser-398, Ser-402, Thr-404, and Ser-405 residues were replaced by alanine (5A), as were the three residues Ser-402, Thr-404, and Ser-405 (3A) and the two residues Ser-396 and Ser-398 (2A). Transfection of these plasmids into 293 cells and subsequent virus infection revealed that full-length, wild-type IRF-3 was phosphorylated (Fig. 4B, lanes 4 and 8), whereas the IRF-3 proteins containing the 2A and 5A mutations were no longer phosphorylated in virus-infected cells (Fig. 4B, lanes 6 and 10). Interestingly, IRF-3-3A was also very weakly phosphorylated as a consequence of virus infection, thus implicating Ser-402, Thr-404, and Ser-405 as potential secondary sites of phosphorylation. With the ΔN IRF-3 protein and the relevant point mutations, phosphorylation was detected with ΔN (Fig. 4B, lane 12) but not with ΔN-2A and ΔN-5A (Fig. 4B, lanes 14 and 18); likewise, ΔN-3A displayed very weak phosphorylation (Fig. 4B, lane 16). These experiments thus implicate Ser-396 and Ser-398 as critical sites of virus-induced phosphorylation of IRF-3; however, residues Ser-402, Thr-404, and Ser-405 also contribute to the observed phosphorylation, since the migration of phosphorylated ΔN-3A is significantly faster than that of ΔN and the phosphorylation level is decreased (Fig. 4B, lanes 12 and 16). Another study suggested the involvement of the Ser residues at aa 385 and 386 as potential phosphoacceptor sites (67); in studies with the S385A and S386A mutation, we found no evidence for inducible phosphorylation at these sites (40a). However, since these sites represent consensus sites for CKI and CKII, constitutive phosphorylation is a possibility.
FIG. 3.
Analysis of IRF-3 deletion mutants in Sendai virus-induced phosphorylation. (A) Schematic representation of four IRF-3 deletions. Thick solid lines and thin dashed lines indicate included and excluded sequences, respectively. The N-terminal IRF homology domain, the NES, and the C-terminal IRF association domain are indicated. (B) Expression plasmids (5 μg each) encoding wild type and deletion mutants of IRF-3 (as indicated above the lanes) were transiently transfected into 293 cells; at 24 h posttransfection, cells were infected with Sendai virus for 16 h (lanes 2, 4, 6, 8, and 10) or were left uninfected (lanes 1, 3, 5, 7, and 9). Whole-cell extracts (20 μg) were prepared from infected and control cells and were analyzed by immunoblotting for IRF-3 forms I and II and for the presence of phosphorylated IRF-3 (P-IRF-3) with anti-IRF-3 antibody.
FIG. 4.
Analysis of IRF-3 point mutations in Sendai virus-induced phosphorylation. (A) Schematic representation of IRF-3 point mutations. The N-terminal IRF homology domain, the NES element, and the C-terminal IRF association domain are indicated. aa 382 to 414 and 141 to 147 are shown. The amino acids targeted for alanine or aspartic acid substitution are shown in larger letters. The point mutations are indicated below the sequence: 2A, S396A and S398A; 3A, S402A, T404A, and S405A; 5A, S396A, S398A, S402A, T404A, and S405A); 5D, S396D, S398D, S402D, T404D, and S405D; J2A, S385A and S386A; NES, S145A and S146A). (B) Expression plasmids (5 μg each) encoding wild type (WT) and point mutants of IRF-3 (as indicated above the lanes) were transiently transfected into 293 cells; at 24 h posttransfection, the cells were infected with Sendai virus for 16 h (lanes 2, 4, 6, 8, 10, 12, 14, 16, and 18) or were left uninfected (lanes 1, 3, 5, 7, 9, 11, 13, 15, and 17). Whole-cell extracts (20 μg) were prepared from infected and control cells and analyzed by immunoblotting for IRF-3 forms I and II and for the presence of phosphorylated IRF-3 (P-IRF-3) with anti-IRF-3 antibody.
IRF-3 phosphorylation induces cytoplasmic to nuclear translocation of IRF-3.
