Genetic Control of Natural Variation in Arabidopsis Glucosinolate Accumulation (original) (raw)
Abstract
Glucosinolates are biologically active secondary metabolites of the Brassicaceae and related plant families that influence plant/insect interactions. Specific glucosinolates can act as feeding deterrents or stimulants, depending upon the insect species. Hence, natural selection might favor the presence of diverse glucosinolate profiles within a given species. We determined quantitative and qualitative variation in glucosinolates in the leaves and seeds of 39 Arabidopsis ecotypes. We identified 34 different glucosinolates, of which the majority are chain-elongated compounds derived from methionine. Polymorphism at only five loci was sufficient to generate 14 qualitatitvely different leaf glucosinolate profiles. Thus, there appears to be a modular genetic system regulating glucosinolate profiles in Arabidopsis. This system allows the rapid generation of new glucosinolate combinations in response to changing herbivory or other selective pressures. In addition to the qualitative variation in glucosinolate profiles, we found a nearly 20-fold difference in the quantity of total aliphatic glucosinolates and were able to identify a single locus that controls nearly three-quarters of this variation.
The glucosinolates, a large group of naturally occurring plant defense compounds, are almost exclusively limited to the order Capparales. These nitrogen- and sulfur-containing secondary metabolites are derived from a variety of protein amino acids (Met, Leu, iso-Leu, Val, Trp, and Phe) through a three-part biosynthetic pathway (Halkier and Du, 1997) comprising: the elongation of the amino acid carbon chain, the formation of the basic glucosinolate skeleton, and further side chain modification (Fig. 1). Elongation of the carbon chain occurs via a threestep process that is similar to elongation of 2-oxoisovalerate in Leu biosynthesis (Chisholm and Wetter, 1964; Graser et al., 2000). First, the 2-oxo acid formed by de-amination of the amino acid is condensed with acetyl-coenzyme A by an isopropylmalate synthase-like enzyme (GS-elongase; de Quiros et al., 2000). Then, isomerization of the resulating alkylmalate followed by oxidative decarboxylation leads to a new 2-oxo acid with an additional methylene group. This new 2-oxo acid can either proceed through an additional carbon elongation cycle or undergo transamination to form a chain-extended amino acid (Fig. 1).
Figure 1.
Glucosinolate biosynthetic pathway. I, 2-Alkylmalic acid; II, 3-alkylmalic acid; III, 2-oxo acid. A, Basic glucosinolate structure. B, Outline of the pathway which can be divided into three parts: elongation of the amino acid side chain, formation of the basic glucosinolate skeleton, and further side chain modification. Each chain elongation cycle adds an additional methylene group (Graser et al., 2000).
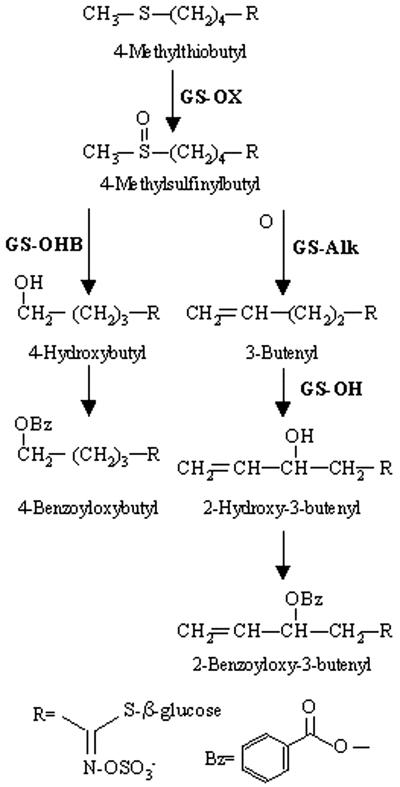
The committed step in formation of the basic glucosinolate skeleton is conversion of the amino acid (chain extended or unmodified) into the corresponding oxime by an amino-specific cytochrome P450 mono-oxygenase (Wittstock and Halkier, 2000). The oxime is then converted to a thiohydroximate intermediate, followed by sequential Glc and sulfate transfer to complete the basic glucosinolate skeleton (Fig. 1; Halkier and Du, 1997). The initially formed glucosinolate can undergo a variety of subsequent transformations that modify the side chain. These side-chain modifications are specific for the precursor amino acid utilized in the formation of the glucosinolate. Figure 2 shows the proposed biosynthetic sequence for the modifications of the chain-elongated Met-derived glucosinolates, which are the major glucosinolates in Arabidopsis and many other Brassicaceae species.
Figure 2.
Side chain modifications of Met-derived glucosinolates in Arabidopsis. Potential side chain modifications for the elongated Met derivative, C4 dihomo-Met, are shown. Steps with natural variation identified in this study are shown in bold to the right or left of each enzymatic arrow.
The principal biological activities of glucosinolates are mediated by hydrolysis products formed when tissue disruption brings glucosinolates into contact with myrosinase, a thioglucosidase. After Glc cleavage, the resulting unstable aglycone generates numerous compounds (isothiocyanates, nitriles, epithiocyanates, and thiocyanates) with diverse biological activities. Myrosinase hydrolysis products can serve as oviposition and feeding stimulants for insects specialized on glucosinolate-containing plants, but act as toxins or feeding deterrents toward generalist insect herbivores (Giamoustaris and Mithen, 1995). Thus, any given glucosinolate may have positive or negative impacts on plant fitness depending upon the insect herbivores present. Previous research has shown that such heterogenous selection due to insect herbivory occurs on glucosinolate concentration in both Arabidopsis and other Brassicaceae (Mauricio and Rausher, 1997; Mauricio, 1998; Stowe, 1998a, 1998b). In the face of such heterogeneous selection pressures, it is not surprising that glucosinolates show extensive genetic variation within and among plant species (Rodman, 1980; Daxenbichler et al., 1991).
We analyzed glucosinolate profiles from the leaves and seeds of 39 different Arabidopsis ecotypes representing a diverse sample of the geographical and environmental range of this species. Extensive variation was found in both the composition and total concentration of glucosinolates in these Arabidopsis ecotypes. The structural variety can be explained by polymorphism at only five genetic loci, creating a modular system for generation of biosynthetic diversity that may be a response to heterogeneous natural selection.
RESULTS
Genetic Control of Natural Variation in Glucosinolate Profiles and Concentrations in Arabidopsis Leaves
To survey natural variation in the glucosinolates of Arabidopsis, we identified and quantified 22 different glucosinolates present in the leaves of 39 different ecotypes (Tables I and II; Hogge et al., 1988). Analysis of the presence or absence of specific glucosinolates allowed us to identify several genetic polymorphisms regulating the composition of leaf glucosinolates. Four loci have been described previously: GS-Elong controls production of glucosinolates with three carbon or four carbon side chains. GS-Alk is responsible for production of alkenyl glucosinolates. GS-OHP catalyzes production of 3-hydroxypropyl glucosinolates. Finally, GS-OH controls production of 2-hydroxy-3-butenyl glucosinolate (Figs. 1 and 2; Magrath et al., 1994; Mithen et al., 1995). In addition to these four loci, evidence for a previously unknown Arabidopsis locus was found in this collection of ecotypes. This locus, designated GS-OX, controls the conversion of methylthioalkyl to methylsulfinylalkyl glucosinolates (Fig. 2 and Table I; Giamoustaris and Mithen, 1996). Most ecotypes carry out this conversion efficiently and typically contain at least twice as much methylsulfinylalkyl as methylthioalkyl glucosinolate in the leaves. However, the Bla-10, Can-0, and Su-0 ecotypes all have higher concentrations of methylthioalkyl than methylsulfinylalkyl glucosinolates, indicating that they are impaired in this conversion and presumably contain a different GS-OX allele than the other ecotypes (Fig. 3A).
