Regulation of Vibrio cholerae Genes Required for Acid Tolerance by a Member of the “ToxR-Like” Family of Transcriptional Regulators (original) (raw)
Abstract
The ability of the intestinal pathogen Vibrio cholerae to undergo an adaptive stress response, known as the acid tolerance response (ATR), was previously shown to enhance virulence. An essential component of the ATR is CadA-mediated lysine decarboxylation. CadA is encoded by the acid- and infection-induced gene cadA. Herein, cadA is shown to be the second gene in an operon with cadB, encoding a lysine/cadaverine antiporter. cadC, which is 5′ of cadB, encodes an acid-responsive, positive transcriptional regulator of cadBA. Unlike in Escherichia coli, V. cholerae cadB and cadA are also transcribed monocistronically. Of note, bicistronic cadBA is transcribed at low constitutive levels in an acid- and CadC-independent manner. CadC represents a new member of the “ToxR-like” family of transcriptional regulators in V. cholerae and, in addition, exhibits extensive amino acid and functional similarity to E. coli CadC. The amino-terminal, putative DNA binding domains of ToxR and CadC are highly conserved, as are the putative promoter elements recognized by these transcription factors.
Vibrio cholerae is the causative agent of the endemic and epidemic diarrheal disease cholera. Since the natural environmental reservoir for this intestinal pathogen is aquatic, it stands to reason that ingestion by a human and subsequent colonization of the relatively sterile small intestine involve the expression of genes that are crucial for survival and adaptation in this new environment. Several research strategies designed to identify pathogen genes that are upregulated during infection have been used to show that adaptation/stress systems are often induced upon entry of a pathogen into its host environment (5, 13, 21, 40, 42). Although much emphasis has been placed on the identification and characterization of V. cholerae virulence factors, such as those within the ToxR/ToxT regulon (37), very little is known about the survival and adaptation systems employed by this gram-negative bacterium during infection.
We recently used recombinase-based in vivo expression technology to identify V. cholerae genes that are transcriptionally induced within two separate animal models of cholera. One such gene was cadA, which encodes a lysine decarboxylase; cadA was subsequently shown to be essential for V. cholerae's ability to undergo an adaptive stress response known as the acid tolerance response (ATR) (25). This stress response has been well characterized in the two closely related enteric pathogens, Escherichia coli and Salmonella enterica serovar Typhimurium, and has been shown to be necessary for pathogenicity of the latter (4, 23, 24, 29). We have found that V. cholerae cells that are acid adapted are more virulent than cells grown at a neutral pH. This finding suggests that the V. cholerae ATR may play an important role in the fitness of this pathogen, with respect to both infectivity of a single host and rapid epidemic spread within populations (25).
Here we extend our characterization of the V. cholerae cadA locus and show that, as in E. coli, it is the downstream gene in and acid-inducible operon, cadBA. However, in contrast to what is observed in E. coli, the V. cholerae cadA also possesses an independent promoter. In addition, we have identified a positive transcriptional regulator of the cadBA operon, CadC, which shows significant homology to both ToxR and the E. coli CadC protein. Mutational analyses revealed that CadC is essential for acid induction of cadBA but is not needed for basal-level transcription of cadBA. The basal-level expression of cadBA was insufficient for a functional ATR, as cadC mutants were unable to mount either an inorganic or organic ATR.
MATERIALS AND METHODS
Sequence analysis.
DNA sequences were analyzed for open reading frames (ORFs) by using DNA Strider version 1.2 (C. Marck). E. coli CadB, CadC, and Lcd amino acid sequences were used to search for putative homologs in the V. cholerae genome sequence (The Center for Genomic Research [TIGR]) using the BLAST algorithm (1). Multiple alignments of conserved protein domains and hydropathy profile predictions were performed using MacVector 6.0.1 (Oxford Molecular). cadC, -B, and -A are located on the V. cholerae large chromosome (replicon 1) beginning at positions 306411, 308670, and 310117, respectively (TIGR database).
Strain and plasmid construction.
All strains, plasmids, and primers used in this study are listed in Tables 1 and 2. Mutagenesis of cadB and cadC was by insertional inactivation of a suicide plasmid, pGP704. The mutagenic plasmids were constructed as follows. A 253-bp internal fragment of cadB was amplified with Pfu polymerase (Stratagene) by PCR using primers BF1 and BR1. _Sfi_I adapters were ligated onto the PCR product, and the resulting DNA fragment was ligated into the _Sfi_I-digested pAC212 backbone as previously described (25) to generate pDSM373. A 288-bp internal fragment of cadC was amplified with Taq polymerase using primers CadCF1 and CadCR1 and was subsequently cloned into pCR-Script Amp SK(+) (Stratagene) according to the manufacturer's directions, generating pDSM582. The cadC fragment was liberated from pDSM582 by _Sac_I/_Sal_I double digestion and cloned into similarly digested pGP704 (26) to generate pDSM588. A recombinant PCR method, gene splicing by overlap extension (15) using primers BAΔF1, BAΔR1, BAΔF2, and BAΔR2, was used to generate strain DSM-V783, which contains a deletion of the entire cadBA coding sequence.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and phenotype | Source or reference(s) |
|---|---|---|
| E. coli | ||
| SM10λ_pir_ | thi recA thr leu tonA lacY supE | Laboratory strain |
| RP4-2-Tc::Mu λ::pir | ||
| DH5αλ_pir_ | F− Δ(lacZYA-argF)U169 recA1 | 11, 18 |
| endA1 hsdR17 supE44 | ||
| thi-1 gyrA96 relA1 λ::pir | ||
| V. cholerae | ||
| C6709-1 | El Tor biotype, Smr | 33 |
| AC-V66 | C6709-1 lacZ::res1-tet-res1 Tcr LacZ− | 5 |
| AC-V168 | C6709-1 lacZ::pGP704, LacZ− | 5 |
| DSM-V376 | AC-V66 cadA::pGP704 | 25 |
| DSM-V387 | C6709-1 cadB::pGP704 | This work |
| DSM-V591 | C6709-1 cadC::pGP704 | This work |
| DSM-V783 | C6709-1 Δ_cadBA_ | This work |
| Plasmids | ||
| pAC212 | pGP704::_Sfi_I-_neo-Sfi_I | 25 |
| pDSM-373 | pGP704::_cadB_′ | This work |
| pDSM582 | pCR-script Amp SK(+)::′_cadC_′ | This work |
| pDSM588 | pGP704::′_cadC_′ | This work |
| pDSM667 | pGemT::cadBA | This work |
| pDSM673 | pGemT::_cadC_′ | This work |
TABLE 2.