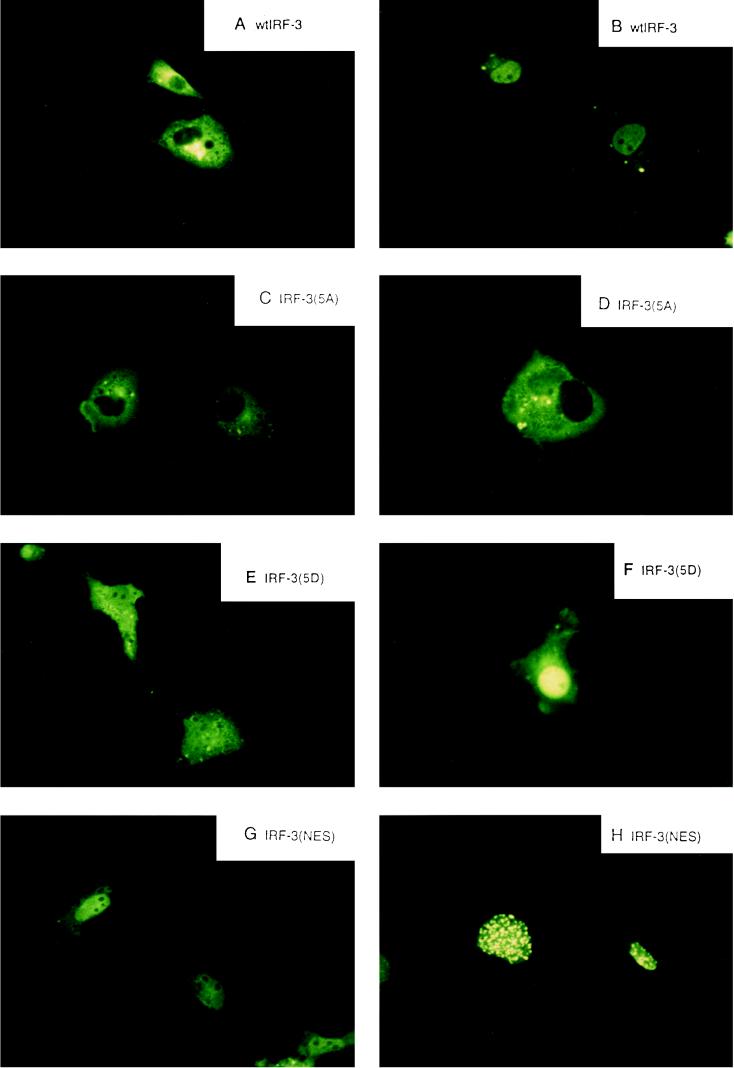
Initial studies indicated that IRF-3 was localized in the cytoplasm of uninfected cells (67); to investigate the role of phosphorylation on IRF-3 localization, wild-type and point-mutated forms of IRF-3 were linked to GFP, transfected into COS-7 cells, and examined for Sendai virus-induced changes in subcellular localization (Fig. 5). In uninfected cells, GFP–IRF-3 localized exclusively to the cytoplasm; Sendai virus infection resulted in translocation of IRF-3 to the nucleus within 8 h in 90 to 95% of the cells (Fig. 5A and B). Mutation of the Ser-Thr cluster in GFP–IRF-3(5A) completely abrogated virus-induced cytoplasm-to-nucleus translocation (Fig. 5C and D). Interestingly, the substitution of the Ser-Thr cluster with the phosphomimetic Asp in GFP–IRF-3(5D) likewise altered subcellular localization. IRF-3(5D) localized both to the nucleus and to the cytoplasm in uninfected cells (Fig. 5E), while virus infection resulted in an intense nuclear pattern of IRF-3(5D) fluorescence (Fig. 5F). Point mutation of a putative nuclear export signal in IRF-3, the L145A-L146A modification—termed IRF–3(nuclear export signal [NES])—also changed subcellular localization of IRF-3. In uninfected cells, GFP–IRF-3(NES) was localized to the nucleus and cytoplasm, with a homogeneous, extranucleolar pattern of nuclear staining. After virus infection, GFP–IRF-3(NES) localized to the nucleus with an intense speckled pattern of nuclear fluorescence in >95% of the cells, suggesting that IRF-3(NES) may be trapped in the nucleus associated with the nuclear pore complex.
FIG. 5.
Virus-dependent cytoplasm-to-nucleus translocation of IRF-3. The subcellular localization of the GFP–IRF-3 (A and B), GFP–IRF-3(5A) (C and D), GFP–IRF-3(5D) (E and F), and GFP–IRF-3(NES) (G and H) in uninfected (A, C, E, and G) and Sendai virus-infected COS-7 cells at 8 h after infection was analyzed. GFP fluorescence in living cells was analyzed with a Leica fluorescence microscope by using a ×40 objective.
Transactivation of PRDI-PRDIII and ISRE promoters by IRF-3.
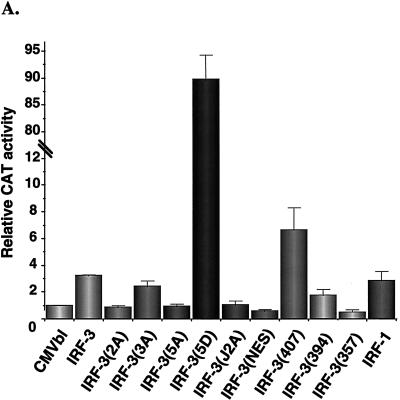
Next, the capacity of IRF-3 to regulate gene expression was analyzed by transient transfection in human 293 and murine NIH 3T3 cells by using the IFN-β and ISG-15 promoters in reporter gene assays. Expression of NF-κB RelA(p65), IRF-1, and IRF-3 alone minimally induced IFN-β promoter activity by between three- and fourfold (Fig. 6A and B), as shown previously (24, 56, 61). Introduction of the C-terminal point mutants—IRF-3(2A), IRF-3(3A), and IRF-3(5A)—reduced the low transactivation capacity of IRF-3 to control levels (Fig. 6A). Interestingly, deletion of the C-terminal 20 aa of IRF-3 to IRF-3(407) stimulated IFN-β activity by about 6-fold, which was indicative of the removal of an inhibitory domain in IRF-3. However, further deletion to 394, 357, or 240 abrogated transactivation potential (Fig. 6A). Mutation of the NES element was not sufficient to stimulate IFN-β activity. Strikingly, the substitution of the Ser-Thr cluster at aa 396 to 405 in IRF-3 with the phosphomimetic Asp generated a very strong, constitutive transactivator protein that alone stimulated the IFN-β promoter by 90-fold.
FIG. 6.