Table I.
Glucosinolates in the leaves of arabidopsis ecotypes
| Ecotype | Genotype | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 11 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | A | OH | OX | ||||||||||
| Ag-0 | 4 | 3 | 3 | 2 | – | – | – | 1.55 | – | 4.69 | 1.56 | 8.10 | |
| Bla-10 | 4 | 3 | 3 | 1 | – | – | – | 0.40 | 0.83 | 1.54 | 0.19 | 0.36 | 2.19 |
| Bs1 | 4 | 3 | 3 | 2 | – | – | – | 1.43 | – | 4.29 | 1.24 | 5.65 | |
| Cal | 4 | 3 | 3 | 2 | – | – | – | 0.32 | – | 1.01 | 4.90 | 11.99 | |
| Cnt | 4 | 3 | 3 | 2 | – | – | 0.31 | 2.47 | – | 7.17 | 0.28 | 12.46 | |
| Ema-1 | 4 | 3 | 3 | 2 | – | – | 0.13 | 1.93 | – | 5.83 | 0.21 | 8.28 | |
| Pog-0 | 4 | 3 | 3 | 2 | 0.06 | – | 0.32 | 2.73 | – | 7.98 | 0.21 | 9.39 | |
| Tac | 4 | 3 | 3 | 2 | – | 0.05 | 0.15 | 2.27 | – | 6.77 | 0.35 | 7.38 | |
| Kas | 4 | 3 | 2 | 2 | – | – | – | – | – | – | 5.31 | 11.64 | |
| Sorbo | 4 | 3 | 2 | 2 | – | – | – | – | – | – | 4.55 | 20.96 | |
| Cvi | 4 | 3 | 1 | 2 | – | – | – | – | – | – | 10.03 | 24.40 | |
| Di-1 | 4 | 2 | – | 2 | 1.79 | 0.03 | – | – | 3.62 | – | – | – | |
| Aa-0 | 4 | 1 | – | 2 | – | – | – | – | 1.31 | – | 0.07 | 0.45 | |
| Col | 4 | 1 | – | 2 | – | 0.37 | – | – | 4.96 | – | 0.03 | – | |
| Ma-0 | 4 | 1 | – | 2 | – | – | – | – | 2.39 | – | – | – | |
| Mt-0 | 4 | 1 | – | 2 | – | – | – | – | 3.98 | – | – | – | |
| Can | 3 | 3 | – | 1 | – | 0.61 | – | – | – | – | 18.43 | 0.40 | |
| Kondara | 3 | 3 | – | 2 | 0.11 | – | – | – | – | – | 25.99 | 0.47 | |
| Ei-2 | 3 | 3 | – | 2 | – | – | – | – | – | 0.04 | 16.79 | 0.19 | |
| Hodja | 3 | 3 | – | 2 | – | – | – | – | – | – | 13.10 | 0.18 | |
| Ita-0 | 3 | 3 | – | 2 | – | – | – | – | – | – | – | – | |
| Kil-0 | 3 | 3 | – | 2 | – | – | – | – | – | – | 14.57 | 0.19 | |
| Mr-0 | 3 | 3 | – | 2 | – | – | – | – | – | – | – | – | |
| Mrk-0 | 3 | 3 | – | 2 | 0.15 | – | – | – | – | – | 20.27 | 0.29 | |
| Rsch-0 | 3 | 3 | – | 2 | – | – | – | – | – | – | 10.02 | 0.22 | |
| Su-0 | 3 | 3 | – | 1 | – | 0.26 | – | – | – | – | 12.01 | 0.07 | |
| Wl-0 | 3 | 3 | – | 2 | 0.06 | – | – | – | – | – | 14.38 | 0.16 | |
| Bl-1 | 3 | 2 | – | 2 | 7.32 | – | – | – | – | – | 0.20 | 0.03 | |
| Di-g | 3 | 2 | – | 2 | – | – | – | – | – | – | – | – | |
| Ka-0 | 3 | 2 | – | 2 | 14.32 | 0.04 | – | – | – | – | – | – | |
| Ler | 3 | 2 | – | 2 | 9.07 | 0.17 | – | – | 0.02 | – | – | – | |
| Lip-0 | 3 | 2 | – | 2 | 5.14 | 0.68 | – | – | – | – | – | – | |
| No-0 | 3 | 2 | – | 2 | 7.22 | 0.34 | – | – | 0.08 | – | – | – | |
| Pet | 3 | 2 | – | 2 | 8.55 | 0.69 | – | – | 0.03 | – | – | – | |
| Pi-0 | 3 | 2 | – | 2 | 2.07 | – | – | – | 0.08 | – | – | – | |
| Sei-0 | 3 | 2 | – | 2 | 3.76 | 0.99 | – | – | 0.07 | – | – | – | |
| Tsu-1 | 3 | 2 | – | 2 | 5.19 | 0.62 | – | – | 0.05 | – | – | 0.02 | |
| Yo-0 | 3 | 2 | – | 2 | 5.40 | 0.49 | – | – | 0.20 | – | – | – | |
| Oy-0 | 3 | 1 | – | 2 | – | – | – | – | – | – | – | – | |
| Average | – | – | – | – | 4.68 | 0.41 | 0.23 | 1.64 | 1.36 | 4.37 | 7.60 | 5.44 | |
| sd | – | – | – | – | 4.15 | 0.30 | 0.10 | 0.90 | 1.78 | 2.89 | 7.96 | 7.08 | |
| Minimum | – | – | – | – | 0.06 | 0.03 | 0.13 | 0.32 | 0.02 | 0.04 | 0.03 | 0.02 | |
| Maximum | – | – | – | – | 14.32 | 0.99 | 0.32 | 2.73 | 4.96 | 7.98 | 25.99 | 24.40 |
| 13 | 14 | 15 | 16 | 18 | 19 | 20 | 21 | 22 | 25 | 26 | 30 | 32 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| – | – | 0.04 | 0.21 | 0.46 | – | 1.11 | 3.19 | 0.36 | 0.02 | 0.57 | 0.10 | 0.71 |
| – | 0.84 | 0.02 | 0.17 | 0.05 | 4.95 | 1.12 | 1.70 | 0.42 | – | 0.43 | 0.10 | 0.41 |
| 0.02 | – | 0.04 | 0.22 | 0.36 | – | 1.07 | 2.53 | 0.31 | – | 0.21 | 0.12 | 0.36 |
| 0.09 | 0.16 | 0.03 | 0.36 | 0.51 | – | 1.77 | 2.43 | 0.47 | 0.09 | 0.28 | 0.21 | 0.52 |
| 0.12 | – | 0.03 | 0.42 | 0.83 | 0.05 | 3.00 | 3.14 | 0.32 | 0.01 | 0.39 | 0.09 | 0.56 |
| 0.10 | – | 0.01 | 0.34 | 0.60 | – | 1.84 | 3.24 | 0.20 | 0.01 | 0.33 | 0.11 | 0.42 |
| 0.11 | – | 0.03 | 0.37 | 0.73 | – | 1.87 | 2.98 | 0.31 | 0.03 | 0.76 | 0.06 | 0.36 |
| 0.05 | – | 0.01 | 0.25 | 0.47 | 0.04 | 1.41 | 3.16 | 0.09 | 0.01 | 0.33 | 0.06 | 0.26 |
| – | – | 0.02 | 0.06 | 0.09 | 0.02 | 0.49 | 1.49 | 0.35 | 0.02 | 0.96 | 0.04 | 0.29 |
| – | – | 0.02 | 0.17 | 0.30 | – | 1.21 | 1.73 | 0.31 | 0.02 | 0.45 | 0.07 | 0.29 |
| 0.09 | – | 0.01 | 0.29 | 0.29 | – | 2.24 | 1.08 | 0.19 | – | 0.39 | 0.09 | 0.38 |
| 0.01 | – | 0.04 | 0.24 | – | 0.51 | 1.24 | 2.82 | 0.44 | 0.03 | 0.56 | 0.05 | 0.20 |
| – | – | 0.02 | 0.11 | – | 0.53 | 1.08 | 1.87 | 0.28 | – | 0.04 | – | 0.21 |
| – | – | 0.02 | 0.16 | – | 0.97 | 0.99 | 2.88 | 0.31 | – | 0.62 | 0.03 | 0.19 |
| – | – | 0.02 | 0.10 | – | 0.15 | 0.86 | 1.73 | 0.32 | – | 0.52 | – | 0.07 |
| – | – | 0.05 | 0.15 | – | 0.30 | 1.13 | 2.81 | 0.52 | – | 0.78 | – | 0.15 |
| – | 3.66 | 0.01 | 0.18 | – | 0.27 | 1.77 | 2.14 | 0.47 | – | 0.38 | 0.15 | 1.00 |
| – | 0.04 | – | 0.11 | – | – | 1.89 | 2.62 | 0.42 | – | 1.01 | 0.03 | 0.53 |
| – | – | 0.01 | 0.12 | – | – | 1.76 | 2.33 | 0.53 | – | 0.48 | 0.08 | 0.86 |
| – | – | – | – | – | – | 0.61 | 1.16 | 0.16 | – | 0.49 | 0.03 | 0.30 |
| – | – | – | – | – | – | – | – | – | – | – | – | – |
| – | 0.06 | 0.01 | 0.14 | – | – | 1.82 | 2.52 | 0.25 | – | 0.34 | 0.08 | 0.58 |
| – | – | – | – | – | – | – | – | – | – | – | – | – |
| – | – | 0.01 | 0.12 | – | – | 2.12 | 2.09 | 0.16 | – | 1.07 | 0.06 | 0.56 |
| – | – | – | 0.09 | – | – | 1.34 | 1.88 | 0.19 | – | 0.71 | 0.01 | 0.24 |
| – | 2.02 | 0.01 | 0.07 | – | 0.06 | 1.36 | 1.97 | 0.25 | – | 0.64 | 0.06 | 0.79 |
| 0.02 | 0.01 | 0.01 | 0.15 | 0.05 | – | 1.22 | 2.00 | 0.18 | – | 0.78 | 0.02 | 0.18 |
| – | 0.08 | 0.02 | 0.10 | – | 0.05 | 1.68 | 1.76 | 0.33 | – | 0.94 | 0.03 | 0.67 |
| – | – | – | – | – | – | – | – | – | – | – | – | – |
| – | – | 0.01 | 0.08 | – | – | 1.54 | 1.78 | 0.26 | – | 0.90 | 0.04 | 0.45 |
| – | – | 0.02 | 0.07 | – | – | 1.32 | 1.94 | 0.21 | – | 0.71 | 0.03 | 0.33 |
| – | 0.04 | 0.01 | 0.08 | – | – | 1.04 | 1.73 | 0.33 | – | 0.76 | 0.02 | 0.27 |
| – | 0.09 | 0.03 | 0.03 | – | 0.03 | 0.88 | 1.77 | 0.18 | – | 0.32 | 0.04 | 0.52 |
| – | 0.14 | 0.03 | 0.08 | – | 0.06 | 1.69 | 2.82 | 0.26 | – | 1.13 | 0.04 | 0.50 |