Sequences of primers used in this study
| Primer | DNA sequence (5′-3′) |
|---|---|
| AF1 | AAGATCGTCGGTATGTTC |
| AR1 | GTCGAAGTGGATGTGTTT |
| BAΔF1 | CCTATTCCCCAAGTCAAA |
| BAΔF2 | GATGTTATGTAAGCCAGCTTTACCCCATA |
| BAΔR1 | GCTGGCTTACATAACATCTCCAAATCG |
| BAΔR2 | CACCCAAGCCAGCACCAA |
| BF1 | ACTGCACTAGCGTGTTTC |
| BR1 | GTCAGCAGAACCGCATCG |
| Cad8F1 | CTCATTGGCATTCTGTGC |
| Cad8R1 | GACGCTTTTCACCTAAAT |
| CadAOR | GTAACCGGCTTTTTCTAA |
| CadAP2 | GCAGTTGGCGAACAGGCTCTTCTTTAAAGA |
| CadAPE | ACACACCCATGTGGTTCAAGATAGCGAAAA |
| CadBOF | GGACTGCGCCATTCGTGT |
| CadBPE | CCACTGCCCATCATGTTGCCAGCCACCACA |
| CadCCF | TGAAGAGCTGACCCGTAT |
| CadCF1 | TCGCCAAGATAGGGAGGTC |
| CadCR1 | CTTGGCGAGAAACATAGGG |
| CadCPE | CTATCTTGGCGATACAGCTTATTCTCATCA |
All plasmids used for construction of insertion mutations were mobilized into V. cholerae from E. coli SM10λ_pir_ as previously described (25), and all integration mutations were subsequently verified by Southern blot analysis (data not shown). The loss of the ability to decarboxylate lysine was shown by utilization of lysine decarboxylation indicator broth (Difco) as previously described (25).
Growth conditions.
All strains were maintained at −80°C in Luria-Bertani (LB) broth containing 30% glycerol. All strains were grown at 37°C in LB broth. The pH of the medium was adjusted with HCl. Ampicillin and streptomycin were used at concentrations of 100 μg ml−1. RNA was harvested from strains grown in the following manner. Overnight cultures of each test strain were grown in LB broth containing ampicillin and then diluted 1:150 into 30 ml of fresh medium plus ampicillin. The diluted cultures were grown with aeration until they reached an optical density (600 nm) of approximately 0.16 to 0.20. At this point, cells were pelleted at 5,000 × g for 5 min at 24°C, and the supernatants were removed by aspiration. Cells were resuspended in 1 ml of LB broth (pH 7.0), and then 10 and 90% of the cells were placed into two microcentrifuge tubes. The cells were pelleted at 12,000 × g for 1 min at 24°C, and the supernatants were removed by aspiration. The 10% cell pellet was resuspended in 1 ml of LB broth (pH 7.0), and the 90% cell pellet was resuspended in 1 ml of LB broth (pH 5.7); then both were transferred to culture tubes and grown at 37°C with aeration for 1 h. After 1 h, all of the pH 7.0-grown cells and half of the pH 5.7-grown cells were pelleted and then flash frozen in a −80°C isopropanol bath. The remainder of each pH 5.7 culture was resuspended in LB (pH 4.5) and incubated at 37°C for 15 min. These cells were then pelleted and flash frozen as above. Strains which were exposed to organic acids were grown exactly as above except that pH 5.7 LB and pH 4.5 LB were supplemented with 0.075× and 0.1× organic acid cocktail, respectively (1× cocktail was 87 mM acetic acid, 25 mM butyric acid, and 37 mM propionic acid). Cell pellets were then used to collect total RNA.
RPAs and primer extension.
RNase protection assays (RPAs) were conducted on total RNA isolated from AC-V168 and DSM-V591 as previously described (25). A 318-bp probe 1 and 732-bp probe 2 fragment were generated by PCR using Taq polymerase and primer pairs AF1-AR1 and CadBOF-CadAOR, respectively. The amplification products were ligated to pGemT (Promega), proper orientation was confirmed, and riboprobes were synthesized using a Maxiscript kit (Ambion) and 50 μCi of [32P]UTP (NEN) as previously described (25). In each case, riboprobes transcribed from the pGemT SP6 or T7 promoters contain pGemT-specific sequence that results in riboprobes that are slightly larger than the original probe 1 or probe 2 _V. cholerae_-specific PCR fragments. Therefore, hybridization of the probe with its target mRNA followed by digestion with RNase results in a smaller protected fragment as the nonhybridizing pGemT-specific transcript is cleaved. RPAs were conducted with an RPAII kit using 1 to 3 μg of RNA as described by the manufacturer (Ambion). The products of RNase protection were separated on 5% denaturing polyacrylamide gels and exposed to phosphor screens (Kodak). Quantification and peak analysis of bands was conducted using a PhosphorImager and the ImageQuant program (Molecular Dynamics).