Transactivation of PRDI-PRDIII- and ISRE-containing promoters by IRF-3. 293 cells were transfected with IFN-β–CAT (A and B) or ISG15-CAT (C) reporter plasmids and the various expression plasmids as indicated below the bar graph. CAT activity was analyzed at 48 h posttransfection with 100 μg (IFN-β–CAT) or 10 μg (ISG15-CAT) of total protein extract for 1 to 2 h at 37°C. Relative CAT activity was measured as fold activation (relative to the basal level of reporter gene in the presence of CMVBI vector alone after normalization with cotransfected β-Gal activity); values are the averages of three experiments, with variability shown by the error bars. WT, wild type.
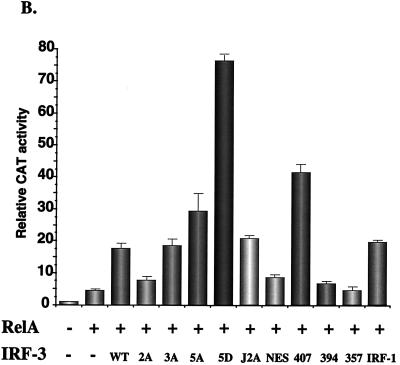
As shown previously, high-level induction of the IFN-β promoter requires synergistic activation by NF-κB and IRF proteins (24, 61). To analyze the properties of IRF-3 in synergistic activation of the IFN-β promoter, coexpression studies were performed with RelA(p65) expression plasmid and different wild-type and mutant forms of IRF-3 (Fig. 6B). Coexpression of RelA and IRF-1 or RelA and IRF-3 stimulated IFN-β–CAT activity by 20- to 25-fold. IRF-3(407) and RelA(p65) stimulated IFN-β activity by about 40-fold, supporting the idea of the removal of an inhibitory domain in IRF-3, whereas both IRF-3(394) and the IRF-3(NES) failed to synergize with RelA in the activation of the IFN-β promoter. RelA and IRF-3(NES) produced a relatively weak 8-fold induction of IFN-β expression, indicating that nuclear localization is not sufficient for IRF-3 activation. The combination of RelA and IRF-3(5D) produced an 80-fold stimulation of IFN-β promoter activity (Fig. 6B); together with the data above, IRF-3(5D) alone appears to be capable of full stimulation of the IFN-β promoter and further synergy with RelA is not observed (Fig. 6; compare panels A and B). Surprisingly, IRF-3(5A) and RelA produced a 30-fold stimulation, suggesting that 5A can still synergize with RelA, despite mutation of the Ser-Thr cluster.
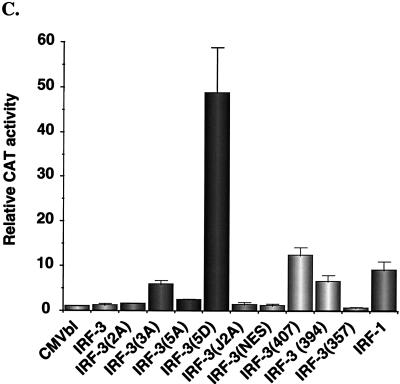
The transactivation potential of IRF-3 was also analyzed by using the ISG-15 promoter, an ISRE-containing regulatory element (Fig. 6C). As shown previously (2) and as described above for the IFN-β promoter, IRF-3 alone weakly activated the ISG-15 promoter; in the context of this regulatory element, IRF-3 was weaker than IRF-1, which produced a 9-fold stimulation. Again, deletion of the C-terminal 20 aa of IRF-3 generated a protein that stimulated gene expression; with the ISG-15 promoter, a 12-fold induction was observed; IRF-3(357) did not stimulate gene expression but rather slightly repressed ISG-15. Again remarkably, IRF-3(5D) produced a 50-fold induction of the ISG-15 promoter (Fig. 6C), thus demonstrating that substitution of the Ser-Thr sites with the phosphomimetic Asp generated a constitutively active form of IRF-3 that functioned as a very strong activator of promoters containing PRDI-PRDIII or ISRE regulatory elements.
Inhibition of IRF-3 degradation.
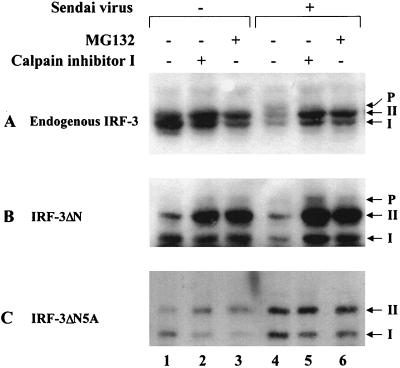
Another consequence of virus infection is the degradation of the IRF-3. Since phosphorylation of proteins is functionally associated with the process of protein degradation via the ubiquitin-dependent proteasome pathway (53, 57, 60), the effect of proteasome inhibitors on virus-induced turnover of IRF-3 was examined. In cells transfected with the ΔN and ΔN5A forms of IRF-3, virus-induced degradation of full-length (endogenous) forms of IRF-3 (Fig. 7A, lanes 1 and 4) and the truncated ΔN (Fig. 7B, lanes 1 and 4) was detected. Addition of the protease inhibitor calpain inhibitor I or the proteasome inhibitor MG132 blocked virus-induced IRF-3 degradation (Fig. 7A and 7B, lanes 4 to 6). Particularly with the ΔN protein, accumulation of the phosphorylated form of ΔN was also detected in virus-infected cells (Fig. 7B, lanes 5 and 6), suggesting that phosphorylation of IRF-3 may represent a signal for subsequent degradation by the proteasome pathway. To confirm this idea, the 5A point-mutated form of IRF-3 was analyzed; the IRF-3-ΔN5A protein was resistant to virus-induced degradation (Fig. 7C, lanes 1 and 4); no further stabilization of IRF-3-ΔN5A occurred with calpain inhibitor I or MG132 addition, and no phosphorylated IRF-3 was detected (Fig. 7C, lanes 4 to 6). These experiments demonstrate that virus-dependent phosphorylation of the C-terminal end of IRF-3 represents a signal for subsequent proteasome-mediated degradation.
