Structure and Genome Organization of AFV2, a Novel Archaeal Lipothrixvirus with Unusual Terminal and Core Structures (original) (raw)
Abstract
A novel filamentous virus, AFV2, from the hyperthermophilic archaeal genus Acidianus shows structural similarity to lipothrixviruses but differs from them in its unusual terminal and core structures. The double-stranded DNA genome contains 31,787 bp and carries eight open reading frames homologous to those of other lipothrixviruses, a single tRNALys gene containing a 12-bp archaeal intron, and a 1,008-bp repeat-rich region near the center of the genome.
The double-stranded (ds) DNA viruses of hyperthermophilic Crenarchaeota exhibit remarkably diverse morphotypes and genome structures and, on the basis of these, they have been assigned to five new viral families: spindle-shaped Fuselloviridae, filamentous Lipothrixviridae, rod-shaped Rudiviridae, droplet-shaped Guttaviridae, and spherical Globuloviridae (8, 16, 17).
The Lipothrixviridae is, so far, the most diverse family with respect to phenotype and genotype, and it has been subdivided into three genera (22): Alphalipothrixvirus, containing TTV1, TTV2, and TTV3 from Iceland (10), Betalipothrixvirus with SIFV from Iceland (2), and Gammalipothrixvirus represented by AFV1 from Yellowstone National Park (3). Here we describe the structural and genomic properties of a novel member of the lipothrixviral family, AFV2, isolated from a hot, acidic spring (93°C, pH 2) in a solfataric field in Italy. We propose assignment to a new genus of this family.
Virus purification.
A sample collected at Pozzuoli was enriched at 75°C, pH 3, as described earlier for thermophilic cultures (18, 26). A novel species was isolated from the enrichment which belongs to the crenarchaeal genus Acidianus, Acidianus sp. strain F28, by plating on Gelrite (Kelco, San Diego, Calif.) containing colloidal sulfur (3). A filamentous virus was observed in the late exponential culture using transmission electron microscopy. Virions were purified from the supernatant of a cell-free culture by precipitating with polyethylene glycol 6000 (3). They yielded a sharp, bluish-white, opalescent band with a buoyant density of about 1.3 g/ml in a CsCl gradient. The virus was termed Acidianus filamentous virus 2, AFV2.
Virion structure.
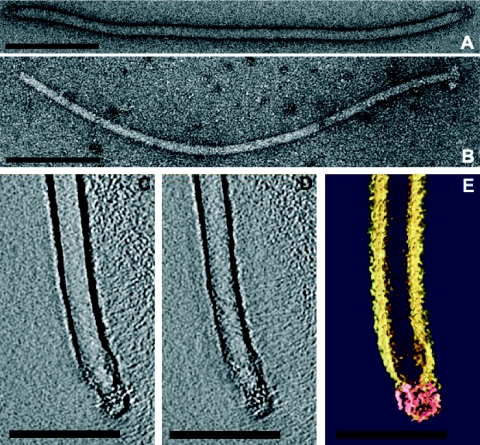
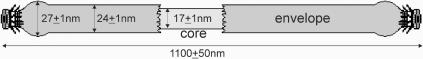
Filamentous virions of AFV2 are 1,100 ± 50 nm long and 24 ± 1 nm wide (Fig. 1A), and morphotypically they resemble members of the family Lipothrixviridae. Moreover, on treatment with 0.1% sodium dodecyl sulfate (SDS) for 1 min, the virion width was reduced to 17 ± 1 nm (Fig. 1B) and an envelope was observed encasing a viral core, as occurs for all known lipothrixviruses. However, the structure of the viral core exhibited no regular structure on its surface and, thereby, differs from both the nucleoprotein complex of the alphalipothrixvirus TTV1 (27) and the nucleosome-like arrangement of the betalipothrixvirus SIFV (2).
FIG. 1.
Electron micrographs of virions of AFV2 negatively stained with 3% uranyl acetate. A, Native virion. B, Virion after treatment with 0.1% SDS for 1 min. C to E, 3D reconstruction of the termini of a native virion. A and B, Two different horizontal slices (0.7 nm) through the 3D data set. C, Color-coded representation of 3D structure. Pink, virion termini; yellow, negative stain encasing the virion. Bars, 200 nm (A and B) or 100 nm (C to E).