| – | – | – | 0.04 | – | – | 0.64 | 0.93 | 0.05 | – | 0.46 | – | – |
| – | 0.02 | 0.05 | 0.10 | – | – | 1.11 | 2.06 | 0.06 | – | 0.38 | – | 0.07 |
| – | 0.21 | 0.03 | 0.09 | – | 0.04 | 1.13 | 2.77 | 0.15 | – | 0.51 | 0.05 | 0.40 |
| – | 0.12 | 0.02 | 0.07 | – | 0.01 | 0.81 | 2.91 | 0.11 | – | 0.77 | 0.04 | 0.35 |
| – | – | – | – | – | – | – | – | – | – | – | – | – |
| 0.07 | 0.54 | 0.02 | 0.16 | 0.40 | 0.50 | 1.38 | 2.23 | 0.28 | 0.03 | 0.58 | 0.06 | 0.41 |
| 0.04 | 1.05 | 0.01 | 0.10 | 0.26 | 1.22 | 0.52 | 0.63 | 0.13 | 0.03 | 0.26 | 0.04 | 0.22 |
| 0.01 | 0.01 | 0.01 | 0.03 | 0.05 | 0.01 | 0.49 | 0.93 | 0.05 | 0.01 | 0.04 | 0.01 | 0.07 |
| 0.12 | 3.66 | 0.05 | 0.42 | 0.83 | 4.95 | 3.00 | 3.24 | 0.53 | 0.09 | 1.13 | 0.21 | 1.00 |
| Totals | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aliph | Indole | MT | MSO | AOP | C3 | C4 | C7 | C8 | C4Per | C8Per |
| 18.49 | 4.16 | 0.81 | 1.32 | 16.36 | 1.56 | 14.34 | 0.31 | 1.82 | 0.902 | 0.854 |
| 13.15 | 2.57 | 6.30 | 2.12 | 4.37 | 1.03 | 9.91 | 0.27 | 1.53 | 0.906 | 0.850 |
| 14.76 | 3.09 | 0.48 | 1.31 | 12.97 | 1.24 | 11.37 | 0.34 | 1.43 | 0.902 | 0.808 |
| 21.84 | 3.21 | 0.89 | 2.22 | 18.73 | 5.06 | 13.32 | 0.57 | 2.29 | 0.725 | 0.801 |
| 27.76 | 3.88 | 0.70 | 3.54 | 23.52 | 0.28 | 22.46 | 0.51 | 3.56 | 0.988 | 0.875 |
| 19.79 | 3.78 | 0.53 | 2.28 | 16.98 | 0.21 | 16.17 | 0.45 | 2.26 | 0.987 | 0.834 |
| 24.19 | 4.08 | 0.42 | 2.35 | 21.42 | 0.27 | 20.42 | 0.43 | 2.23 | 0.987 | 0.838 |
| 19.51 | 3.59 | 0.36 | 1.76 | 17.39 | 0.40 | 16.61 | 0.31 | 1.67 | 0.967 | 0.843 |
| 17.94 | 2.82 | 0.35 | 0.55 | 17.04 | 5.31 | 11.66 | 0.10 | 0.78 | 0.687 | 0.886 |
| 27.55 | 2.51 | 0.36 | 1.38 | 25.81 | 4.55 | 20.96 | 0.24 | 1.50 | 0.822 | 0.862 |
| 37.81 | 1.67 | 0.47 | 2.62 | 34.72 | 10.03 | 24.40 | 0.38 | 2.62 | 0.709 | 0.873 |
| 7.69 | 3.86 | 0.76 | 5.14 | 1.79 | 1.82 | 4.13 | 0.29 | 1.44 | 0.694 | 0.832 |
| 3.76 | 2.21 | 0.74 | 2.50 | 0.52 | 0.07 | 2.29 | 0.11 | 1.29 | 0.970 | 0.921 |
| 7.70 | 3.83 | 1.19 | 6.48 | 0.03 | 0.40 | 5.93 | 0.19 | 1.18 | 0.937 | 0.861 |
| 3.57 | 2.59 | 0.22 | 3.35 | 0.00 | 0.00 | 2.54 | 0.10 | 0.93 | 1.000 | 0.903 |
| 5.71 | 4.16 | 0.45 | 5.26 | 0.00 | 0.00 | 4.28 | 0.15 | 1.28 | 1.000 | 0.895 |
| 26.47 | 3.00 | 5.08 | 2.56 | 18.83 | 22.70 | 0.67 | 0.33 | 2.77 | 0.029 | 0.894 |
| 29.17 | 4.05 | 0.60 | 2.00 | 26.57 | 26.14 | 0.47 | 0.14 | 2.42 | 0.018 | 0.945 |
| 19.84 | 3.35 | 0.94 | 1.88 | 17.02 | 16.79 | 0.23 | 0.20 | 2.62 | 0.014 | 0.929 |
| 14.22 | 1.81 | 0.33 | 0.61 | 13.28 | 13.10 | 0.18 | 0.03 | 0.91 | 0.014 | 0.968 |
| – | – | – | – | – | – | – | – | – | – | |
| 17.44 | 3.12 | 0.72 | 1.96 | 14.76 | 14.63 | 0.19 | 0.22 | 2.40 | 0.013 | 0.916 |
| – | – | – | – | – | – | – | – | – | – | – |
| 23.57 | 3.33 | 0.62 | 2.24 | 20.71 | 20.42 | 0.29 | 0.18 | 2.68 | 0.014 | 0.937 |
| 11.92 | 2.78 | 0.25 | 1.43 | 10.24 | 10.02 | 0.22 | 0.10 | 1.58 | 0.021 | 0.940 |
| 16.70 | 2.87 | 2.93 | 1.69 | 12.08 | 14.29 | 0.13 | 0.13 | 2.15 | 0.009 | 0.943 |
| 16.25 | 2.97 | 0.21 | 1.39 | 14.65 | 14.45 | 0.16 | 0.17 | 1.40 | 0.011 | 0.892 |
| 10.16 | 3.05 | 0.83 | 1.78 | 7.55 | 7.60 | 0.08 | 0.13 | 2.35 | 0.010 | 0.948 |
| – | – | – | – | – | – | – | – | – | – | – |
| 16.47 | 2.95 | 0.49 | 1.66 | 14.32 | 14.36 | 0.00 | 0.12 | 1.99 | 0.000 | 0.943 |
| 11.01 | 2.88 | 0.36 | 1.58 | 9.07 | 9.24 | 0.02 | 0.10 | 1.65 | 0.002 | 0.943 |
| 7.27 | 2.83 | 0.33 | 1.80 | 5.14 | 5.86 | 0.00 | 0.10 | 1.31 | 0.000 | 0.929 |
| 9.23 | 2.30 | 0.68 | 1.33 | 7.22 | 7.65 | 0.11 | 0.07 | 1.40 | 0.014 | 0.952 |
| 11.78 | 4.24 | 0.74 | 2.49 | 8.55 | 9.38 | 0.09 | 0.12 | 2.19 | 0.010 | 0.948 |
| 2.83 | 1.44 | 0.00 | 0.76 | 2.07 | 2.07 | 0.08 | 0.04 | 0.64 | 0.037 | 0.841 |
| 6.12 | 2.55 | 0.09 | 2.27 | 3.76 | 4.77 | 0.07 | 0.10 | 1.18 | 0.014 | 0.922 |
| 7.80 | 3.46 | 0.70 | 1.89 | 5.21 | 6.02 | 0.11 | 0.14 | 1.53 | 0.018 | 0.916 |
| 7.49 | 3.81 | 0.52 | 1.57 | 5.40 | 6.01 | 0.21 | 0.11 | 1.16 | 0.034 | 0.913 |
| – | – | – | – | – | – | – | – | – | – | – |
| 15.34 | 3.11 | 0.90 | 2.20 | 12.23 | 7.36 | 5.83 | 0.21 | 1.78 | 0.413 | – |
| 8.43 | 0.73 | 1.30 | 1.26 | 8.66 | 7.10 | 7.92 | 0.14 | 0.66 | 0.448 | – |
| 2.83 | 1.44 | 0.00 | 0.55 | 0.00 | 0.00 | 0.00 | 0.03 | 0.64 | 0.000 | – |
| 37.81 | 4.24 | 6.30 | 6.48 | 34.72 | 26.14 | 24.40 | 0.57 | 3.56 | 1.000 | – |
Table II.
Glucosinolates in the seeds of Arabidopsis ecotypes
| Genotypes | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | A | OH | OX | OHB | ||||||||
| Ag-0 | 4 | 3 | 3 | 2 | 2 | – | – | 1.02 | – | – | 2.40 | 0.15 |
| Bla-10 | 4 | 3 | 3 | 1 | 2 | – | – | 0.41 | – | – | 1.12 | – |
| Bs1 | 4 | 3 | 3 | 2 | 2 | 0.31 | – | 1.18 | 2.02 | – | 8.17 | 0.29 |
| Cal | 4 | 3 | 3 | 2 | 1 | – | – | – | – | – | 0.46 | 0.21 |
| Cnt | 4 | 3 | 3 | 2 | 1 | – | – | – | 4.20 | – | 13.66 | 0.30 |
| Ema-1 | 4 | 3 | 3 | 2 | 2 | – | – | 0.58 | 0.65 | – | 2.38 | – |
| Pog-0 | 4 | 3 | 3 | 2 | 1 | – | – | – | 2.91 | – | 7.67 | – |
| Tac | 4 | 3 | 3 | 2 | 1 | 0.19 | 0.15 | – | 2.47 | – | 5.84 | 0.13 |
| Kas | 4 | 3 | 2 | 2 | 2 | 0.46 | – | 0.49 | 0.42 | – | 1.43 | 0.98 |
| Sorbo | 4 | 3 | 2 | 2 | 2 | 0.37 | – | 0.89 | 0.25 | – | 1.58 | – |
| Cvi | 4 | 3 | 1 | 2 | 1 | 0.36 | – | – | – | – | – | 0.96 |
| Di-1 | 4 | 2 | – | 2 | 1 | 0.93 | – | – | – | 0.74 | – | 0.12 |
| Aa-0 | 4 | 1 | – | 2 | 2 | 0.26 | – | 0.81 | – | 0.52 | – | – |
| Col | 4 | 1 | – | 2 | 2 | 0.69 | – | 2.20 | – | 1.87 | – | – |
| Ma-0 | 4 | 1 | – | 2 | 2 | 0.47 | – | 1.11 | – | 0.22 | – | 0.21 |
| Mt-0 | 4 | 1 | – | 2 | 2 | 0.37 | – | 1.43 | – | 0.71 | – | – |
| Can | 3 | 3 | – | 1 | – | 0.39 | – | – | – | – | – | 1.64 |
| Kondara | 3 | 3 | – | 2 | – | 4.19 | 0.90 | – | – | – | – | 6.96 |
| Ei-2 | 3 | 3 | – | 2 | – | 2.53 | – | – | – | – | – | 4.44 |
| Hodja | 3 | 3 | – | 2 | – | 2.27 | 1.41 | – | – | – | – | 7.60 |
| Ita-0 | 3 | 3 | – | 2 | – | 1.83 | 1.40 | – | – | – | – | 1.36 |
| Kil-0 | 3 | 3 | – | 2 | – | 1.36 | 0.55 | – | – | – | – | 2.50 |
| Mr-0 | 3 | 3 | – | 2 | – | 0.84 | – | – | – | – | 0.13 | 1.63 |
| Mrk-0 | 3 | 3 | – | 2 | – | 1.47 | 0.80 | – | – | – | – | 1.54 |
| Rsch-0 | 3 | 3 | – | 2 | – | – | – | – | – | – | – | – |
| Su-0 | 3 | 3 | – | 1 | – | 0.71 | – | – | 0.13 | – | 0.64 | 0.99 |
| Wl-0 | 3 | 3 | – | 2 | – | 2.17 | 0.33 | – | – | – | – | 3.87 |
| Bl-1 | 3 | 2 | – | 2 | – | 1.88 | – | – | – | – | – | – |
| Di-g | 3 | 2 | – | 2 | – | 4.17 | – | – | – | – | – | – |
| Ka-0 | 3 | 2 | – | 2 | – | 10.91 | – | – | – | – | – | 0.29 |
| Ler | 3 | 2 | – | 2 | – | 7.86 | 0.19 | – | – | – | – | 0.12 |
| Lip-0 | 3 | 2 | – | 2 | – | 3.26 | – | – | – | – | – | – |
| No-0 | 3 | 2 | – | 2 | – | 14.16 | 0.50 | – | – | 0.28 | – | – |
| Pet | 3 | 2 | – | 2 | – | 8.47 | – | – | – | – | – | – |
| Pi-0 | 3 | 2 | – | 2 | – | 3.24 | – | – | – | – | – | – |