FIG. 7.
Stabilization of IRF-3 by proteasome inhibitors. IRF-3 ΔN (Δ9-133) (B) or IRF-3 ΔN2A (C) expression plasmids were transiently transfected into 293 cells; at 24 h posttransfection, cells were infected with Sendai virus and treated for 12 h with calpain inhibitor I (100 μM [lanes 2 and 5]) or MG132 proteasome inhibitor (40 μM [lanes 3 and 6]). Ethanol, the solvent for calpain inhibitor I and MG132, was added to the cells as a control (lanes 1 and 4). Endogenous (A) and transfected (B and C) IRF-3 proteins were detected in whole-cell extracts (20 μg) by immunoblotting.
Interaction between IRF-3 and CBP in virus-infected cells.
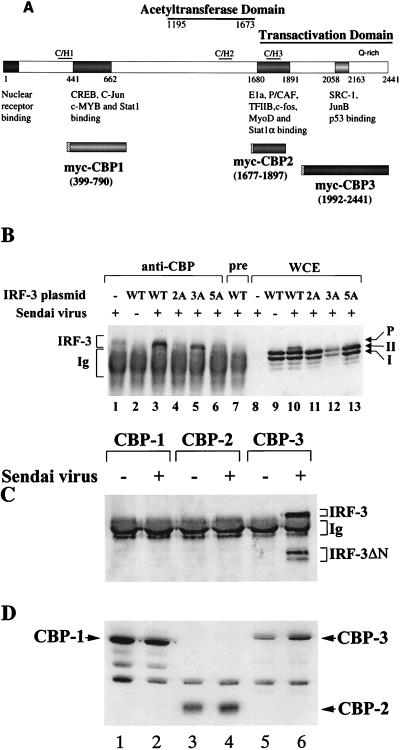
To examine the possibility that IRF-3 associated with the coactivator CBP/p300 (Fig. 8A) following Sendai virus infection, CBP was immunoprecipitated from virus-infected cells with anti-CBP antibody; an immunoblot for IRF-3 revealed that IRF-3 was coprecipitated from virus-infected cells but not from uninfected cells (Fig. 8B, lanes 2 and 3). This interaction was observed clearly in cells cotransfected with the IRF-3 expression plasmid (Fig. 8B, lane 3) but was not seen when the immunoprecipitation was performed with preimmune serum (Fig. 8B, lane 7). The endogenous IRF-3 also coprecipitated from virus-infected cells (Fig. 8B, lane 1). However, mutation of the Ser-Thr residues identified as the virus-inducible phosphorylation sites abrogated the association of IRF-3 with CBP. In particular, IRF-3(2A) and IRF-3(5A) were detected in whole-cell extract immunoblot but not in the CBP immunoprecipitate (Fig. 8B; compare lanes 4 and 6 with lanes 11 and 13). With the IRF-3(3A) mutant, interaction with CBP was still observed (Fig. 8B, lane 5). The high background in all lanes represents secondary antibody reactivity with rabbit immunoglobulin G from the immunoprecipitation. Immunoblot analysis of the whole-cell extracts revealed that phosphorylated IRF-3, as well as forms I and II, was present in virus-infected cells (Fig. 8B, lane 10), and in cells transfected with 2A, 3A, and 5A the forms I and II but not the phosphorylated form of IRF-3 were observed (Fig. 8B, lanes 11 to 13). CBP has several domains that bind transcription factors, which were designated CBP1, CBP2, and CBP3 (Fig. 8A [for a review, see reference 29). To determine which domain of CBP interacts with IRF-3, the three specific subdomains were myc tagged at the 5′ end by subcloning into the pCDNA3 vector (Fig. 8A). 293 cells were cotransfected with these myc-tagged CBP expression plasmids together with the IRF-3 ΔN (Δ9-133) expression plasmid. At 24 h after transfection, the cells were infected with Sendai virus, coimmunoprecipitated with anti-myc antibody 16 h later (21) and then immunoblotted for IRF-3. Endogenous IRF-3 and transfected IRF-3 ΔN proteins coprecipitated with CBP-3 from virus-infected cells but not from uninfected cells (Fig. 8C, lane 6). In cells cotransfected with CBP-1 and CBP-2, no endogenous or transfected ΔN IRF-3 was detected (Fig. 8C, lanes 1 to 4). Immunoblot analysis of the whole-cell extracts revealed that all three myc-tagged CBP proteins were efficiently expressed in uninfected and virus-infected cells (Fig. 8D). These results demonstrate that IRF-3 binds to the C-terminal domain of CBP in virus-infected cells and that interaction with CBP requires phosphorylation of IRF-3 at Ser-396 and Ser-398 but not at Ser-402, Thr-404, and Ser-405.
FIG. 8.