In order to characterize the termini, virions were analyzed by electron tomography (7). For this, negatively stained samples were mounted in a high-tilt grid holder (6) and a tilt series was recorded at a calibrated (19) magnification of ×34,000 or ×44,000 at about −1 μm defocus, at room temperature under low-dose conditions, with a tilt range of −70° to +70° with a 2° increment. After aligning the tilted projections, the three-dimensional (3D) reconstruction was performed by weighted back-projection using an EM software package (9). Visualization of the 3D volume was done using the Amira software package (Mercury Computer Systems, Düsseldorf, Germany). The results showed that each virion end constitutes a complex collar with two sets of filaments, resembling a bottle brush with a solid round cap 17 nm in diameter on its top (Fig. 1C to E). This terminal structure, which is modeled schematically on the whole virion in Fig. 2, has not been previously observed in a virus.
FIG. 2.
Scheme of the structure of an AFV2 virion.
It seems likely that the virion termini participate in cellular adsorption and, since the host cells are devoid of pilus-like structures, it is likely they attach directly to the cell surface (2). However, extensive efforts to provide evidence for the direct attachment of AFV2 virions to host cells using electron microscopy failed.
Virion composition.
Seven proteins with apparent molecular masses of 6, 26, 35, 40, 45, 50, and 65 kDa were detected in AFV2 virions using SDS-polyacrylamide gel electrophoresis (21) (not shown). No lipids were detected after extracting with chloroform-methanol (volume ratio, 1:1) and subjecting to thin-layer chromatography using conditions identical to those that yielded lipids from virions from other crenarchaeal viruses (2, 3, 8, 20).
Host range and virus-host relationships.
The host range of AFV2 was tested by attempting to propagate the virus in species related to its natural host. Purified virus was added to cultures of Acidianus infernus, Acidianus brierleyi, Acidianus ambivalens, “_Acidianus hospitalis_” (3), Sulfolobus solfataricus, and Sulfolobus islandicus, but no viral propagation was observed by electron microscopy in supernatants of these cultures.
On production of AFV2, neither cell debris nor a decrease in cell density was observed in the growing culture of Acidianus sp. strain F28, and we inferred, therefore, that replication of the virus did not produce cell lysis. Moreover, infected host cells were not cured of the virus after five successive transfers into fresh medium (dilution, 1:1,000) and continuous growth for 2 months. Furthermore, treatment of infected cells with mitomycin C (0.5 μg/ml) or UV irradiation did not increase virus production when monitored by electron microscopy. Thus, in its virus-host relationship, AFV2 resembles other known viruses of hyperthermophilic and acidophilic archaea which exist intracellularly in a stable carrier state and are nonlytic.
Genome organization.
Nucleic acid isolated from the virions (3) was insensitive to RNase A but digestible by type II restriction endonucleases, consistent with it being dsDNA. Therefore, a clone library was prepared in pUC18 and the genome was sequenced with an approximately fivefold coverage, as described earlier (8). Remaining sequence ambiguities were resolved by generating and sequencing PCR fragments amplified from appropriate regions of the viral DNA. Terminal regions of the viral genome were not represented in the clone library, as we have observed for other linear viral genomes (3, 8, 13, 14). Given the low amounts of genomic DNA that were available (<1 μg purified DNA), the ends (1 to 2 kb at each end) were sequenced by primer walking on AFV2 DNA amplified with the GenomePhi kit (Amersham Biotech), and the termini were defined as the points at which sequence reads reproducibly ended. No inverted terminal repeats or other symmetrical sequences were detected. The total sequence was 31,787 bp with an A+T content of 64%.
An open reading frame (ORF) map was constructed using MUTAGEN (5), and start codons (79% AUG, 11% GUG, and 9% UUG), TATA-like promoter motifs, and/or Shine-Dalgarno motifs were assigned as described earlier (8) (see Table S1 in the supplemental material).
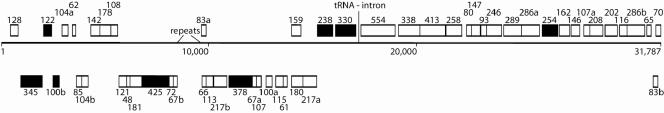
A map for the whole viral genome containing 51 putative ORFs (>40 amino acids) is presented in Fig. 3. About 70% of these are arranged in putative operons (see Table S1 in the supplemental material), and about 40% of the genes are predicted to generate transcripts that are leaderless or carry very short leaders (3, 23, 24). Searches against public sequence databases (1) revealed eight ORFs which are homologous to other lipothrixviral ORFs from the genera Sulfolobus and Acidianus (Fig. 3; see also the supplemental material), but only three genes were assigned functions on the basis of sequence matches with public databases (Table 1).
FIG. 3.
Genome map of AFV2 showing the location and size of the putative genes present on the two DNA strands. Genes are expressed from left to right in the upper row and from right to left in the lower row. ORF homologs that are shared with other Acidianus or Sulfolobus viruses AFV1, SIFV, SIRV1, and SIRV2 are represented by filled rectangles.