| Sei-0 | 3 | 2 | – | 2 | – | 2.58 | 0.20 | – | – | – | – | – |
| Tsu-1 | 3 | 2 | – | 2 | – | 3.61 | 5.58 | – | – | – | – | – |
| Yo-0 | 3 | 2 | – | 2 | – | 2.56 | – | – | – | – | – | 0.16 |
| Oy-0 | 3 | 1 | – | 2 | – | 1.23 | 0.62 | – | – | – | – | – |
| Average | – | – | – | – | – | 2.69 | 1.05 | 1.01 | 1.63 | 0.72 | 3.79 | 1.66 |
| sd | – | – | – | – | – | 3.29 | 1.49 | 0.53 | 1.50 | 0.60 | 4.18 | 2.18 |
| Minimum | – | – | – | – | – | 0.19 | 0.15 | 0.41 | 0.13 | 0.22 | 0.13 | 0.12 |
| Maximum | – | – | – | – | – | 14.16 | 5.58 | 2.20 | 4.20 | 1.87 | 13.66 | 7.60 |
| 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| – | 0.25 | 0.47 | – | 0.07 | – | – | 0.56 | – | 0.26 | 1.72 | 4.75 |
| – | 0.16 | 0.14 | – | 0.18 | 0.80 | – | 0.28 | 0.18 | 0.07 | 3.74 | 2.47 |
| – | 0.34 | 0.52 | – | – | – | – | 0.33 | – | 0.11 | 6.86 | 2.02 |
| – | – | 0.47 | 0.25 | 0.05 | – | – | 0.51 | 0.11 | 0.17 | 1.95 | 3.41 |
| – | 0.49 | 2.87 | 0.15 | 0.15 | – | – | 0.85 | – | 0.58 | 2.26 | 4.55 |
| – | 0.46 | 0.17 | – | 0.25 | – | 0.02 | 0.92 | – | 0.22 | 1.52 | 6.01 |
| – | 0.55 | 0.62 | – | 0.10 | 0.08 | – | 0.77 | – | 0.29 | 2.93 | 5.99 |
| – | 0.42 | 0.65 | 0.10 | – | 0.23 | – | 0.50 | – | 0.28 | 4.66 | 4.56 |
| – | – | 1.51 | – | – | 0.10 | 0.03 | 0.22 | – | 0.07 | 0.61 | 2.99 |
| – | – | 12.52 | – | – | 0.12 | 0.02 | 0.34 | – | 0.31 | 3.34 | 2.72 |
| – | – | 3.31 | 0.13 | – | 0.09 | – | 0.32 | – | 0.13 | 4.01 | 4.53 |
| – | – | – | – | 0.07 | 0.30 | – | 0.65 | – | – | 13.09 | 5.28 |
| – | – | – | – | – | – | 0.02 | 0.22 | – | – | 11.70 | 1.90 |
| 0.19 | – | – | – | 0.07 | 0.08 | 0.06 | 0.55 | – | – | 18.95 | 2.76 |
| – | – | – | – | – | 0.22 | 0.03 | 0.16 | – | – | 7.66 | 2.49 |
| – | – | – | – | – | 0.10 | 0.01 | 0.31 | – | – | 12.74 | 2.49 |
| – | – | – | – | – | 0.15 | – | 0.29 | 0.06 | – | 0.26 | 6.81 |
| – | – | 0.14 | – | – | 3.20 | – | 0.15 | – | 0.07 | 0.27 | 3.29 |
| – | – | – | – | – | 0.89 | – | 0.19 | – | – | 0.17 | 4.80 |
| – | – | 0.13 | – | – | 6.85 | 0.02 | 0.11 | – | – | 0.26 | 2.61 |
| – | – | – | – | – | 9.04 | – | 0.34 | – | 0.15 | 0.72 | 6.43 |
| – | – | – | – | – | 3.33 | – | 0.28 | – | – | 0.11 | 6.07 |
| – | – | – | – | – | 0.66 | 0.02 | 0.28 | – | – | 0.25 | 8.19 |
| – | – | – | – | – | 5.90 | – | 0.28 | – | – | 0.36 | 5.83 |
| – | – | – | – | – | – | – | – | – | – | – | – |
| – | – | – | 0.10 | – | 0.94 | 0.02 | 0.17 | – | – | 0.48 | 4.17 |
| – | – | – | – | – | 4.07 | – | 0.12 | – | – | 0.21 | 2.46 |
| – | – | – | – | – | 2.09 | 0.02 | 0.16 | – | – | 0.48 | 4.60 |
| – | – | – | – | – | 0.69 | – | 0.20 | – | – | 0.41 | 4.93 |
| – | – | – | – | – | 0.52 | – | 0.14 | – | – | 0.07 | 3.78 |
| – | – | – | – | – | 2.76 | – | 0.11 | – | – | 0.36 | 3.19 |
| – | – | – | – | – | 0.41 | – | 0.09 | – | – | 0.55 | 1.97 |
| – | – | – | – | – | 6.81 | – | 0.16 | – | – | 0.52 | 2.97 |
| – | – | – | – | – | 1.20 | 0.02 | 0.08 | – | – | 0.48 | 2.92 |
| – | – | – | – | – | 0.38 | 0.02 | 0.08 | – | – | 0.70 | 2.30 |
| – | – | – | – | – | 1.48 | – | 0.17 | – | – | 0.30 | 3.38 |
| – | – | – | – | – | 14.54 | – | 0.26 | – | 0.34 | 0.68 | 3.70 |
| – | – | – | – | – | 1.67 | 0.02 | 0.12 | – | – | 0.84 | 1.93 |
| – | – | – | – | – | 10.78 | 0.04 | 0.13 | – | – | 0.37 | 2.71 |
| – | 0.38 | 1.81 | 0.15 | 0.12 | 2.52 | 0.02 | 0.30 | 0.12 | 0.22 | 2.81 | 3.89 |
| – | 0.14 | 3.38 | 0.06 | 0.07 | 3.57 | 0.01 | 0.22 | 0.06 | 0.14 | 4.43 | 1.58 |
| – | 0.16 | 0.13 | 0.10 | 0.05 | 0.08 | 0.01 | 0.08 | 0.06 | 0.07 | 0.07 | 1.90 |
| – | 0.55 | 12.52 | 0.25 | 0.25 | 14.54 | 0.06 | 0.92 | 0.18 | 0.58 | 18.95 | 8.19 |
| 21 | 23 | 24 | 25 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.41 | – | 0.08 | 0.19 | 0.18 | 0.86 | 0.11 | 3.89 | – | 12.40 | – | 0.03 |
| 0.08 | 0.06 | 0.22 | 0.04 | 0.11 | 0.12 | 0.07 | 2.03 | 0.06 | 6.54 | 0.02 | 0.01 |
| 0.12 | – | 0.88 | 0.06 | 0.22 | 0.56 | 0.30 | 2.12 | – | 4.78 | – | 0.02 |
| 0.24 | 0.06 | 0.08 | 0.11 | 0.25 | 0.11 | 0.08 | 3.09 | 0.06 | 8.31 | 0.03 | 0.04 |
| 0.41 | 0.08 | 0.26 | 0.03 | 0.07 | 0.60 | 0.04 | 2.48 | 0.05 | 7.56 | 0.01 | 0.02 |
| 0.29 | 0.10 | 0.46 | 0.05 | 0.16 | 0.61 | 0.22 | 5.35 | – | 14.82 | – | 0.03 |
| 0.10 | – | 0.53 | 0.06 | 0.09 | 1.09 | 0.21 | 2.96 | 0.01 | 9.15 | – | 0.04 |
| 0.29 | – | 0.64 | 0.05 | 0.12 | 0.87 | 0.22 | 2.25 | – | 6.80 | – | 0.04 |
| 0.59 | – | 0.09 | 0.07 | 0.60 | 0.15 | 0.16 | 1.57 | – | 6.24 | – | 0.03 |
| 0.17 | – | 0.29 | 0.05 | 0.25 | 0.24 | – | 1.62 | – | 4.07 | – | 0.03 |
| 0.13 | – | 0.09 | 0.06 | 0.56 | – | 0.04 | 1.95 | 0.02 | 9.69 | – | 0.02 |
| 0.25 | – | 0.37 | 0.14 | 0.75 | – | 0.02 | 2.23 | 0.01 | 6.78 | – | 0.03 |
| 0.80 | – | 0.87 | 0.04 | 0.34 | – | 0.73 | 1.75 | – | 3.41 | – | 0.02 |
| 0.80 | – | 1.05 | 0.11 | 0.60 | – | 0.76 | 1.36 | – | 2.69 | – | 0.05 |
| 1.08 | – | 0.48 | 0.04 | 0.23 | – | 0.75 | 1.58 | – | 4.55 | 0.01 | 0.04 |
| 0.57 | – | 0.47 | 0.04 | 0.48 | – | 0.64 | 1.15 | – | 2.65 | – | 0.07 |
| 0.19 | 0.09 | – | 0.04 | 0.50 | – | 0.01 | 2.08 | – | 14.21 | 0.01 | 0.02 |
| 0.59 | 0.06 | 0.06 | 0.08 | 1.33 | – | 0.01 | 1.01 | – | 6.55 | – | 0.02 |
| 0.52 | 0.10 | – | 0.07 | 1.84 | 0.01 | 0.01 | 1.48 | – | 11.57 | – | 0.04 |
| 0.67 | 0.07 | – | 0.04 | 1.55 | – | 0.01 | 0.56 | – | 3.79 | – | 0.04 |
| 0.35 | 0.14 | 0.08 | 0.08 | 1.22 | – | 0.01 | 1.82 | – | 16.27 | – | 0.04 |
| 0.14 | 0.10 | – | 0.07 | 1.36 | – | 0.01 | 1.39 | – | 9.24 | – | 0.04 |
| 0.57 | 0.13 | – | 0.08 | 0.90 | 0.02 | – | 1.47 | – | 13.63 | 0.01 | 0.03 |
| 0.40 | 0.06 | 0.07 | 0.07 | 1.14 | 0.01 | 0.01 | 1.46 | – | 12.40 | – | 0.05 |
| – | – | – | – | – | – | – | – | – | – | – | – |
| 0.39 | 0.10 | 0.10 | 0.09 | 0.88 | 0.03 | 0.02 | 1.67 | – | 11.29 | – | 0.04 |
| 0.09 | 0.08 | – | 0.07 | 1.42 | 0.01 | – | 0.66 | – | 4.89 | – | 0.05 |
| 0.30 | 0.09 | 0.07 | 0.08 | 1.97 | – | – | 1.39 | – | 12.62 | – | 0.02 |
| 0.06 | 0.08 | 0.07 | 0.09 | 1.16 | – | – | 2.18 | 0.01 | 13.87 | – | 0.03 |
| 0.35 | 0.17 | 0.07 | 0.10 | 2.89 | – | 0.01 | 1.17 | 0.01 | 7.56 | – | 0.06 |
| 0.09 | 0.07 | – | 0.09 | 1.97 | – | – | 0.78 | – | 7.47 | – | 0.05 |
| 0.16 | 0.08 | 0.07 | 0.09 | 1.85 | – | 0.01 | 1.22 | 0.01 | 7.29 | – | 0.05 |
| 0.29 | 0.10 | 0.10 | 0.09 | 2.39 | 0.03 | 0.01 | 1.06 | 0.01 | 5.39 | – | 0.06 |
| 0.35 | 0.10 | 0.07 | 0.09 | 1.66 | – | 0.01 | 1.14 | 0.01 | 9.23 | 0.02 | 0.03 |
| 0.84 | 0.09 | 0.07 | 0.10 | 1.79 | – | 0.02 | 0.92 | – | 7.66 | – | 0.05 |
| 0.15 | 0.07 | 0.16 | 0.06 | 1.45 | – | – | 1.11 | – | 6.69 | – | 0.04 |
| 0.32 | 0.07 | – | – | 1.11 | – | – | 1.17 | – | 7.16 | – | 0.04 |
| 0.70 | 0.10 | 0.09 | 0.05 | 0.77 | 0.01 | 0.01 | 1.24 | – | 7.56 | – | 0.03 |
| 1.35 | – | – | 0.12 | 0.65 | – | – | 1.17 | – | 7.12 | – | 0.03 |
| 0.40 | 0.09 | 0.27 | 0.08 | 0.97 | 0.31 | 0.16 | 1.72 | 0.02 | 8.26 | 0.02 | 0.04 |