Primer extension to map the 5′ ends of cadA, cadB, and cadC mRNA transcripts produced after acid challenge was conducted using primers CadAPE (and CadAP2), CadBPE, and CadCPE, respectively. Approximately 10 pmol of each primer was end labeled and used for primer extensions with a primer extension system-avian myeloblastosis virus reverse transcriptase kit as described by the manufacturer (Promega). pDSM567, which contains the entire cadBA promoter region and coding sequences, and pDSM673, which contains the cadC promoter region, were generated by PCR using primer pairs Cad8F1-Cad8R1 and CadCCF-CadCRI, respectively. Amplification products were ligated into pGemT, and the plasmids were used as templates for sequencing reactions using the appropriate end-labeled primer. Sequencing reactions were conducted by denaturation of template DNA as described elsewhere (16), followed by utilization of the denatured template in manual sequencing reactions using a Sequenase DNA sequencing kit (version 2.0) as described by the manufacturer (Amersham).
Products of primer extensions and sequencing reactions were resolved in 7% denaturing polyacrylamide gels, dried, and exposed to phosphor screens. Products were analyzed using a PhosphorImager.
ATR and competition assays.
ATR and competition assays were conducted as previously described (25). The output ratio from each in vivo and in vitro competition was corrected for any deviations in the inoculum ratio from a value of 1:1. The competitive index was calculated by dividing the in vivo output ratio by the in vitro output ratio.
RESULTS
Identification of CadB and CadC homologs.
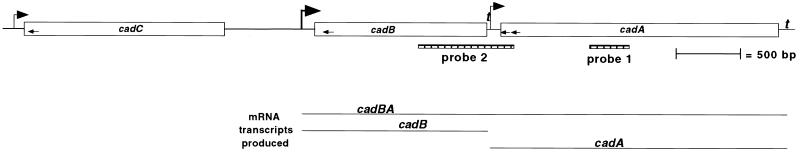
In an earlier study, cadA was identified as an infection-induced gene of V. cholerae and shown to encode an inducible lysine decarboxylase (25). Searches against the V. cholerae genome sequence revealed the presence of V. cholerae DNA sequences 5′ of cadA on the large chromosome coding for putative polypeptides with 67 and 27% identity to E. coli CadB and CadC, respectively. Due to the high degree of identity of the putative CadB and the similar location of its gene immediately upstream of cadA, we designated this gene cadB.
In E. coli, CadB acts as an antiporter that is responsible for the transport of cadaverine, the by-product of lysine decarboxylation, from the bacterial cell, concomitant with import of lysine (24, 30). The V. cholerae cadB ORF consists of 1,338 bp and encodes a predicted protein of 446 amino acids (46.8 kDa). It lies upstream of, and in a putative bicistronic operon with, the previously identified cadA (Fig. 1). To verify the requirement of the cadBA operon for lysine decarboxylation and cadaverine export, a plasmid insertion mutation within cadB was constructed; the resulting mutant was unable to decarboxylate lysine in a lysine decarboxylase indicator medium. Although this result does not distinguish between loss of CadB function and a polar effect of the mutation on cadA, given that cadB represents the only lysine/cadaverine antiporter homolog in the V. cholerae genome, and given data below that substantiate bicistronic transcription of cadBA, it is likely that both effects occur.
FIG. 1.
Schematic depiction of the V. cholerae cadCBA locus. Arrows above the designated ORFs depict transcriptional orientation and represent promoters based on 5′ end mapping of mRNA species by primer extension. The locations of primers used for primer extension analysis are depicted by small arrows within the ORFs. Riboprobes used for RPAs and corresponding to the _V. cholerae_-specific component of the probe are represented by hatched boxes labeled probe 1 and probe 2. Relative sizes of cadBA, cadB, and cadA transcripts are represented by black lines beneath the ORFs. The locations of putative transcriptional terminators sequences (t) are also shown.
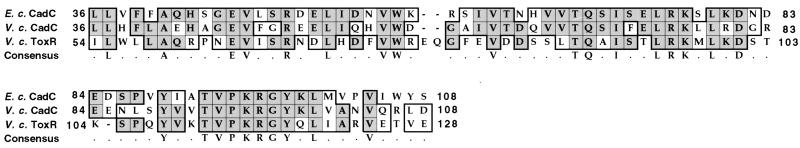
CadC has been shown to act as a positive transcriptional regulator of the cadBA operon in E. coli (24, 41). The putative V. cholerae cadC ORF consists of 1,560 bp and encodes a predicted protein of 519 amino acids (58.6 kDa). CadC lies upstream of, and in the same transcriptional orientation as, cadB but is separated from the cadB open reading frame by region of 699 bp (Fig. 1). The V. cholerae CadC also shares a high degree of identity with the N-terminal portion of V. cholerae ToxR (Fig. 2).
FIG. 2.
Multiple sequence alignment of amino-terminal portions of E. coli (E. c.) CadC and V. cholerae (V. c.) CadC and ToxR. Identical amino acids are highlighted by a gray background; conservative changes are outlined in black. A consensus sequence is depicted below the multiple alignment.
ToxR is a transcriptional activator localized within the inner membrane that is required for proper expression of virulence factors such as cholera toxin and the toxin coregulated pilus (reviewed in reference 37). The N-terminal domain of ToxR is cytoplasmic and encodes an OmpR-like DNA binding domain that is necessary for both positive and negative transcriptional regulation of ToxR-regulated genes. In addition, ToxR possesses a transmembrane segment and periplasmic domain that are believed to be involved in receiving and transmitting proper signals to elicit ToxR-mediated regulation (26, 27, 31). Based on amino acid sequence similarity at the N-terminal end of V. cholerae ToxR and CadC and a conserved hydropathy profile throughout (data not shown), CadC is predicted to be a transmembrane transcriptional activator with topological and localization features similar to those of ToxR.
Transcription of cadA is regulated by CadC.