TABLE 1.
Functions assigned to ORFs of AFV2
| ORF | GenBank best match | e-value | Database match | e-value |
|---|---|---|---|---|
| 104b | pNOB8, ORF72 | 1e-02 | CopG family | 7.2e-04 |
| zf-C2H2 | 6.3e-01 | |||
| 330 | AFV1, ORF313 | 4e-18 | Glycosyl transferase 1 | 3.1e-03 |
| 378 | SIFV, ORF267 | 7e-12 | Glycosyl transferase 1 | 6.4e-10 |
A tRNALys gene is present in the central part of the genome (Fig. 3), the first to be detected in a crenarchaeal viral genome, and it contains a 12-bp archaeal intron which can generate a “bulge-helix-bulge” splicing junction, where host-encoded splicing enzymes act (11). This intron is smaller than other known tRNA introns (11) but has the same size as putative introns discovered in ORFs of the rudiviruses SIRV1 and SIRV2 (14). The AAG codon of tRNALys is one of two used by the Acidianus viruses for lysine. Moreover, it is used approximately 40% more frequently in AFV2 than in the other known Acidianus virus, ARV1, with the same host specificity (25), suggesting a possible rationale for the presence of this tRNALys gene in the AFV2 genome.
The genome contains an unusual 1,008-bp region extending from positions 8608 to 9615 flanked by ORF67 upstream and by ORF66 and ORF83 downstream. It carries two large 46-bp direct repeats and multiple imperfect short repeats throughout the region, and its base composition is strongly biased, with one DNA strand containing only 7% guanosines. The region is bordered by the inverted repeat GTCACTGACATAATA.
The lack of detectable sequence symmetry between the terminal regions of the linear AFV2 genome contrasts with the genomes of the crenarchaeal rudiviruses, which carry large inverted terminal repeats and apparently replicate by a self-priming mechanism generating head-to-head and tail-to-tail intermediates (13). A very short, 11-bp-long inverted repeat, CG10, is present at the termini of the gammalipothrixvirus AFV1 (3). The terminal repeat at each end is preceded by a 300-bp region consisting of many direct repeats of the pentanucleotide TTGTT or close variants thereof, resembling the arrangement of telomeric ends of linear eukaryal chromosomes (3). The replication mechanism for this virus remains unclear. For the alphalipothrixvirus TTV1 (12) and the betalipothrixvirus SIFV (2), the terminal sequences of their genomes were not determined, although recent partial sequencing of the SIFV genomic ends provided evidence for a large inverted terminal repeat (13).
Considering the likely diversity of replication mechanisms for linear double-stranded DNA genomes of crenarchaeal viruses, we cannot exclude that replication of AFV2 is initiated internally in the genome, as has been observed for some bacterial linear genomes (15). If so, then the 1,008-bp region containing extensive sequence repeats, near the center of the viral genome (Fig. 3), may constitute a replication initiation site.
In conclusion, we propose classifying AFV2 in a new genus, “Deltalipothrixvirus,” of the family Lipothrixviridae. The unusual terminal and core structures, apparent lack of lipids, and exceptional genomic structure all help to distinguish it from the other genera. Moreover, these novel properties provide new evidence for the broad diversity of dsDNA viruses from the hyperthermophilic crenarchaea (17, 18), and the implications of these observations promise to enhance our understanding of the origin and evolution of viruses in general.
Nucleotide sequence accession number.
Sequence data from this article have been deposited with the EMBL/GenBank Data libraries under accession no. AJ854042.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Xu Peng for help in generating the first clone library, Hien Phan for amplifying and sequencing viral DNA, and Ariane Briegel for visualization using the Amira software package. Patrick Forterre is thanked for helpful discussions.
The research was supported by grants from the Deutsche Forschungsgemeinschaft (PR663/2-1), the Danish Natural Science Research Council, and the European Union (QLK3-2000-00649). K.B. received a Ph.D. grant from Copenhagen University.
Footnotes
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. Kristjansson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267**:**252-266. [DOI] [PubMed] [Google Scholar]
- 3.Bettstetter, M., X. Peng, R. A. Garrett, and D. Prangishvili. 2003. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology 315**:**68-79. [DOI] [PubMed] [Google Scholar]
- 4.Blum, H., W. Zillig, S. Mallock, H. Domday, and D. Prangishvili. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281**:**6-9. [DOI] [PubMed] [Google Scholar]
- 5.Brügger, K., P. Redder, and M. Skovgaard. 2003. MUTAGEN: Multi User Tool for Annotating GENomes. Bioinformatics 19**:**2480-2481. [DOI] [PubMed] [Google Scholar]
- 6.Chalcroft, J., and C. L. Davey. 1984. A simply constructed extreme-tilt holder for the Philips eucentric goniometer stage. J. Microsc. 134**:**41-48. [Google Scholar]
- 7.Frank, J. 1992. Electron tomography. Plenum Press, New York, N.Y.