| 0.30 | 0.03 | 0.29 | 0.03 | 0.73 | 0.37 | 0.24 | 0.92 | 0.02 | 3.57 | 0.01 | 0.01 |
| 0.06 | 0.06 | 0.06 | 0.03 | 0.07 | 0.01 | 0.01 | 0.56 | 0.01 | 2.65 | 0.01 | 0.01 |
| 1.35 | 0.17 | 1.05 | 0.19 | 2.89 | 1.09 | 0.76 | 5.35 | 0.06 | 16.27 | 0.03 | 0.07 |
| Totals | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aliph | Indole | Benzyl | MT | MSO | AOP | Benzoxy | C3 | C4 | C7 | C8 | C4Per | C8Per |
| 29.20 | 0.41 | 0.19 | 18.09 | 5.38 | 4.30 | 1.18 | 0.18 | 6.58 | 4.45 | 17.15 | 0.973 | 0.794 |
| 18.79 | 0.08 | 0.04 | 13.39 | 2.93 | 1.74 | 0.39 | 0.91 | 5.60 | 2.31 | 9.01 | 0.860 | 0.796 |
| 31.03 | 0.12 | 0.06 | 14.64 | 2.35 | 12.60 | 1.10 | 0.53 | 19.61 | 2.45 | 6.80 | 0.974 | 0.735 |
| 19.70 | 0.24 | 0.11 | 13.49 | 3.97 | 1.31 | 0.57 | 0.25 | 3.07 | 3.60 | 11.72 | 0.925 | 0.765 |
| 41.23 | 0.41 | 0.03 | 12.64 | 5.55 | 21.61 | 0.79 | 0.07 | 23.63 | 3.33 | 12.11 | 0.997 | 0.784 |
| 34.91 | 0.31 | 0.05 | 22.25 | 7.18 | 4.00 | 1.02 | 0.16 | 6.13 | 6.27 | 20.83 | 0.975 | 0.769 |
| 35.99 | 0.10 | 0.06 | 15.65 | 6.86 | 11.49 | 1.44 | 0.17 | 15.43 | 3.73 | 15.14 | 0.989 | 0.802 |
| 31.12 | 0.29 | 0.05 | 14.58 | 5.21 | 9.56 | 1.25 | 0.69 | 14.71 | 2.75 | 11.36 | 0.855 | 0.805 |
| 18.12 | 0.62 | 0.07 | 8.61 | 3.21 | 5.36 | 0.94 | 1.16 | 4.77 | 1.79 | 9.23 | 0.804 | 0.838 |
| 28.94 | 0.19 | 0.05 | 9.44 | 3.06 | 15.92 | 0.52 | 0.74 | 18.82 | 1.96 | 6.79 | 0.962 | 0.776 |
| 26.21 | 0.13 | 0.06 | 15.83 | 4.85 | 4.76 | 0.64 | 1.01 | 7.36 | 2.27 | 14.22 | 0.879 | 0.862 |
| 31.37 | 0.25 | 0.14 | 22.77 | 6.74 | 1.05 | 0.81 | 1.98 | 13.85 | 2.88 | 12.06 | 0.875 | 0.807 |
| 22.53 | 0.82 | 0.04 | 17.73 | 2.64 | 1.07 | 1.09 | 0.60 | 13.76 | 1.97 | 5.31 | 0.958 | 0.729 |
| 33.87 | 0.86 | 0.11 | 24.13 | 5.44 | 2.89 | 1.41 | 1.37 | 23.78 | 1.91 | 5.45 | 0.946 | 0.740 |
| 20.18 | 1.11 | 0.04 | 14.49 | 2.87 | 1.79 | 1.03 | 0.92 | 9.74 | 1.74 | 7.04 | 0.914 | 0.802 |
| 23.61 | 0.58 | 0.04 | 17.11 | 3.51 | 1.80 | 1.19 | 0.95 | 15.52 | 1.46 | 5.14 | 0.942 | 0.779 |
| 26.52 | 0.19 | 0.04 | 16.79 | 7.10 | 2.03 | 0.54 | 1.04 | 0.27 | 2.37 | 21.02 | 0.206 | 0.899 |
| 28.21 | 0.59 | 0.08 | 11.15 | 4.34 | 11.36 | 1.36 | 9.62 | 0.42 | 1.16 | 9.84 | 0.042 | 0.895 |
| 28.07 | 0.52 | 0.07 | 14.21 | 4.99 | 6.97 | 1.90 | 5.26 | 0.19 | 1.67 | 16.37 | 0.035 | 0.907 |
| 27.26 | 0.69 | 0.04 | 11.53 | 4.13 | 10.00 | 1.60 | 12.08 | 0.40 | 0.67 | 6.40 | 0.032 | 0.905 |
| 40.85 | 0.35 | 0.08 | 28.07 | 8.17 | 3.34 | 1.27 | 13.49 | 0.73 | 2.16 | 22.70 | 0.051 | 0.913 |
| 26.34 | 0.14 | 0.07 | 14.17 | 6.90 | 3.86 | 1.41 | 6.60 | 0.12 | 1.67 | 15.31 | 0.018 | 0.902 |
| 28.17 | 0.59 | 0.08 | 16.14 | 8.47 | 2.60 | 0.96 | 2.40 | 0.40 | 1.75 | 21.82 | 0.143 | 0.926 |
| 31.38 | 0.40 | 0.07 | 20.25 | 6.91 | 3.01 | 1.21 | 9.31 | 0.38 | 1.74 | 18.23 | 0.039 | 0.913 |
| – | – | – | – | – | – | – | – | – | – | – | – | – |
| 22.46 | 0.41 | 0.09 | 14.58 | 4.34 | 2.47 | 0.97 | 2.53 | 1.30 | 1.84 | 15.46 | 0.339 | 0.894 |
| 20.34 | 0.09 | 0.07 | 9.91 | 2.91 | 6.04 | 1.48 | 7.99 | 0.22 | 0.78 | 7.35 | 0.027 | 0.904 |
| 25.37 | 0.32 | 0.08 | 16.74 | 4.76 | 1.88 | 1.99 | 5.94 | 0.48 | 1.55 | 17.22 | 0.075 | 0.917 |
| 27.80 | 0.06 | 0.09 | 17.30 | 5.13 | 4.17 | 1.20 | 6.02 | 0.41 | 2.38 | 18.80 | 0.064 | 0.888 |
| 27.65 | 0.35 | 0.10 | 9.56 | 3.92 | 11.20 | 2.97 | 14.32 | 0.08 | 1.31 | 11.34 | 0.006 | 0.896 |
| 24.93 | 0.09 | 0.09 | 11.44 | 3.49 | 7.98 | 2.02 | 12.78 | 0.36 | 0.89 | 10.66 | 0.027 | 0.923 |
| 16.86 | 0.16 | 0.09 | 9.62 | 2.06 | 3.26 | 1.92 | 5.52 | 0.56 | 1.31 | 9.26 | 0.092 | 0.876 |
| 34.55 | 0.29 | 0.09 | 13.98 | 3.91 | 14.16 | 2.50 | 23.86 | 0.84 | 1.22 | 8.36 | 0.034 | 0.873 |
| 25.42 | 0.37 | 0.09 | 12.22 | 3.00 | 8.47 | 1.73 | 11.33 | 0.49 | 1.22 | 12.15 | 0.041 | 0.909 |
| 17.30 | 0.86 | 0.10 | 9.82 | 2.38 | 3.24 | 1.86 | 5.41 | 0.72 | 1.00 | 9.96 | 0.117 | 0.909 |
| 17.63 | 0.15 | 0.06 | 9.81 | 3.75 | 2.58 | 1.49 | 5.71 | 0.30 | 1.28 | 10.07 | 0.050 | 0.887 |
| 38.26 | 0.32 | 0.00 | 23.62 | 9.54 | 3.95 | 1.15 | 24.84 | 0.68 | 1.43 | 10.86 | 0.027 | 0.884 |
| 17.09 | 0.72 | 0.05 | 11.50 | 2.05 | 2.72 | 0.82 | 5.00 | 0.86 | 1.36 | 9.49 | 0.147 | 0.875 |
| 24.81 | 1.39 | 0.12 | 19.44 | 3.46 | 1.23 | 0.68 | 13.28 | 0.37 | 1.30 | 9.83 | 0.027 | 0.883 |
| 26.95 | 0.41 | 0.07 | 15.18 | 4.67 | 5.73 | 1.27 | 5.68 | 5.60 | 2.02 | 12.15 | 0.436 | 0.852 |
| 6.57 | 0.31 | 0.03 | 4.67 | 1.92 | 4.83 | 0.56 | 6.34 | 7.44 | 1.10 | 4.92 | 0.435 | 0.062 |
| 16.86 | 0.06 | 0.00 | 8.61 | 2.05 | 1.05 | 0.39 | 0.07 | 0.08 | 0.67 | 5.14 | 0.006 | 0.729 |
| 41.23 | 1.39 | 0.19 | 28.07 | 9.54 | 21.61 | 2.97 | 24.84 | 23.78 | 6.27 | 22.70 | 0.997 | 0.926 |
Figure 3.
Effect of GS-OX on conversion of methylthioalkyl to methylsulfinylalkyl glucosinolates. Bar diagrams show the ratio of methylthioalkyl (MT) glucosinolate to methylsulfinylalkyl (MSO) glucosinolate content in leaf (A) and seeds (B) for each ecotype analyzed.
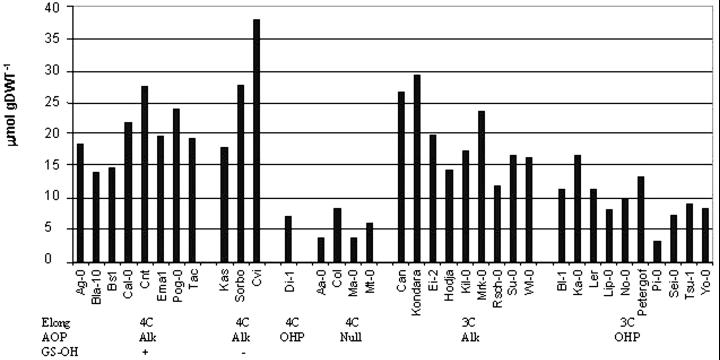
In addition to the variation in glucosinolate profile, there were dramatic differences in the total concentration of the Met-derived glucosinolates in leaves of the 39 ecotypes. There was a nearly 20-fold range between the ecotype with the highest accumulation, Cvi, with about 37.81 μmol g dry weight−1, and the lowest accumulating ecotype, Pi-0, with 2.83 μmol g dry weight−1 (Table I). Comparing the accumulation of aliphatic glucosinolates in conjunction with the allelic status of the inferred genes indicated that only the GS-Alk and GS-Ohp loci (collectively referred to as Alkohp) had significant control over accumulation of glucosinolates in Arabidopsis (Fig. 4). These two loci are tightly linked and are responsible for converting methylsulfinylalkyl glucosinolates into alkenyl (GS-Alk) or hydroxyalkyl (GS-Ohp) glucosinolates. Ecotypes that accumulate significant levels of the precursor methylsulfinylalkyl glucosinolate are null for GS-Alk and GS-Ohp reactions, and therefore can be classified as containing a third AOP allele (Kliebenstein et al., 2001). Further, mapping in mul-tiple ecotypes suggested that all three variants are due to different alleles at a single genetic locus (Kliebenstein et al., 2001). Therefore, we contrasted the GS-Alk, GS-Ohp, and _GS-AOP_null variants by ANOVA. Ecotypes with GS-Alk had aliphatic glucosinolates concentrations 2.5 times higher than ecotypes with GS-Ohp, and 4.0 times that of the _GS-AOP_null (F = 25.9, dffactor = 2, dferror = 32, and P = 2.1 × 10−7 by ANOVA). In addition, this locus or closely linked loci explained 61% of the variation among ecotypes for accumulation of leaf aliphatic glucosinolates. Previous reports have also shown that other enzymes involved in modifying glucosinolate chain structure can control glucosinolate accumulation (de Quiros, et al., 2000).