Since CadC has previously been shown to be a positive transcriptional regulator of the cadBA operon in E. coli (24, 41), we sought to determine if CadC was similarly required for cadA expression in V. cholerae. V. cholerae strain DSM-V591, which contains an insertion mutation in cadC, was unable to decarboxylate lysine upon inoculation into lysine decarboxylase indicator medium whereas the wild-type parental strain did, suggesting that cadC was necessary for proper expression of cadBA.
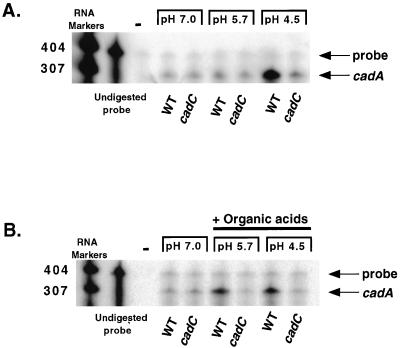
To determine whether the inability of the cadC strain to decarboxylate lysine was due to an inability to properly regulate transcription of cadA, RPAs were conducted with probe 1, a riboprobe specific for cadA transcripts (Fig. 1). Total RNA was collected from isogenic strains AC-V168 and DSM-V591 after growth in a rich medium at pH 7.0, 5.7, or 4.5. As previously reported (25), a large induction of cadA transcription occurs in wild-type V. cholerae cells upon exposure to pH 4.5 (Fig. 3A). In contrast, the cadC strain showed no induction of cadA transcription upon exposure to low pH. Of note, a basal level of cadA transcript is seen which appears both pH and CadC independent (Fig. 3A and B).
FIG. 3.
RPA for cadA transcription in wild-type (WT) and cadC mutant strains. Total RNA was prepared from bacteria grown in either low-pH media (A) or low-pH media supplemented with organic acids (B). The lanes corresponding to undigested probe represent the size of probe 1 used in the assays. Negative controls lacking bacterial RNA (−) show no protected bands. The protected cadA transcript is indicated. Relative molecular sizes are indicated on the left.
To determine if CadC was also necessary for the previously reported induction of cadA transcription upon exposure to low pH plus organic acids (25), similar RPAs were conducted using total RNA collected from AC-V168 and DSM-V591 grown in the presence of organic acids. As expected, AC-V168 showed an increase in cadA transcription when grown in medium at pH 5.7 and 4.5 that had been supplemented with organic acids (Fig. 3B and reference 25). In contrast, no increase in cadA transcript was evident in samples prepared from DSM-V591, indicating that CadC is required for organic acid-induced transcription of cadA at pH 5.7.
Transcription of the cadBA operon.
Since cadA was shown to play an essential role in the ATR of V. cholerae (25) but not of E. coli (34), we sought to develop a better understanding of the differences in regulation of cadA expression in these two organisms. Previous studies of E. coli have shown that cadA exists as part of a bicistronic operon with cadB. Transcription of this operon occurs from a promoter that has been designated pCad (41). pCad has been shown to be stimulated in a CadC-dependent manner by low pH, oxygen limitation, and high lysine concentrations (2, 23, 29, 35, 39, 41). To determine if cadB and cadA were cotranscribed in V. cholerae, RPA analysis was performed using probe 2, a riboprobe designed to span the entire cadB-cadA intergenic region and portions of cadB and cadA (Fig. 1). Three possibilities for potential transcripts were considered: (i) bicistronic cadB and cadA transcripts, (ii) specific cadB and cadA transcripts, and (ii) both bicistronic and single gene-specific transcripts. Based on probe 2, we expected that bicistronic cadBA would result in protection of a 732-bp fragment. A monocistronic cadB transcript would result in protection of a fragment that spanned the 3′ end of the cadB ORF (532 bp) as well as downstream sequences, depending on the site(s) of transcriptional termination. A monocistronic cadA transcript would result in a protected fragment that encompassed the 5′ end of the cadA ORF (101 bp) as well as upstream sequences, depending on the site of transcriptional initiation.
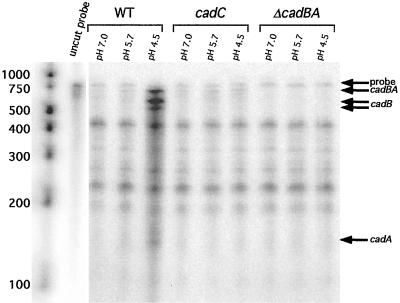
Total RNA was prepared from AC-V168 grown at pH 7.0, 5.7, and 4.5, and RPAs using probe 2 revealed protected fragments that correspond to sizes expected for a bicistronic transcript. Therefore, cadB and cadA are transcribed as a bicistronic operon in V. cholerae (Fig. 4; depicted in Fig. 1). Upon shift to pH 4.5, a condition known to increase transcription of cadA, an increase in the amount of bicistronic cadBA message was seen. In addition, two protected fragments that correspond to the sizes expected of transcripts terminated at the end of the cadB ORF appeared. A minor band that would correspond to the expected size of a _cadA_-specific transcript also appeared (Fig. 4).
FIG. 4.
RPA for cadBA transcription in wild-type, (WT) cadC mutant, and Δ_cadBA_ strains. Total RNA was prepared from strains grown at the pH values indicated. The lanes corresponding to undigested probe represents the size of synthesized probe 2 used in the assays. Bands corresponding to undigested full-length probe and cadBA, cadB, and cadA transcripts are indicated. Molecular weight markers are shown with relative sizes. The undigested probe and molecular weight markers are shown from a shorter exposure, which was necessary to prevent overexposure in comparison to protected fragments.