IRF-3 interacts with CBP in virus-infected cells. (A) Schematic representation of CBP, illustrating the domains involved in interaction with host or viral proteins (modified from reference 29) and the myc-tagged CBPs (CBP1, CBP2, and CBP3) used for immunoprecipitation. (B) 293 cells were transfected with wild type (WT) and point mutants of IRF-3 expression plasmid (5 μg, as indicated above the lanes) or were left untransfected (lanes 1 and 8). At 24 h after transfection, the cells were infected with Sendai virus for 16 h (lanes 1, 3 to 8, and 10 to 13) or were left uninfected (lanes 1 and 9). Whole-cell extracts (300 μg, except lane 1, which was 600 μg) were immunoprecipitated with anti-CBP antibody A22 (lanes 1 to 6) or with preimmune serum (lane 7). The immunoprecipitated complexes (lanes 1 to 7) or 30 μg of whole-cell extracts (lanes 8 to 13) was run on SDS–5% PAGE gels and subsequently probed with anti-IRF-3 antibody. (C) 293 cells were cotransfected with myc-tagged CBP expression plasmids (as indicated above the lanes) and the IRF-3 ΔN (Δ9-133) expression plasmid. At 24 h after transfection, the cells were infected with Sendai virus (lanes 2, 4, and 6) or were left uninfected (lanes 1, 3, and 5). Whole-cell extracts (300 μg) were immunoprecipitated with monoclonal anti-myc-tag antibody 9E10. The immunoprecipitated complexes were run on SDS–5% PAGE gels, and different forms of IRF-3 in the precipitates were analyzed by immunoblotting with anti-IRF-3 antibody. (D) Whole-cell extracts (30 μg) from panel C were also analyzed directly for the expression of myc-tagged CBP by immunoblotting using anti-myc antibody 9E10.
DISCUSSION
Several novel aspects of posttranslational regulation of the IRF-3 transcription factor are highlighted in the present study. (i) Sendai virus-dependent phosphorylation of IRF-3 occurring in a cluster of Ser and Thr sites in the carboxyl-terminal end of the protein was detected. The residues implicated in this regulatory phosphorylation event are Ser-396, Ser-398, Ser-402, Thr-404, and Ser-405, particularly the Ser-396 and Ser-398 residues. (ii) Phosphorylation of the IRF-3 in the Ser-Thr cluster resulted in cytoplasm-to-nucleus translocation of IRF-3; nuclear translocation was blocked by mutation of the phosphorylated amino acids. (iii) Sendai virus infection induced the DNA binding and transactivation potential of IRF-3. Furthermore, IRF-3 containing the phosphomimetic Asp at the sites of C-terminal phosphorylation was an exceptionally strong transactivator of PRDI-PRDIII- and ISRE-containing promoters. (iv) Phosphorylation was also required for the association of IRF-3 with the CBP coactivator protein. (v) Sendai virus infection resulted in IRF-3 degradation; again, phosphorylation was required as a signal for inducer-mediated degradation, since mutation of the Ser-Thr cluster also blocked virus induced degradation.
Cytoplasm-to-nucleus translocation of IRF-3 as a consequence of virus infection was inhibited by mutation of the Ser-Thr cluster, indicating an important regulatory role for C-terminal phosphorylation in the activation of IRF-3. Also strikingly, the conversion of the phosphorylation sites to the phosphomimetic Asp altered the subcellular localization of IRF-3 in uninfected cells. A proportion of IRF-3(5D) was localized to the nuclei of uninfected cells, suggesting that some IRF-3 may shuttle to and from the nucleus constitutively; this observation is consistent with the identification of a nuclear export signal in IRF-3. Mutation of L144A and L145A in the NES element produced the most impressive alterations in subcellular localization. In uninfected cells, IRF-3 was partitioned in both the nucleus and the cytoplasm; virus infection changed the nuclear pattern of staining from extranucleolar homogeneous staining as observed for wild-type IRF-3 to an intense nuclear speckling. At this stage, the nature of the subnuclear changes in IRF-3 localization are not explained, although it is possible that IRF-3(NES) translocates efficiently into the nucleus but becomes trapped in the nuclear pore complex during the export process.
One of the striking results of the mutagenesis of the C-terminal domain of IRF-3 was the generation of IRF-3(5D), an exceptionally strong activator of IFN-β and ISG-15 gene expression. The phosphomimetic form of IRF-3 alone was able to stimulate IFN-β expression as strongly as virus infection, a level of stimulation not previously been observed in coexpression experiments (24, 61). In previous experiments, we demonstrated that IRF-3 was able to bind the ISRE element of ISG-15, as well as the PRDIII-PRDI and IE regions of the IFN-β and IFN-α promoters, respectively (2, 56). Virus induction results in the appearance of two new protein-DNA complexes; supershift experiments confirmed that both complexes contain IRF-3 (32a); it is not clear at this stage whether the upper complex also contains other proteins, such as in the VIC (10, 25) and DRAF (16) complexes, or whether the lower complex represents a breakdown product of IRF-3. Strikingly, the same complexes appeared following cotransfection of IRF-3(5D) expression plasmid in the absence of virus induction, indicating that IRF-3(5D) represented a constitutive DNA binding form of IRF-3. Thus, in uninfected cells, IRF-3(5D) localized in part to the nucleus (Fig. 5), interacted with DNA constitutively, and was a strong activator of gene expression (Fig. 6).