Figure 4.
GS-AOP regulates the accumulation of aliphatic glucosinolates in the leaves of Arabidopsis. The bars depict the average total aliphatic glucosinolate accumulation in leaves of 39 ecotypes. The ecotypes are classified based on the inferred genotype at three biosynthetic loci: GS-Elong, either C3- or C4-accumulating ecotypes; GS-AOP, alkenyl-, hydroxypropyl-, or methylsulfinylalkyl-containing ecotypes; and GS-OH, functional or nonfunctional alleles.
Genetic Control of Natural Variation in Glucosinolate Profiles and Concentrations in Arabidopsis Seeds
We analyzed the same collection of 39 ecotypes for accumulation of seed glucosinolates and identified 34 different glucosinolates (Table II). This included the 22 glucosinolates present in the leaves, plus benzoate esters of the three to eight carbon hydroxyalkyl and 2-hydroxy-3-butenyl glucosinolates (Hogge, et al., 1998). In addition to the polymorphisms previously described for the leaf tissue, we identified two new polymorphisms. In the ecotypes producing significant levels of four carbon aliphatic glucosinolates, there was variation for the accumulation of 4-hydroxybutyl glucosinolate. We named this new polymorphism GS-OHB.
We also identified a new allele of the GS-OH locus. The Kas and Sorbo ecotypes contain significant amounts of 2-hydroxy-3-butenyl glucosinolate in the seeds, but accumulate only minimal levels in the leaves, whereas Cvi produces no detectable amounts of this glucosinolate in either tissue. This suggests that there are three tissue-specific alleles of GS-OH: positive in leaf and seed, positive in seed only, and negative in all tissues (as displayed by Cvi). This is further supported by mapping data which indicates that all three phenotypic states of GS-OH are in fact due to alleles either at a single locus or closely linked loci (D.J. Kliebenstein, unpublished data).
Another difference in glucosinolate profiles between leaves and seeds is the ratio of methylthioalkyl to methylsulfinylalkyl glucosinolates. The leaves of most ecotypes typically contained at least twice the level of methylsulfinylalkyl as methylthioalkyl, except for three ecotypes (Bla-10, Can-0, and Su-0) that reversed this ratio (Fig. 3A). It is interesting that seeds of all ecotypes contained more methylthioalkyl than methylsulfinylalkyl, like the leaves of Bla-10, Can-0, and Su-0 (Fig. 3B).
Coordinate Control of Glucosinolate Accumulation
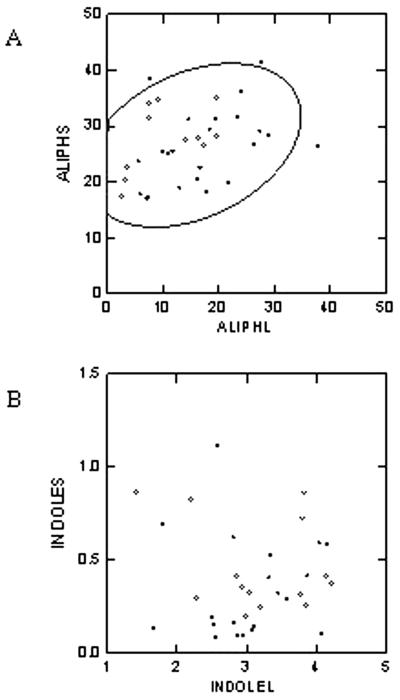
Although GS-AOP explained 61% of the variation in leaf aliphatic glucosinolates, this locus exhibited no significant association with concentration of seed aliphatic glucosinolates. This indicates that there may be different control mechanisms for the accumulation of glucosinolates in the two tissues. To test for coordinate regulation of seed and leaf glucosinolate accumulation, we compared the total accumulation of indolic or aliphatic glucosinolates between the two tissues. The levels of aliphatic glucosinolates in leaves and seeds of the different ecotypes showed a significant positive correlation (r = 0.35, P = 0.04, and n = 34; Fig. 5A). This suggests coordinate regulation of glucosinolate accumulation in these organs, which is presumably under the control of loci other than GS-AOP. One possible explanation is the transport of leaf glucosinolates into the seed. In contrast, the indolic glucosinolates showed no significant correlation between seed and leaf accumulation (r = −0.06, P = 0.70, and n = 34; Fig. 5B). Further, we observed no such negative correlations among the levels of the different glucosinolate classes (aliphatic, indolic, and aromatic) in either seeds or leaves (data not shown).
Figure 5.
Correlation of glucosinolate accumulation between the leaves and seeds. A, Scatter plot depicting the relationship between aliphatic glucosinolate accumulation in leaves (ALIPHL) and seeds (ALIPHS) of the ecotypes tested. The 90% confidence ellipse interval is drawn for reference. The values are in μmol g dry weight−1. B, Scatter plot depicting the relationship between indolic glucosinolate accumulation in the leaves (INDOLEL) and seeds (INDOLES) of the ecotypes tested. The values are in μmol g dry weight−1.
Genetics of Side-Chain Modification in Arabidopsis
We identified six aliphatic side chain modifications with apparent natural variation. These are: GS-OX (conversion of methylthioalkyl to methylsulfinylalkyl glucosinolate), GS-Alk (production of alkenyl glucosinolates), GS-Ohp (production of 3-hydroxypropyl glucosinolate), GS-null (absence of GS-Alk or GS-Ohp leading to the accumulation of methylsulfinylalkyl glucosinolates), GS-OHB (production of 4-hydroxybutyl glucosinolate), and GS-OH (production of 2-hydroxy-3-butenyl glucosinolate; Fig. 2). Previous analysis had shown that GS-Alk, GS-Ohp, and GS-AOPnull are allelic variants of a single genetic locus and that the three putative alleles of GS-OH are also all variants of a single locus (Parkin et al., 1994; Mithen et al., 1995; Giamoustaris, et al., 1996; Kliebenstein et al., 2001). We examined the newly described GS-OX polymorphism to see if the variation was caused by segregation at a single genetic locus. We used the ecotype, Wei-0, which has a GS-OX phenotype similar to Bla-10, Can-0, and Su-0 (data not shown). Wei-0 was crossed to L_er_, which is able to efficiently convert methylthioalkyl to methylsulfinylalkyl glucosinolates. HPLC analysis of the F1 progeny showed that _GS-OX_Wei-0 is recessive to _GS-OX_Ler (data not shown). HPLC analysis of 92 random F2 progeny showed that 71 individuals had the L_er_-like variant and 21 individuals had the Wei-0 phenotype. This is indistinguishable from a 3:1 segregation pattern and suggests that the GS-OX variation is due to segregation of a single genetic locus in this cross. The GS-OX locus mapped to the bottom of chromosome I, 3 cM teleomeric of the microsatellite AthGeneA and 20 cM centromeric from nga692 (Fig. 6). Further work is required to identify the gene responsible for this variation. In combination with the previously published data, this suggests that most of the natural variation in side-chain modifications in Arabidopsis can be explained by segregation of a small number of loci with large, discrete effects on glucosinolate profiles.
Figure 6.
Map of GS-OX on Chromosome I. Ninety-two F2 progeny were scored for the microsatellites indicated and for the GS-OX biochemical phenotype. The distance from the AthGeneA and nga692 markers to GS-OX is shown in cM to the right of the arrows. The approximate location of GS-OX is shown to the left of the chromosome.
Genetics of Chain Elongation in Arabidopsis
The elongation of the carbon side chain of the base amino acid is the first step in the biosynthesis of a majority of Arabidiopsis glucosinolates. Met-derived glucosinolates in Arabidopsis have side-chain lengths of three to eight carbon atoms (one to six additional carbon atoms) with the three- (C3) and four- (C4) carbon chain lengths being predominant (Hogge et al., 1988). Previous work showed that a single genetic locus (GS-Elong) determines the conversion of C3 to C4 aliphatic glucosinolates in Arabidopsis (Magrath et al., 1994). Map-based cloning studies have suggested that the gene responsible for this variation is a homolog of isopropylmalate synthase, which condenses acetyl-coenzyme A with an oxo acid (Fig. 1; de Quiros et al., 2000). It is interesting that plants with the C3 GS-Elong allele also accumulate glucosinolates with side chains of seven (C7) and eight carbon (C8) atoms (Tables I and II). This is unexpected because the biosynthesis of C7 and C8 glucosinolates is assumed to require the full series of shorter-chain intermediates (C3, C4, C5, and C6), one of which could also give rise to C4 glucosinolates (Fig. 1). Two models could account for this phenomenon: (a) The C3 GS-Elong allele does not block the chain elongation process, but resists diversion of metabolic flow at the C4 stage, and so channels intermediates toward longer chain length products; or (b) the C3 GS-Elong allele does block the chain elongation process prior to the formation of C4 intermediates, requiring the C7 and C8 glucosinolates to be produced in a different pathway.
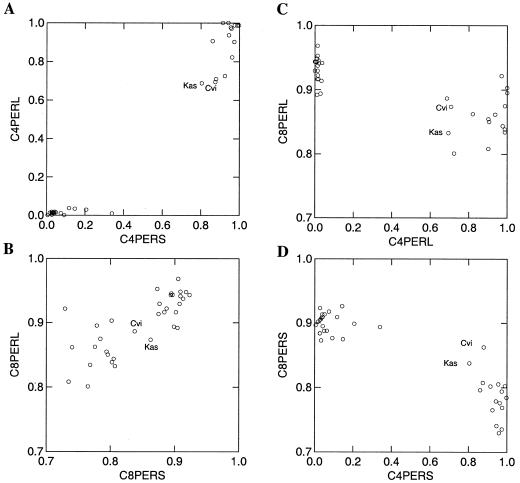
To differentiate between these two models, we compared the efficiency of conversion of C3 with C4 and C7 with C8 intermediates in glucosinolate biosynthesis. This was done by generating functions that measured the efficiency of each reaction. For the three- to four-carbon reaction, the function is C4per = C4/(C3 + C4). For the seven- to eight-carbon reaction, the function is C8per = C8/(C7 + C8). Both of these functions utilize the ratio of the accumulation of the precursor and product chain lengths as a gauge of reaction efficiency. As shown in Figure 7A, the efficiency of the C3 to C4 reaction is highly correlated between the leaf and seed tissues of the same ecotype (r = 0.98, P < 10−8, and n = 34). The same is also true for the C7 to C8 conversion (r = 0.77, P < 10−8, and n = 34; Fig. 7B). This implies that these reactions are under strict genetic control with minimal differences between the two tissues.