Since additional fragments of unexpected sizes were protected in each lane, we wished to confirm the identities of the indicated transcripts. An isogenic V. cholerae strain that contained a deletion of the entire cadBA coding sequence was constructed and used in similar RPAs. As shown in Fig. 4, deletion of the cadBA coding sequence resulted in loss of bicistronic cadBA and of monocistronic cadB and cadA bands, thus confirming that the indicated bands are specific for cad transcripts.
In Fig. 4, the protected fragments corresponding to the _cadB_-specific transcript, which would result from termination or processing downstream of the cadB ORF, was shown to be visible only upon shift to pH 4.5. Analysis of the nucleotide sequence immediately downstream of cadB revealed two large inverted repeats, designated as IR 1 and IR 2, which could potentially code for factor-independent transcriptional terminators (see Fig. 6). IR 1 is a perfect inverted repeat whose predicted stem and loop structure is followed by three uracil nucleotides. IR 2 is an imperfect repeat; the spacing of the run of uracil nucleotides is four positions distal to the base of the predicted hairpin structure. Termination at IR 1 would result in a 562-bp cadB transcript, while termination at IR 2 would result in a 621-bp cadB transcript. These sizes are in good agreement with the sizes for cadB transcripts indicated in Fig. 4. The presence of only three uracil nucleotides at the base of IR 1 and the spacing of uracil nucleotides distal to the stem of IR 2 would be consistent with low efficiency termination at these sites (8).
FIG. 6.
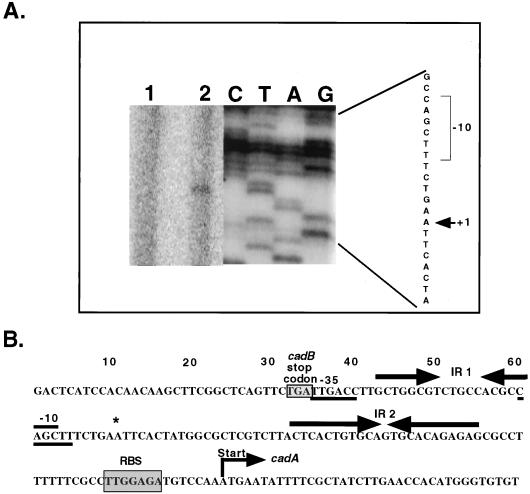
(A) Primer extension analysis of cadA transcript. The primer extension product obtained with primer CadAPE is depicted in lane 2. Lane 1 represents a negative control where no RNA was added. The sequencing ladder obtained with the same primer is shown to the right. The +1 site is indicated by an arrow, and a likely −10 box is shown. The length of exposure of the sequencing ladder was reduced to prevent overexposure in comparison to primer extension products. (B) Nucleotide sequence of the cadA promoter region. The transcription start site is indicated by an asterisk, and a likely −10 and a −35 site are underlined. IR 1 and IR 2 are indicated by arrows. The probable ribosomal binding site (RBS) is shaded, and the translational start is indicated.
Since we had shown that CadC was required for low-pH-induced transcription of cadA (Fig. 3), we investigated whether regulation by CadC occurred at the bicistronic cadBA promoter, the internal cadA promoter, or both. Total RNA was collected from DSM-V591, and RPA analysis using probe 2 was carried out. No significant increase in cadBA (or cadB or cadA) transcript was seen upon shift of DSM-V591 to lower pHs (Fig. 4). Furthermore, basal-level transcription of cadBA is CadC independent (Fig. 4). These results indicate that CadC regulation of both cadA and cadB expression occurs at the cadBA and cadA promoters (Fig. 4).
Identification of the transcription start sites of cadC, cadB, and cadA.
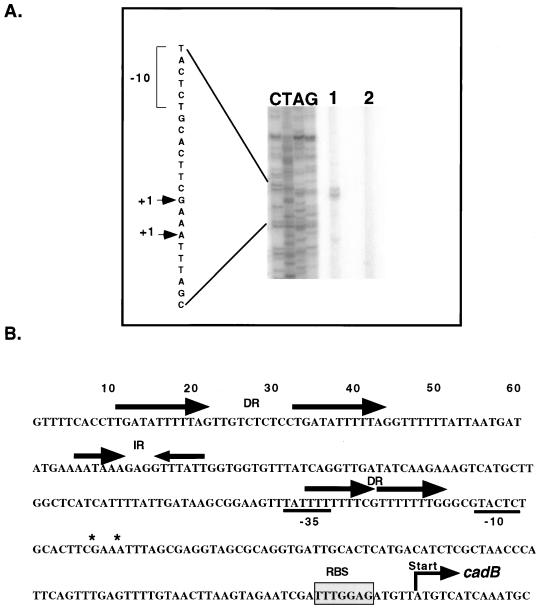
Primer extension analysis was carried out to map the 5′ termini of the cadB, cadA, and cadC transcripts to begin to define the promoter elements within this region. Total RNA was extracted and used for primer extension as described in Materials and Methods. For cadB, two primer extension products separated by three nucleotides were detected using RNA collected after acid challenge (Fig. 5A). This suggests a transcriptional start site(s) that is located 96 to 99 bp upstream of the translational start codon. Putative promoter elements were deduced according to the apparent transcriptional start site(s). The predicted −35 element (TATTTT) bears little resemblance to the E. coli ς70 consensus sequence (TTGACA). In addition, the −10 region (TACTCT) varies from the consensus (TATAAT) by three bases (Fig. 5B). Interestingly this −10 region shows greater similarity to the ςS consensus sequence (TATACT) in that it varies by only two nucleotides. A mutation in rpoS that encodes ςS, though, has previously been shown not to affect cadA transcription under the tested conditions (25). The predicted weakness of these −10 and −35 elements may account for the low level of basal transcription seen in the absence of the positive regulator CadC.
FIG. 5.