The recent crystal structure of the related IRF-1 protein bound to PRDI provides evidence for a novel helix-turn-helix motif that latches onto a GAAA core sequence via three of the five conserved tryptophan amino acids of the DNA binding domain (20). By analogy with IRF-3, two GAAANN sequences present in PRDIII of IFN-β and another GAAANN element present in the PRDI may serve as DNA contacts for multiple IRF-3(5D) proteins with strong activating potential. Similarly, the ISRE element of the ISG-15 promoter also contains several GAAANN anchors for potential IRF binding. Given the range of promoters that possess this hexameric sequence (48), it will be of interest to determine the capacity of IRF-3(5D) to stimulate expression of different cytokine and chemokine genes.
IRF-3 joins a growing list of cellular and viral proteins that functionally interact with CBP/p300 proteins, highly homologous proteins originally identified through their interactions with adenovirus E1A and CREB proteins (1, 13). As a critical determinant of its global transcriptional coactivator activity, CBP/p300 possesses histone acetyltransferase activity (5, 50). Acetylation of histones is involved in the destabilization and remodeling of nucleosomes, which is a crucial step in permitting the accessibility of transcriptional factors to DNA templates. Several studies have now demonstrated that CBP/p300 participates in the transcriptional process by providing a scaffold for different classes of transcriptional regulators on specific chromatin domains (12, 50). A growing body of biochemical and genetic evidence also implicates CBP/p300 as a negative regulator of cell growth, based on its interactions with adenovirus E1a, simian virus 40 large T antigen, and the tumor suppressor p53 among others. With regard to p53-CBP/p300 complex formation, functional interaction between these two important growth regulatory proteins accounts for several of the known activities of p53 (3, 29, 40); interestingly, CBP/p300 was shown recently to acetylate p53 and to stimulate its transactivation potential (28). It will be of interest to determine whether IRF-3 is similarly modified by CBP association. The functional consequences of IRF-3 interaction with CBP/p300 remain to be elucidated, although recent studies demonstrated that CBP/p300 also functionally interacts with STAT1 (68) and STAT2 (7) and may contribute to IFN-α and IFN-γ nuclear signalling. Elegant studies published during review of the manuscript demonstrated that synergistic activation of the IFN-β promoter requires recruitment of CBP/p300 to the enhanceosome, via a new activating surface assembled from the activation domains of all of the transcription factors in the enhanceosome (37, 45). Alterations in any of the activation domains decreased both CBP recruitment and transcriptional synergy. By analogy, recruitment of CBP/p300 to DNA-bound IRF-3 is likely required for maximal transcriptional activation. Association requires the interaction of the C-terminal domain of IRF-3 and the C-terminal interaction domain of CBP, a region previously shown to associate with the p53 tumor suppressor, whereas STAT1 and STAT2 associate with different regions of CBP (7, 68).
Virus-induced phosphorylation of IRF-3 also represents a signal for proteasome-mediated degradation of IRF-3, since mutation of Ser-396 and Ser-398 or the use of proteasome inhibitors prevented postinfection degradation of IRF-3. Virus-induced degradation of IRF-3 is reminiscent of the virus-induced turnover of another member of the IRF family, IRF-2. In response to double-stranded RNA (dsRNA) or viral induction, the 50-kDa IRF-2 protein is proteolytically processed into a smaller, 24- to 27-kDa protein (51) comprising the 160-aa DNA-binding domain (DBD) of IRF-2, which was termed TH3 (14) or In4 (65). Although TH3 has been shown to bind DNA and to repress transcription more efficiently than the full-length IRF-2 protein (42), its physiological role is not clear. Since the induction kinetics of TH3 are slower than that of IFN-β in response to dsRNA or viral infection (14), it has been suggested that the IRF-2 cleavage product is a postinduction repressor of IFN-β gene expression (65).
Virus-induced phosphorylation of IRF-3 at the C-terminal Ser and Thr residues and its subsequent degradation by a proteasome-dependent pathway are also similar to the well-studied phosphorylation and degradation of IκBα, which leads to activation of NF-κB binding activity (reviewed in references 4 and 6). In unstimulated cells, NF-κB heterodimers are retained in the cytoplasm by inhibitory IκB proteins. Upon stimulation by many activating agents, including cytokines, viruses, and dsRNA, IκBα is rapidly phosphorylated and degraded, resulting in the release and nuclear translocation of NF-κB. The amino terminus of IκBα represents a signal response domain for activation of NF-κB, and substitution of alanine for either Ser-32 or Ser-36 completely abolished the signal-induced phosphorylation and degradation of IκBα and blocked activation of NF-κB. These mutations also blocked in vitro ubiquitination of the IκBα protein. The amino terminus of IκBα is necessary for signal-induced phosphorylation and ubiquitination, but for degradation to occur, there is an absolute requirement for the C-terminal PEST domain (reviewed in references 4 and 6).