Figure 7.
Correlation of C7 to C8 and C3 to C4 intermediates in the biosynthesis of chain-elongated Met-derived glucosinolates. A, Scatter plot showing the correlation of the conversion of C3 to C4 in the seeds and leaves. B, Scatter plot showing the correlation of the conversion of C7 to C8 in the seeds and leaves. C, Scatter plot showing the correlation of the conversion of C3 to C4 with C7 to C8 in the leaves. D, Scatter plot showing the correlation of the conversion of C3 to C4 with C7 to C8 in the seeds.
Furthermore, the efficiency of C3 to C4 conversion showed a strong negative correlation with the C7 to C8 conversion efficiency (for leaf, r = −0.78, P < 10−8, and n = 34; for seed, r = −0.92, P < 10−8, and n = 34; Fig. 7, C and D). Ecotypes with higher C3 ratios had lower C7 ratios (Fig. 7C and D). Thus, ecotypes containing the GS-Elong C3 allele are more efficient at elongating C7 intermediates to C8 intermediates than ecotypes containing the GS-Elong C4 allele (Fig. 7, C and D). This correlation is independent of the total level of C3, C4, C7, or C8 glucosinolates. The involvement of the GS-Elong C3 allele in altering the production of C7 and C8 glucosinolates supports the hypothesis that this allele does not block chain elongation after C3 but instead prevents the diversion of metabolites at the C4 chain length.
DISCUSSION
Modular Variation in Glucosinolate Profiles
One possible effect of heterogeneous natural selection on plant defense products is the rapid evolution of new compounds, new mixtures of compounds, or new patterns of gene regulation controlling compound accumulation. New compounds may increase resistance to herbivores that have become adapted to existing defenses, whereas mixtures may provide a unique complement of defenses, retarding counteradaptation of enemies. In this study, the 39 ecotypes sampled could be divided into seven different classes, each with its own unique mixture of leaf glucosinolates (Table III). These seven different classes are created by variation at only three of the five identified genetic loci, GS-Elong, GS-AOP, and GS-OH (Fig. 1 and 2 and Table III). If the GS-OX locus is also used to separate the glucosinolate profiles, Arabidopsis theoretically could have up to 14 different glucosinolate profiles. Thus, a small set of polymorphic loci in Arabidopsis generates a modular alteration in glucosinolate profile. This may enable rapid responses to changing selective pressures and could allow evolution in response to altered herbivore abundance.
Table III.
Major genetic loci controlling biosynthesis of glucosinolates in Arabidopsis
| Ecotype | Elong | AOP | OX | OH | Profile |
|---|---|---|---|---|---|
| Ag-0 | 4 | 3 | 2 | 3 | |
| Bla-10 | 4 | 3 | 1 | 3 | |
| Bs-1 | 4 | 3 | 2 | 3 | Allyl |
| Cal-0 | 4 | 3 | 2 | 3 | Butenyl |
| Cnt | 4 | 3 | 2 | 3 | HydroxyButenyl |
| Ema-1 | 4 | 3 | 2 | 3 | |
| Tac | 4 | 3 | 2 | 3 | |
| Pog-0 | 4 | 3 | 2 | 3 | |
| Sorbo | 4 | 3 | 2 | 2 | |
| Kas | 4 | 3 | 2 | 2 | Allyl |
| Cvi | 4 | 3 | 2 | 1 | Butenyl |
| Di-1 | 4 | 2 | 2 | – | OHPropyl/4MSO |
| Aa-0 | 4 | 1 | 2 | – | |
| Col-0 | 4 | 1 | 2 | – | 4MSO-butyl |
| Ma-0 | 4 | 1 | 2 | – | |
| Mt-0 | 4 | 1 | 2 | – | |
| Can-0 | 3 | 3 | 1 | – | |
| Kondara | 3 | 3 | 2 | – | |
| Ei-2 | 3 | 3 | 2 | – | |
| Hodja | 3 | 3 | 2 | – | |
| Ita-0 | 3 | 3 | 2 | – | Allyl |
| Kil-0 | 3 | 3 | 2 | – | |
| Mr-0 | 3 | 3 | 2 | – | |
| Mrk-0 | 3 | 3 | 2 | – | |
| Su-0 | 3 | 3 | 1 | – | |
| Wi-0 | 3 | 3 | 2 | – | |
| Rsch-0 | 3 | 3 | 2 | – | |
| Bl-1 | 3 | 2 | 2 | – | |
| Di-g | 3 | 2 | 2 | – | |
| Ka-0 | 3 | 2 | 2 | – | |
| Ler | 3 | 2 | 2 | – | |
| Lip-0 | 3 | 2 | 2 | – | OHpropyl |
| No-0 | 3 | 2 | 2 | – | |
| Pet | 3 | 2 | 2 | – | |
| Pi-0 | 3 | 2 | 2 | – | |
| Sei-0 | 3 | 2 | 2 | – | |
| Tsu-1 | 3 | 2 | 2 | – | |
| Yo-0 | 3 | 2 | 2 | – | |
| Oy-0 | 3 | 1 | 2 | – | 3MSOpropyl |
Arabidopsis undoubtedly contains additional glucosinolates (and polymorphic loci controlling their formation) that remain to be discovered. Several minor peaks in our extracts were not identified. Furthermore, because only 39 ecotypes were analyzed and several glucosinolates were only found in a single accession (Tables I and II), much of the variation remains to be sampled. Given the modular nature of glucosinolate profiles in Arabidopsis, a single new locus could produce a whole series of novel glucosinolate chemotypes.
Large Variation in the Level of Glucosinolate Accumulation
In addition to altering the types of glucosinolates, heterogeneous natural selection might also favor a broad range of glucosinolate concentrations. In the leaves of the 39 ecotypes tested, there was a 20-fold difference in the total concentration of aliphatic glucosinolates (Table I and Fig. 3). More than 60% of this variation was due to the GS-AOP locus. However, even after accounting for the effect of GS-AOP, there is still a large amount of variation that is due to other factors. In the seeds of these ecotypes, the high and low ecotypes showed only a 3-fold range of aliphatic glucosinolate concentrations (Table II). This again suggests that independent factors regulate the accumulation of aliphatic glucosinolates in different tissues. Quantitative trait locus mapping of the loci influencing the accumulation of aliphatic glucosinolates in Arabidopsis will enable us to identify the factors regulating the difference between leaf and seed accumulation.
Aliphatic glucosinolate concentration was more variable in leaves than in seeds, whereas indolic glucosinolate variation showed the opposite pattern. The indolic glucosinolates exhibited only a 3-fold quantitative difference from high to low ecotype in the leaf, but had a 20-fold range from high to low ecotype in the seed (Tables I and II). This finding suggests that indolic glucosinolate accumulation in the leaves and seeds is under different modes of regulation. Further, this regulation is separate from that controlling the aliphatic glucosinolates. This could be a possible consequence of the different herbivore pressures on seeds and leaves and the fact that aliphatic and indole glucosinolates have different effects on herbivores (Bartlet et al., 1994).
Multiple Effects of GS-Elong on Aliphatic Glucosinolate Chain Elongation
Previous analysis had identified two different alleles of GS-Elong in Arabidopsis. One allele was associated with the accumulation of C3 side-chain aliphatic glucosinolates and the other with C4 side-chain glucosinolates (Fig. 1). This led to the hypothesis that ecotypes containing the C3 allele are blocked in their ability to elongate the side chain past three carbons, and thus lack the potential to produce C4 glucosinolates (Magrath et al., 1994). However, ecotypes with both alleles are able to produce the C7 and C8 side-chain glucosinolates, which theoretically require the C4 intermediate. In our ecotype analysis, we found that the allelic status at GS-Elong controls the production of C7 and C8 glucosinolates in addition to C3 and C4 glucosinolates. Ecotypes with the C3 allele contain a higher ratio of C8 to C7 glucosinolates than the C4 allele (Fig. 7). This close correlation between C3 and C4 glucosinolates and C7 and C8 glucosinolates suggests that all of these glucosinolates are formed in the same pathway. This linkage could be explained alternatively by the biosynthesis of the different carbon length glucosinolates occurring via separate pathways employing some of the same enzymatic machinery or employing enzymes encoded by closely linked genes. It is interesting that the Kas and Cvi ecotypes are intermediate in all of the distributions examined (Fig. 7). This could be explained by either an intermediate GS-Elong allele or a modifying second locus. Further map-based cloning of this function will help to elucidate this difference.
Future Work
Our survey of the natural variation in glucosinolate composition of 39 ecotypes of Arabidopsis revealed variation in seven distinct side-chain modifications of aliphatic glucosinolates representing five polymorphic loci.We have already begun to use this variation to clone the GS-OX and GS-OH loci as well as to identify and clone quantitative trait loci controlling glucosinolate concentration. In addition, we are investigating the susceptibility of these 39 ecotypes to insects and pathogens to explore the importance of glucosinolates in plant defense. The study of natural variation in Arabidopsis provides a valuable set of tools for answering questions about the biosynthesis, evolution and function of these interesting natural products.
MATERIALS AND METHODS
Plant Growth
One hundred plants were grown in 3.25- × 3.25- × 2.25-inch pots at 18 pots to a flat for 3 weeks in a standard soil/vermiculite mixture. They were placed 10 inches from 4 × 60-W cool-white bulbs and 4 × 60-W wide-spectrum bulbs (GE, Fairfield, CT) in a 16-h-light, 8-h-dark photoperiod. At 3 weeks, the tissue was harvested and the leaf material was lyophilyzed. Plant growth continued for seed production, and seeds were harvested after 2 months. Ten milligrams of leaf material and 5 mg of seeds were used for the glucosinolate extraction. All samples and ecotypes were done in triplicate.
Ecotypes
This study analyzed the following ecotypes (with abbreviation, location, and Nottingham Stock Center no.): Aa-0, Rhon, Germany, N900; Ag-0, Argentat, France, N901; Bla-10, Bl-1, Bologna, Italy, N968; Bs-1, Basel, N996; Cal, Calver, UK, N1062; Can, Canary Islands, N1064; Cnt, Canterbury, UK, N1635; Col-0, Columbia, MO, N1092; Cvi, Cape Verde Islands, N1096; Di-1, Dijon, France, N1108; Di-g, Dijon, France, N910; Ei-2, Eifel, Germany, N1124; Ema-1, East Malling, UK, N1637; Hodja, Khurmatov, Tadjikistan, N922; Ita-0, Ibel Tazekka, Morocco, N1244; Ka-0, Karnten, Austria, N1266; Kas, Kasmir, India, N903; Kil-0, Killean, UK, N1270; Kondara, Khurmatov, Tadjikistan, N916; L_er_, Landsberg, Germany, N1642; Lip-0, Lipowiec, Poland, N1336; Ma-0, Marburg, Germany, N1356; _M_r-0, Monte Tosso, Italy, N1372; Mrk-0, Markt, Germany, N1374; Mt, Martuba, Libya, N1380; No, N1394; Oy-0, Oystese, Norway, N1643; Petergof, Russia, N926; Pi-0, Pitztal, Austria, N1456; Pog-0, Point Gray, BC, N1476; Rsch-0, Rschew, Russia, N1490, Sei-0, Seis am Schlern, Italy, N1504; Su-0, Southport, UK, N1540;Tsu-1, Tsu, Japan, N1640; Wl-0, Wildbad, Germany, N1630; and Yo-0, Yosemite, CA, N1622.