- 8.Häring, M., X. Peng, K. Brügger, R. Rachel, K. O. Stetter, R. A. Garrett, and D. Prangishvili. 2004. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology 323**:**233-242. [DOI] [PubMed] [Google Scholar]
- 9.Hegerl, R. 1996. The EM program package: a platform for image processing in biological electron microscopy. J. Struct. Biol. 116**:**30-34. [DOI] [PubMed] [Google Scholar]
- 10.Janekovic, D., S. Wunderl, I. Holz, W. Zillig, A. Gierl, and H. Neumann. 1983. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic sulfur reducing archaebacterium Thermoproteus tenax. Mol. Gen. Genet. 192**:**39-45. [Google Scholar]
- 11.Lykke-Andersen, J., C. Aagaard, M. Semionenkov, and R. A. Garrett. 1997. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci. 22**:**326-331. [DOI] [PubMed] [Google Scholar]
- 12.Neumann, H. 1988. Struktur, Funktion und Variabilität eines archaebakterialen Genomes. Ph.D. dissertation. Ludwig-Maximilians-Universität of Munich, Munich, Germany.
- 13.Peng, X., H. Blum, Q. She, S. Mallok, K. Brügger, R. A. Garrett, W. Zillig, and D. Prangishvili. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291**:**226-234. [DOI] [PubMed] [Google Scholar]
- 14.Peng, X., A. Kessler, H. Phan, R. A. Garrett, and D. Prangishvili. 2004. Multiple variants of the archaeal DNA rudivirus SIRV1 in a single host and a novel mechanism of genome variation. Mol. Microbiol. 54**:**366-375. [DOI] [PubMed] [Google Scholar]
- 15.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. 1999. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol. 32**:**437-445. [DOI] [PubMed] [Google Scholar]
- 16.Prangishvili, D., K. M. Stedman, and W. Zillig. 2001. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 9**:**39-42. [DOI] [PubMed] [Google Scholar]
- 17.Prangishvili, D., and R. A. Garrett. 2004. Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses. Biochem. Soc. Trans. 32**:**204-208. [DOI] [PubMed] [Google Scholar]
- 18.Rachel, R., M. Bettstetter, B. P. Hedlund, M. Häring, A. Kessler, K. O. Stetter, and D. Prangishvili. 2002. Remarkable morphological diversity of viruses and virus-like particles in terrestrial hot environments. Arch. Virol. 147**:**2419-2429. [DOI] [PubMed] [Google Scholar]
- 19.Reilein, A. 1998. Preparation of catalase crystals. [Online.] http://www.itg.uiuc.edu/publications/techreports/98-009.
- 20.Rettenberger, M. 1990. Das Virus TTV1 des extrem thermophilen Schwefel-Archaebakteriums Thermoproteus tenax: Zusammensetzung und Struktur. Ph.D. thesis. University of Munich, Munich, Germany.
- 21.Schägger, H., and G. Jagow. 1987. Tricine dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166**:**368-379. [DOI] [PubMed] [Google Scholar]
- 22.Stedman, K., D. Prangishvili, and W. Zillig. 2004. Lipothrixviridae, p. 95-102. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 23.Tolstrup, N., C. W. Sensen, R. A. Garrett, and I. G. Clausen. 2000. Two different and highly organised mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4**:**175-179. [DOI] [PubMed] [Google Scholar]
- 24.Torarinsson, E., H.-P. Klenk, and R. A. Garrett. 2005. Divergent transcriptional and translational signals in Archaea. Environ. Microbiol. 7**:**47-54. [DOI] [PubMed] [Google Scholar]
- 25.Vestergaard, G., M. Haering, X. Peng, R. Rachel, R. A. Garrett, and D. Prangishvili. Virology, in press. [DOI] [PubMed]
- 26.Zillig, W., A. Kletzin, C. Schleper, I. Holz, D. Janekovic, H. Hain, M. Lanzendörfer, and J. K. Kristjansson. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 16**:**609-628. [Google Scholar]
- 27.Zillig, W., W.-D. Reiter, P. Palm, F. Gropp, H. Neumann, and M. Rettenberger. 1988. Viruses of archaebacteria, p. 517-558. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]