Similarities and differences between the observed degradation of IRF-3 and the mechanism of IκBα degradation exist. The C-terminal phosphorylation of IRF-3 as a consequence of virus infection is required for its subsequent degradation based on the deletion and point mutation analysis of the region -ISNSHPLSLTSDQ- between aa 395 and 407. Minimally, phosphorylation of Ser-396 and Ser-398 are required for subsequent degradation, although Ser-402, Ser-404, and Ser-405 may represent secondary phosphorylation sites. Likewise, in the case of IκBα, phosphorylation of Ser-32 and Ser-36 are required for inducer-mediated degradation. Furthermore, the protease inhibitor calpain inhibitor I and the more specific proteasome inhibitor MG132 block IRF-3 turnover. A major difference in the mechanisms of IκBα and IRF-3 turnover lies in the nature of the inducing stimuli. Multiple inducers—cytokines such as tumor necrosis factor and interleukin 1, viruses, lipopolysaccharide oxidative stress, etc. (6)—all lead to the induction of IκBα phosphorylation and degradation whereas IRF-3 phosphorylation appears to be induced only by virus infection and dsRNA addition; other inducers have not resulted in IRF-3 turnover (40a). A significant temporal difference also exists between IκBα phosphorylation-turnover and IRF-3 phosphorylation-degradation. Many activators of NF-κB stimulate IκBα phosphorylation within minutes, and TNF-induced degradation occurs within the first 15 to 30 min after treatment. In the case of IRF-3, phosphorylation is not detected until 6 to 8 h after infection and continues in a heterogeneous manner over the following 10 to 12 h. Previous experiments, however, have demonstrated that Sendai virus-induced turnover of IκBα also occurs slowly over several hours (24).
Based on the data presented in this study and by analogy with the properties of other IRF family members (48), we propose the following model to explain several observations. IRF-3 exists in a latent state in the cytoplasm of uninfected cells; the C terminus may physically interact with the DNA binding domain in such a way as to obscure both the DBD and the IAD regions of the protein; the presence of an autoinhibitory domain within the C-terminal 20 aa (407 to 427) would explain the activating effect of this deletion, as seen previously with IRF-4 (11, 19). Virus-induced phosphorylation at the Ser-Thr cluster at aa 396 to 405 leads to a conformational change in IRF-3, exposing both the DBD and IAD and relieving C-terminal autoinhibition. Translocation to the nucleus, occurring via an unidentified nuclear localization sequence or in conjunction with a transcriptional partner associating through the IAD region, leads to DNA binding at ISRE- and PRDI-PRDIII-containing promoters. Phosphorylation is also necessary for IRF-3 association with the chromatin-remodeling activity of CBP/p300. The presence of a NES element ultimately shuttles IRF-3 from the nucleus and terminates the initial activation of IFN-responsive promoters. The phosphorylated form of IRF-3 exported from the nucleus may then be susceptible to proteasome-mediated degradation. This scenario shares several features with the protein synthesis-independent activation of NF-κB and further suggests that IRF-3 may represent a component of virus- or dsRNA-inducible complexes such as DRAF (16) or VIC (10, 25) that could play a primary role in the induction of IFN or IFN-responsive genes.
ACKNOWLEDGMENTS
We thank Dimitris Thanos, Stephane Richard, and Illka Julkunen for reagents used in this study. We also thank members of the Molecular Oncology Group, Lady Davis Institute, for helpful discussions.
This research was supported by grants from the Medical Research Council of Canada (J.H. and R.L.), the National Cancer Institute (J.H.), and the National Institutes of Health (P.M.P.). R.L. was supported in part by a Fraser Monat McPherson Fellowship from McGill University, C.H. was supported by a FRSQ/FCAR studentship, and J.H. was supported by an MRC Scientist award.
REFERENCES
- 1.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 2.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Beauparlant P, Hiscott J. Biological and biochemical inhibitors of the NF-κB/Rel proteins and cytokine synthesis. Cytokine Growth Factor Rev. 1996;7:175–190. doi: 10.1016/1359-6101(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 8.Bluyssen H A R, Durbin J E, Levy D E. ISGF3γ p48, a specificity switch for interferon activated transcription factors. Cytokine Growth Factor Rev. 1996;7:11–17. doi: 10.1016/1359-6101(96)00005-6. [DOI] [PubMed] [Google Scholar]
- 9.Bovolenta C, Driggers P H, Marks M S, Medin J A, Politis A D, Vogel S N, Levy D E, Sakaguchi K, Appella E, Coligan J E, Ozato K. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci USA. 1994;91:5046–5050. doi: 10.1073/pnas.91.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bragança J, Génin P, Bandu M-T, Darracq N, Vignal M, Cassé C, Doly J, Civas A. Synergism between multiple virus-induced-factor-binding elements involved in the differential expression of IFN-A genes. J Biol Chem. 1997;272:22154–22162. doi: 10.1074/jbc.272.35.22154. [DOI] [PubMed] [Google Scholar]
- 11.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferases and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 14.Cohen L, Hiscott J. Characterization of TH3, an induction specific protein interacting with the interferon-β promoter. Virology. 1992;191:589–599. doi: 10.1016/0042-6822(92)90234-g. [DOI] [PubMed] [Google Scholar]
- 15.Crepieux P, Coll J, Stehelin D. The Ets family of proteins: weak modulators of gene expression in quest for transcriptional partners. Crit Rev Oncog. 1994;5:615–638. [PubMed] [Google Scholar]
- 16.Daly C, Reich N C. Double-stranded RNA activates novel factors that bind to the interferon stimulated response element. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 18.Driggers P H, Ennist D L, Gleason S L, Mak W-H, Marks M S, Levi B-Z, Flanagan J R, Appella E, Ozato K. An interferon γ-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 20.Escalante C R, Yie J, Thanos D, Aggarwal A K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 21.Evan G I, Lewis G K, Bishop J M. Isolation of monoclonal antibodies specific for products of avian oncogene myb. Mol Cell Biol. 1984;4:2843–2850. doi: 10.1128/mcb.4.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita T, Kimura Y, Miyamoto M, Barsoumian E L, Taniguchi T. Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 23.Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-β gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, Roulston A, Lacoste J, Hiscott J. Viral induction of the human interferon beta promoter: modulation of transcription by NF-κB/rel proteins and interferon regulatory factors. J Virol. 1994;68:4707–4715. doi: 10.1128/jvi.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Génin P, Bragança J, Darracq N, Doly J, Civas A. A novel PRDI and TG binding activity involved in virus-induced transcription of IFN-A genes. Nucleic Acids Res. 1995;23:5055–5063. doi: 10.1093/nar/23.24.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 29.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 30.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 31.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, Tan R S-P, Takasugi T, Kadokawa Y, Schindler C, Schreiber R D, Noguchi S, Taniguchi T. Regulation of IFN-α/β genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-α/β. Genes to Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 32.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:903–913. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 32a.Heylbroeck, C. Unpublished data.