96-Well Glucosinolate Extraction and Purification
This purification technique follows the basic sephadex/sulfatase Arabidopsis protocol previously described (Hogge et al., 1988). Samples were harvested into deep-well micro-titer tubes (either 10 fresh leaf discs frozen in liquid nitrogen, 10 mg freeze-dried leaf material, or 5 mg dried seeds). Four 2.3-mm ball bearings were added and the samples ground into a fine powder in a paint shaker by high speed agitation. To extract glucosinolates, 400 μL of methanol, 10 μL of 0.3 m lead acetate, and either 120 μL of water for seeds and freeze-dried material or 80 μL of water for fresh tissue was added. The samples were mixed for 1 min in the paint shaker and allowed to incubate for 60 min at 180 rpm on a rotary shaker. The tissue and protein were pelleted by centrifuging for 10 min at 2,500_g_ and the supernatant used for anion-exchange chromatography. Ninety-six well filter plates (Millipore, Tempe, AZ, catalogue no. MAHVN4550) were loaded with 45 μL of diethylaminoethyl Sephadex A-25 using the Millipore multiscreen column loader. Then 300 μL of water was added and allowed to equilibrate for 2 to 4 h. After water was removed with 2 to 4 s of vacuum on the vacuum manifold (Qiagen, Valencia, CA), 150 μL of the supernatant was added to the 96-well columns and the liquid removed by 2 to 4 s of vacuum. This step was repeated once to bring the total volume of plant extract to 300 μL. The columns were washed four times with 150 μL of 67% (v/v) methanol, three times with 150 μL of water, and three times with 150 μL of 1 m sodium acetate. To desulfate glucosinolates on the column, 10 μL of water and 10 μL of sulfatase solution was added to each column and the plates incubated overnight at room temperature (Hogge et al., 1988). Desulfo-glucosinolates were eluted by placing a deep-well 2-mL 96-well plate in the bottom of the 96-well vacuum manifold and aligning the 96-well column plate. The DEAE Sephadex was then washed twice with 100 μL of 60% (v/v) methanol and twice with 100 μL of water.
HPLC
Forty microliters of the glucosinolate extract was run on a 5-μm column (Lichrocart 250–4 RP18e, Hewlett-Packard, Waldbronn, Germany) on a Hewlett-Packard 1100 series HPLC. Compounds were detected at 229 nm and separated utilizing either of the two following programs with aqueous acetonitrile. For seeds, the program was an 8-min gradient from 1.5% to 5.0% (v/v) acetonitrile, a 2-min gradient from 5% to 7% (v/v) acetonitrile, a 32-min gradient from 7% to 52% (v/v) acetonitrile, a 2-min gradient from 52% to 92% (v/v) acetonitrile, 5 min at 92% (v/v) acetonitrile, a 3-min gradient from 92% to 1.5% (v/v) acetonitrile, and a final 8 min at 1.5% (v/v) acetonitrile. For leaf material, the program was a 6-min gradient from 1.5% to 5.0% (v/v) acetonitrile, a 2-min gradient from 5% to 7% (v/v) acetonitrile, a 7-min gradient from 7% to 25% (v/v) acetonitrile, a 2-min gradient from 25% to 92% (v/v) acetonitrile, 6 min at 92% (v/v) acetonitrile, a 1-min gradient from 92% to 1.5% (v/v) acetonitrile, and a final 5 min at 1.5% (v/v) acetonitrile.
Glucosinolate Identification and Quantification
All glucosinolates had been previously isolated and identified Arabidopsis (P. Brown and T. Gershenzon, unpublished data; Hogge et al., 1988). The identity of HPLC peaks was based on a comparison of retention time and UV absorption spectrum as deterimined on a diode-array detector with those of standards. Results are given as μmol g dry weight−1 tissue calculated from HPLC peak areas using response factors computed for pure de-sulfo glucosinolate standards at A229 nm (P. Brown and T. Gershenzon, unpublished data). Each ecotype was run in triplicate. The list of compound identities is as follows: 1, 3-hydroxypropyl; 2, 3- methylsulfinylpropyl; 3, 4-hydroxybutyl; 4, 2(S)-hydroxy-3-butenyl; 5, 4-methylsulfinylbutyl; 6, 2(R)-hydroxy-3-butenyl; 7, allyl; 8, 2-methylthioethyl; 9, 5-methylsulfinylpentyl; 10, 2-hydroxy-4-pentenyl; 11, 3-butenyl; 12, 1-methylethyl; 13, 6-methylsulfinylhexyl; 14, 3-methylthiopropyl; 15, 4-hydroxy-indolyl-3-methyl; 16, 7-methylsulfinylheptyl; 17, 2-methylpropyl; 18, 4-pentenyl; 19, 4-methylthiobutyl; 20, 8-methylsulfinyloctyl; 21, indolyl-3-methyl; 22, 4-methoxy-indolyl-3-methyl; 23, 5-methylthiopentyl; 24, 6-methylthiohexyl; 25, benzyl; 26, 1-methoxy-indolyl-3-methyl; 27, 3-benzoyloxypropyl; 28, 2-benzoyloxy-3-butenyl; 29, 4-benzoyloxybutyl; 30, 7-methylthioheptyl; 31, 5-benzoyloxypentyl; 32, 8-methylthiooctyl; 33, 6-benzoyloxyhexyl; and 34, 8-benzoyloxyoctyl.
Statistics
Means are given for each ecotype. Systat (version 7) was utilized for statistical analysis. ANOVA utilized for GS-AOP is as follows: leaf aliphatic glucosinolate = constant + GS-AOP.
Mapping and Microsatellites
DNA was isolated in a 96-well format as previously described (Kliebenstein et al., 2001). Five microliters of the resuspended DNA was added to 20 μL of PCR reaction mixture (2.5 mm MgCl2, 200 pm primers, and 0.5 units TAQ polymerase) containing primers for the microsatellite markers listed. Microsatellite primer sequences were obtained from The Arabidopsis Information Resource (www.Arabidopsis.org). The reactions were run with the following cycle program: 95°C for 3 min; 40 cycles of 95°C at 20 s, 56°C at 20 s, and 72°C for 1 s; and 72°C for 3 min and 4°C final). The polymorphisms were scored on 4% (w/v) agarose. The allelic state at the GS-OX locus was scored by analyzing the production of 3-methylthiopropyl glucosinolate by HPLC as described above. Mapping of the markers and GS-OX utilized Mapmaker (version 3; Lander et al., 1987).
ACKNOWLEDGMENT
We thank Michael Reichelt for assistance with glucosinolate quantification.
LITERATURE CITED
- Bartlet E, Parsons D, Williams I, Clark S. The influence of glucosinolates and sugars on feeding by the cabbage stem flea beetle, Psylliodes chrysocephala. Entomol Exp Appl. 1994;73:77–83. [Google Scholar]
- Chisholm MD, Wetter LR. Biosynthesis of mustard oil glucosides: IV. The administration of methionine-14C and related compounds to horseradish. Can J Biochem. 1964;42:1033–1040. doi: 10.1139/o64-114. [DOI] [PubMed] [Google Scholar]
- Daxenbichler ME, Spencer GF, Carlson DG, Rose GB, Brinkler AM, Powell RG. Glucosinolate composition of seeds from 297 species of wild plants. Phytochemistry. 1991;30:2623–2638. [Google Scholar]
- de Quiros HC, Magrath R, McCallum D, Kroymann J, Schnabelrauch D, Mitchell-Olds T, Mithen R. α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet. 2000;101:429–437. [Google Scholar]
- Giamoustaris A, Mithen R. The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica Napus Ssp Oleifera) on its interaction with specialist and generalist pests. Ann Appl Biol. 1995;126:347–363. [Google Scholar]
- Giamoustaris A, Mithen R. Genetics of aliphatic glucosinolates: 4. Side-chain modification in Brassica oleracea. Theor Appl Genet. 1996;93:1006–1010. doi: 10.1007/BF00224105. [DOI] [PubMed] [Google Scholar]
- Graser G, Schneider B, Oldham N, Gershenzon J. The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae) Arch Biochem Biophys. 2000;378:411–419. doi: 10.1006/abbi.2000.1812. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Du L. The biosynthesis of glucosinolates. Trends Plant Sci. 1997;2:425–431. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Hogge LR, Reed DW, Underhill EW, Haughn GW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chromatography-mass spectometry. J Chromatgr Sci. 1988;26:551–556. [Google Scholar]
- Kliebenstein D, Lambrix V, Reichelt M, Gershnzon J, Mitchell-Olds T. Gene duplication and the diversification of secondary metabolism: side chain modification of glucosinolates in Arabidopsis thaliana. Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Magrath R, Bano F, Morgner M, Parkin I, Sharpe A, Lister C, Dean C, Turner J, Ludiate D, Mithen R. Genetics of aliphatic glucosinolates: I. Side chain elongation in Brassica napus and Arabidopsis thaliana. Heredity. 1994;72:290–299. [Google Scholar]
- Mauricio R. Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat. 1998;151:20–28. doi: 10.1086/286099. [DOI] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution. 1997;51:1435–1444. doi: 10.1111/j.1558-5646.1997.tb01467.x. [DOI] [PubMed] [Google Scholar]
- Mithen R, Clarke J, Lister C, Dean C. Genetics of aliphatic glucosinolates: 3. Side chain structure of aliphatic glucosinolates in Arabidopsis thaliana. Heredity. 1995;74:210–215. [Google Scholar]
- Parkin I, Magrath R, Keith D, Sharpe A, Mithen R, Lydiate D. Genetics of aliphatic glucosinolates: II. Hydroxylation of alkenyl glucosinolates in Brassica napus. Heredity. 1994;72:594–598. [Google Scholar]
- Rodman J. Population variation and hybridization in sea-rockets (Cakile, Cruciferae): seed glucosinolate characters. Am J Bot. 1980;67:1145–1159. [Google Scholar]
- Stowe KA. Experimental evolution of resistance in Brassica rapa: correlated response of tolerance in lines selected for glucosinolate content. Evolution. 1998a;52:703–712. doi: 10.1111/j.1558-5646.1998.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Stowe KA. Realized defense of artificially selected lines of Brassica rapa: effects of quantitative genetic variation in foliar glucosinolate concentration. Env Entomol. 1998b;27:1166–1174. [Google Scholar]
- Wittstock U, Halkier BA. Cytochrome P450CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of L-phenyl alanine to phenylacetaldoxime in the biosynthesis of thebenzylglucosinolate. J Biol Chem. 2000;275:14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]