(A) Primer extension analysis of cadB transcripts. The two primer extension products obtained with primer CadBPE are depicted in lane 1. Lane 2 represents a negative control where no RNA was added. The sequencing ladder obtained with the same primer is shown to the left. The +1 sites are indicated by arrows, and a likely −10 box is shown. The length of exposure of the sequencing ladder was reduced to prevent overexposure in comparison to primer extension products. (B) Nucleotide sequence of the cadB promoter region. Direct (DR) and inverted (IR) repeats are indicated by arrows. Transcription start sites are indicated by asterisks, and a likely −10 and a −35 site are underlined. The probable ribosomal binding site (RBS) is shaded, and the translational start is indicated.
Analysis of the region upstream of the cadB transcription start sites revealed regions rich in A and T nucleotides. Two direct repeats of TGATAT5NG are located starting at position −180, and two smaller direct repeats of T7NG overlap the −35 region (Fig. 5B). These repeats represent potential CadC binding sites and show striking similarity to A/T-rich regions that ToxR has been shown to bind (7, 14, 20, 27). In particular, the TGATAT5NG repeat is similar to the consensus repeat sequence TG(a/T)3TTTNN bound by ToxR within the ompT promoter (20).
Primer extension of cadA transcripts was conducted using two separate primers, and in each case a single primer extension product was detected (Fig. 6A and data not shown). This probable transcriptional start site is located 73 bp upstream of the translational start codon. The predicted −35 element (TTGACC) differs from the E. coli ς70 consensus sequence by only one nucleotide, but the −10 region (CAGCTT) bears little similarity to the consensus (Fig. 6B). Comparison of these regions to consensus sequences utilized by other sigma factors revealed no other significant sigma factor motifs. Two inverted repeats of 8 and 11 bp are found upstream of the cadA translational start site and, as mentioned above, potentially represent the site(s) of inefficient termination of transcripts originating from the cadB promoter.
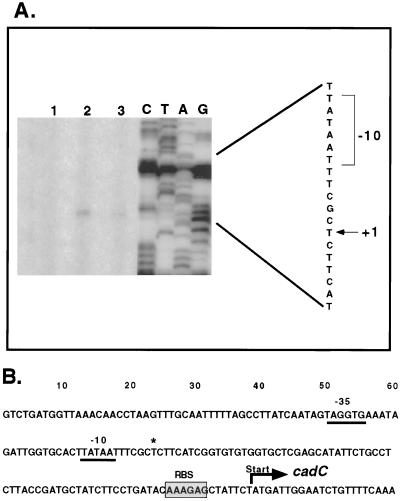
A single primer extension product was detected for cadC (Fig. 7A). This product suggests a transcriptional start site that is located 75 bp upstream of the translational start codon. A poorly conserved −35 element was predicted (TAGGTG), but a consensus −10 was evident (Fig. 7B).
FIG. 7.
(A) Primer extension analysis of cadC transcript. The primer extension products obtained with primer CadCPE are depicted in lanes 2 and 3. Lane 3 represents threefold less RNA than lane 2. Lane 1 represents a negative control where no RNA was added. The sequencing ladder obtained with the same primer is shown to the right. The +1 site is indicated by an arrow, and a likely −10 box is shown. The length of exposure of the sequencing ladder was reduced to prevent overexposure in comparison to primer extension products. (B) Nucleotide sequence of the cadC promoter region. The transcriptional start is indicated by an asterisk, and a likely −10 and a −35 site are underlined. The probable ribosomal binding site (RBS) is shaded, and the translational start is indicated.
CadC does not regulate other genes required for acid tolerance or genes essential for virulence.
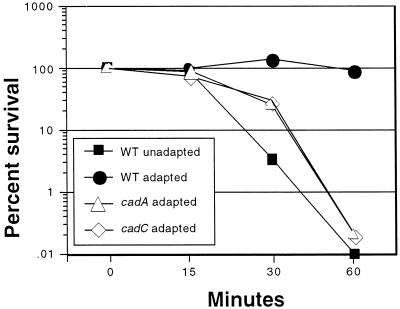
Since CadC acts as a transcriptional regulator of the cadBA operon and shares a high degree of similarity to ToxR, which directly or indirectly regulates as many as 17 genes, we sought to determine whether CadC regulates additional pathways, other than lysine decarboxylation, that are required for acid tolerance. This possibility was tested by assessment of the ability of DSM-V591 to undergo an attenuated ATR. We reasoned that if CadC regulated additional pathways required for ATR, the ATR defect exhibited by this cadC strain should be more severe than the ATR defect exhibited by a cadA strain. ATR assays using AC-V168 (wild type), DSM-V591 (cadC), and DSM-V376 (cadA) were performed as previously described (25). All three strains, when unadapted, were quickly killed when exposed to pH 4.5 (data not shown). Acid-adapted AC-V168 was able to mount a productive ATR and showed complete survival out to 60 min (Fig. 8). However, both DSM-V591 and DSM-V376 exhibited attenuated ATRs, and the rates of acid killing for both of these adapted strains were virtually identical (Fig. 8). Note that the rates of death of adapted DSM-V376 and DSM-V591 were not as severe as that for the unadapted wild-type strain, suggesting that some acid adaptation is occurring in these mutant strains. Similar results were obtained in organic ATR assays (data not shown). The sum of these experiments supports the hypothesis that CadC does not regulate additional genes (besides cadBA) that are required for the ATR of V. cholerae.
FIG. 8.
ATR assays of wild-type (WT; AC-V168), cadA (DSM-V376), and cadC (DSM-V591) V. cholerae strains. Strains were acid adapted or unadapted as described in Materials and Methods, and percent survival was calculated as a function of time after resuspension of bacteria in acid challenge medium, pH 4.5.