- 33.Hiscott J, Nguyen H, Lin R. Molecular mechanisms of interferon beta gene induction. Semin Virol. 1995;6:161–173. [Google Scholar]
- 34.Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch K-P, Gabriele L, Waring J F, Bachmann M F, Zingernagel R M, Morse III H C, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 35.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 36.Kawakami T, Matsumoto M, Sato M, Harada H, Taniguchi T, Kitigawa M. Possible involvement of the transcription factor ISGF3γ in virus-induced expression of the IFN-β gene. FEBS Lett. 1995;358:225–229. doi: 10.1016/0014-5793(94)01426-2. [DOI] [PubMed] [Google Scholar]
- 37.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon β enhanceosome. Mol Cell. 1998;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 38.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan R S-P, Takasugi T, Matsuyama T, Mak T M, Noguchi S, Taniguchi T. Essential and non-redundant roles of p48 (ISGF3γ) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 39.Levy D E. Interferon induction of gene expression through the Jak-Stat pathway. Semin Virol. 1995;6:181–190. [Google Scholar]
- 40.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 40a.Lin, R. Unpublished data.
- 41.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin R, Mustafa A, Nguyen H, Hiscott J. Mutational analysis of interferon (IFN) regulatory factors 1 and 2: effects on the induction of IFN-β gene expression. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- 43.Matsuyama T, Grossman A, Mittrücker H-W, Siderovski D P, Kiefer F, Kawakami T, Richardson C D, Taniguchi T, Yoshinaga S K, Mak T W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuyama T, Kimura T, Kitagawa M, Watanabe N, Kundig T, Amakawa R, Kishihara K, Wakeham A, Potter J, Furlonger C, Narendran A, Suzuki H, Ohashi P, Paige C, Taniguchi T, Mak T. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 45.Merika M, Williams A, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 46.Mittrücker H-W, Matsuyama T, Grossman A, Kündig T M, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi P S, Mak T W. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 47.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to the IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen, H., J. Hiscott, and P. M. Pitha. The growing family of IRF transcription factors. Cytokine Growth Factor Rev., in press. [DOI] [PubMed]
- 49.Nguyen H, Lin R, Hiscott J. Activation of multiple growth regulatory genes following inducible expression of IRF-1 or IRF/RelA fusion proteins. Oncogene. 1997;15:1425–1435. doi: 10.1038/sj.onc.1201318. [DOI] [PubMed] [Google Scholar]
- 50.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 51.Palombella V J, Maniatis T. Inducible processing of interferon regulatory factor-2. Mol Cell Biol. 1992;12:3325–3336. doi: 10.1128/mcb.12.8.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pitha P M, Au W-C. Induction of interferon alpha gene expression. Semin Virol. 1995;6:151–159. [Google Scholar]
- 53.Read M A, Neish A S, Luscinskas F W, Palombella V J, Maniatis T, Collins T. The proteasome pathway is required for cytokine-induced endothelial-leukocyte adhesion molecule expression. Immunity. 1995;2:493–506. doi: 10.1016/1074-7613(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 54.Reis L F L, Harada H, Wolchok J D, Taniguchi T, Vilcek J. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-β and IFN-inducible genes. EMBO J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P. Nucleotide sequence of the kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schafer S, Lin R, Moore P, Hiscott J, Pitha P M. Regulation of type 1 interferon gene expression by interferon regulatory factor 3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 57.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 59.Sharf R, Meraro D, Azriel A, Thornton A M, Ozato K, Petricoin E F, Larner A C, Schaper F, Hauser H, Levi B-Z. Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and to bind DNA. J Biol Chem. 1997;272:9785–9792. doi: 10.1074/jbc.272.15.9785. [DOI] [PubMed] [Google Scholar]
- 60.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 61.Thanos D, Maniatis T. Identification of the rel family members required for virus induction of the human beta interferon gene. Mol Cell Biol. 1995;15:152–164. doi: 10.1128/mcb.15.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 64.Weisz A, Marx P, Sharf R, Appella E, Driggers P H, Ozato K, Levi B-Z. Human interferon consensus sequence binding protein is a negative regulator of enhancer elements common to interferon-inducible genes. J Biol Chem. 1992;267:25589–25596. [PubMed] [Google Scholar]
- 65.Whiteside S T, King P, Goodbourn S. A truncated form of the IRF-2 transcription factor has the properties of a postinduction repressor of interferon-β gene expression. J Biol Chem. 1994;269:27059–27065. [PubMed] [Google Scholar]
- 66.Yamagata T, Nishida J, Tanaka T, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/mcb.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct activation of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E. Two contact regions between STAT1 and CBP/p300 in interferon gamma signalling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein Barr Virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]