The ability of DSM-V591 (cadC) to colonize the infant mouse intestine was tested in a competition assay. DSM-V591 and the virulent, isogenic strain AC-V66 were mixed to attain an input ratio of approximately 1:1, and the mixture was used in competition assays as previously described (25). DSM-V591 exhibited no detectable defect in colonization, giving a competitive index of 0.96, which is essentially identical to the competitive index reported for a cadA mutant (25). These results suggest that in this model system, CadC is not required for regulation of factors essential for V. cholerae intestinal colonization.
DISCUSSION
The cadA gene of V. cholerae was previously identified as an infection-induced gene in two different animal models of V. cholerae (25). In vitro transcription analysis revealed that the gene was induced by low pH, high lysine concentration, and oxygen limitation. It was shown that cadA plays an essential role in the ATR of V. cholerae. Additionally, it was found that V. cholerae cells that were acid adapted prior to intragastric inoculation into animals greatly outcompeted unadapted cells, suggesting interesting implications for ATR in the epidemic spread of V. cholerae. In the present study, we examined the gene arrangement, transcription, and regulation of the cadA locus in more detail. The regulation of cadA has not been investigated in S. enterica serovar Typhimurium, the only other organism known to require lysine decarboxylation by cadA for ATR (30).
We found that V. cholerae contains cadB and cadC homologs, which are linked to cadA and arranged similarly as in E. coli. CadB in E. coli functions as a lysine/cadaverine antiporter, and its structural gene, cadB, is transcribed in a bicistronic operon with cadA. The cadBA operon is positively regulated by CadC (2, 23, 29, 35, 39, 41). RPA analysis revealed that cadB and cadA are also transcribed as a bicistronic operon in V. cholerae. However, unlike in E. coli, cadB- and _cadA_-specific transcripts are also present in V. cholerae.
Neely and Olson (29) previously determined the kinetics of cadBA expression in E. coli and found that a cadBA transcript is not detectable at neutral pH but becomes apparent upon shift to pH 5.8. Similarly, in V. cholerae, cadBA transcription was greatly increased upon shift from neutral to acidic pH, although pH 4.5 was necessary to see induction (or pH 4.5 and 5.7 plus organic acids [data not shown]). In addition, cadA monocistronic transcript appears upon shift to low pH. Together, these results suggest that our previous demonstration of low-pH induction of cadA transcription (25) occurs at the level of increased bicistronic cadBA and monocistronic cadA mRNA. pH-regulated induction of cadBA in E. coli is completely dependent on CadC in that a cadC null mutant does not express cadBA under any conditions tested (28). Unlike the case for E. coli, though, V. cholerae shows basal-level expression of cadBA transcript at neutral pH. In addition, this basal-level transcription is CadC independent, as mutations in cadC resulted in no decrease in transcription at any pH tested.
The location of cadC adjacent to cadBA on the V. cholerae large chromosome, and the similarity of the predicted CadC primary sequence to that of E. coli CadC, suggested that CadC might act as a transcriptional regulator required for low-pH induction of cadBA. Indeed, a greater than fivefold increase in cadBA mRNA was detected upon exposure of wild-type cells to low pH, and this increase was completely ablated in a cadC mutant. Again, however, basal-level transcription of cadBA still occurs in a cadC mutant, suggesting that functional lysine decarboxylase may be produced at a low level.
What might be the role of low constitutive levels of CadA and CadB in V. cholerae? Amino acid decarboxylases contribute to either biosynthetic or biodegradative pathways and have been shown to play important roles in polyamine synthesis or pH homeostasis, respectively (23, 24, 30). Biosynthetic amino acid decarboxylases are generally expressed constitutively, while biodegradative amino acid decarboxylases require low-pH induction for expression (3, 23, 24, 35, 38). E. coli is known to possess both biosynthetic and biodegradative lysine decarboxylases, Lcd and CadA, respectively. Lcd is responsible for the constitutive, low-level biosynthesis of cadaverine from lysine (17, 19). However, after acid induction, the bulk of lysine decarboxylase activity is due to the induction of cadA (23, 29). Extensive searches conducted against the V. cholerae genome sequence reveal no other potential lysine decarboxylase-encoding genes, suggesting that CadA is the sole enzyme responsible for the decarboxylation of lysine by V. cholerae (D. S. Merrell and A. Camilli, unpublished data). We hypothesize that the CadC-independent, basal-level expression of cadBA results in low levels of lysine decarboxylase necessary for biosynthetic decarboxylation, but at a level that is undetectable in our broth indicator medium assay. Upon exposure of V. cholerae to low pH, oxygen limitation, and high lysine concentrations, high levels of CadA and CadB are expressed in a CadC-dependent manner to assist with maintenance of pH homeostasis.
The N-terminal domain of CadC shares a high degree of identity with the N-terminal DNA binding domain of ToxR. ToxR is responsible for transcriptional activation of the ToxR/ToxT virulence gene regulon. Upon receiving unknown signals in the host intestine, ToxR stimulates transcription of toxT and several other genes; ToxT subsequently acts as a major transcription factor for a large set of virulence genes (37). V. cholerae also contains TcpP, an additional inner membrane protein that shares homology with ToxR in both sequence and function (6, 12). Recent studies have shown that both TcpP and ToxR are required for maximal expression of ToxT (12). toxR and tcpP are cotranscribed with toxS and _tcp_H, respectively; ToxS and TcpH are inner membrane proteins required for maximal activity of ToxR and TcpP, respectively (9, 12, 32). The V. cholerae and E. coli CadC proteins, though similar to ToxR in their N-terminal domains and cellular locations, appear not to require a ToxS-like component for activity (20). This suggests that the V. cholerae CadC protein is independently capable of receiving and transmitting signals. However, the possibility remains that an unidentified protein is capable of associating with CadC and facilitating its function.
It has long been known that the ability to respond and adapt to low-pH environments is an important aspect of the life cycle of many pathogenic organisms that colonize within the gastrointestinal tract. For instance, mutant strains of S. enterica serovar Typhimurium, Listeria monocytogenes, and Helicobacter pylori that are compromised in their ability to survive acid exposure have been shown to exhibit attenuated virulence (4, 10, 22, 36). Interestingly, the CadC homolog of S. enterica serovar Typhimurium was previously identified using in vivo expression technology as an infection-induced gene expressed within both the small intestines and spleens of BALB/c mice (13). The fact that two genes within the cad locus have been shown to be upregulated in two pathogens (V. cholerae and S. enterica serovar Typhimurium) upon entry into the host suggests that homologs of these genes may play crucial roles in the survival and adaptation of other pathogenic organisms as well.
Studies of the mechanism of transcriptional regulation by ToxR have shown that it binds to promoter elements exhibiting high degrees of A/T richness (7, 14, 20, 27). Analysis of the region upstream of the cadB promoter revealed the presence of direct repeats that are strikingly similar to ToxR-recognized elements in the ompT, ompU, ctxAB, and toxT promoters (20). Despite this sequence similarity, we found that a toxR null mutant shows normal levels of cadA expression (25), suggesting that ToxR does not regulate transcription of the cad locus under the conditions tested. Instead, these findings suggest that ToxR and CadC recognize very similar A/T-rich sequences but with a high degree of fidelity.
ACKNOWLEDGMENTS
This research was supported by NIH training grant AI 07422 (D.S.M.), NIH grants AI 40262 and Pew Scholars Award P0168SC (A.C.), and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928).
We thank E. Olson for helpful discussion, and we thank E. Joyce and E. Lloyd Angelichio for critical discussion and support. In addition, we thank A. Sonenshein, J. Mecsas, and M. Waldor for critical readings of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auger E A, Bennett G N. Regulation of lysine decarboxylase activity in Escherichia coli K-12. Arch Microbiol. 1989;151:466–468. doi: 10.1007/BF00416608. [DOI] [PubMed] [Google Scholar]
- 3.Auger E A, Redding K E, Plumb T, Childs L C, Meng S Y, Bennett G N. Construction of lac fusions to the inducible arginine- and lysine decarboxylase genes of Escherichia coli K12. Mol Microbiol. 1989;3:609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- 4.Bearson B L, Wilson L, Foster J W. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 7.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 8.d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 9.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 10.Foster J W. When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol. 1999;2:170–174. doi: 10.1016/S1369-5274(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hase C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R. Recombinant PCR. In: Gelfand D H, Innis M A, Sninski J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 16.Kadokami Y, Deike C A, Lewis R V. Precipitation of the template DNA can be omitted after NaOH denaturation in double-stranded DNA sequencing. BioTechniques. 1995;18:40–41. [PubMed] [Google Scholar]
- 17.Kikuchi Y, Kojima H, Tanaka T, Takatsuka Y, Kamio Y. Characterization of a second lysine decarboxylase isolated from Escherichia coli. J Bacteriol. 1997;179:4486–4492. doi: 10.1128/jb.179.14.4486-4492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolter R, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 19.Lemonnier M, Lane D. Expression of the second lysine decarboxylase gene of Escherichia coli. Microbiology. 1998;144:751–760. doi: 10.1099/00221287-144-3-751. [DOI] [PubMed] [Google Scholar]
- 20.Li C C, Crawford J A, Dirita V J, Kaper J B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in vibrio cholerae. Mol Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 21.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 22.Marron L, Emerson N, Gahan C G, Hill C. A mutant of Listeria monocytogenes LO28 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl Environ Microbiol. 1997;63:4945–4947. doi: 10.1128/aem.63.12.4945-4947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng S Y, Bennett G N. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol. 1992;174:2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng S Y, Bennett G N. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J Bacteriol. 1992;174:2670–2678. doi: 10.1128/jb.174.8.2670-2678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrell D S, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 26.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 28.Neely M N, Dell C L, Olson E R. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J Bacteriol. 1994;176:3278–3285. doi: 10.1128/jb.176.11.3278-3285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neely M N, Olson E R. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J Bacteriol. 1996;178:5522–5528. doi: 10.1128/jb.178.18.5522-5528.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park Y K, Bearson B, Bang S H, Bang I S, Foster J W. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 31.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. . (Erratum, 57:660, 1989.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfau J D, Taylor R K. Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J Bacteriol. 1998;180:4724–4733. doi: 10.1128/jb.180.17.4724-4733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, Pearson G D, Mekalanos J J. Proceedings of the 28th Joint Conference, U.S.-Japan Cooperative Medical Science Program on Cholera and Related Diarrheal Diseases. 1992. Cholera vaccine strains derived from a 1991 Peruvian isolate of Vibrio cholerae and other El Tor strains. [Google Scholar]
- 34.Rowbury R J, Goodson M. PhoE porin of Escherichia coli and phosphate reversal of acid damage and killing and of acid induction of the CadA gene product. J Appl Bacteriol. 1993;74:652–661. doi: 10.1111/j.1365-2672.1993.tb05199.x. [DOI] [PubMed] [Google Scholar]
- 35.Sabo D L, Boeker E A, Byers B, Waron H, Fischer E H. Purification and physical properties of inducible Escherichia coli lysine decarboxylase. Biochemistry. 1974;13:662–670. doi: 10.1021/bi00701a005. [DOI] [PubMed] [Google Scholar]
- 36.Scott D R, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 37.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 38.Tabor C W, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabor H, Hafner E W, Tabor C W. Construction of an Escherichia coli strain unable to synthesize putrescine, spermidine, or cadaverine: characterization of two genes controlling lysine decarboxylase. J Bacteriol. 1980;144:952–956. doi: 10.1128/jb.144.3.952-956.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 41.Watson N, Dunyak D S, Rosey E L, Slonczewski J L, Olson E R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992;174:530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]