Vibrio fischeri Uses Two Quorum-Sensing Systems for the Regulation of Early and Late Colonization Factors (original) (raw)
Abstract
Vibrio fischeri possesses two quorum-sensing systems, ain and lux, using acyl homoserine lactones as signaling molecules. We have demonstrated previously that the ain system activates luminescence gene expression at lower cell densities than those required for lux system activation and that both systems are essential for persistent colonization of the squid host, Euprymna scolopes. Here, we asked whether the relative contributions of the two systems are also important at different colonization stages. Inactivation of ain, but not lux, quorum-sensing genes delayed initiation of the symbiotic relationship. In addition, our data suggest that lux quorum sensing is not fully active in the early stages of colonization, implying that this system is not required until later in the symbiosis. The V. fischeri luxI mutant does not express detectable light levels in symbiosis yet initiates colonization as well as the wild type, suggesting that ain quorum sensing regulates colonization factors other than luminescence. We used a recently developed V. fischeri microarray to identify genes that are controlled by ain quorum sensing and could be responsible for the initiation defect. We found 30 differentially regulated genes, including the repression of a number of motility genes. Consistent with these data, ain quorum-sensing mutants displayed an altered motility behavior in vitro. Taken together, these data suggest that the sequential activation of these two quorum-sensing systems with increasing cell density allows the specific regulation of early colonization factors (e.g., motility) by ain quorum sensing, whereas late colonization factors (e.g., luminescence) are preferentially regulated by lux quorum sensing.
The association between the luminescent marine bacterium Vibrio fischeri and the Hawaiian bobtail squid, Euprymna scolopes, is a model system for the study of mutualistic bacterium-host interactions that has been successfully used to investigate the processes underlying the initiation, accommodation, and persistence of symbioses (33, 34). The association is initiated every generation, as newly hatched juvenile squids collect their bacterial symbionts from the surrounding seawater (30). Within the first hour after hatching, bacterial cells present in the ambient seawater trigger the secretion of mucus from the superficial epithelium of the squid light organ. V. fischeri cells attach to this mucus, forming dense aggregates. Between 3 and 6 h after hatching, the bacterial aggregates begin to move towards pores that lead into the interior of the light organ. After entering the pores, bacterial cells travel through ciliated ducts into the deep crypt regions of the light organ. Interestingly, this latter process is V. fischeri specific and requires flagellum-driven motility (30). Once inside the light organ, V. fischeri cells grow on nutrients supplied by the squid host and reach maximal colonization levels at approximately 24 h postinoculation (15, 35). Several genetic factors required for the different stages of this cooperative association have been identified; for example, (i) motility is essential for the initiation of the symbiosis (14, 26-28), (ii) the ability to acquire iron is important for the symbionts to maintain a normal level of colonization (16), (iii) defense against oxidative stress is necessary to efficiently colonize (44), (iv) luminescence expression is required for persistent colonization (41), and (v) the regulation of colonization gene expression is important at each of these stages (22, 28, 41, 45, 46).
The coordinated expression of colonization factors can be controlled by a process termed quorum sensing, which relies on the secretion of small signal molecules that accumulate in the surrounding environment. Once the bacterial population reaches a threshold cell density, the concentration of the quorum-sensing signal is sufficient to induce gene expression either directly by interacting with a transcriptional regulator or indirectly by activation of a signaling cascade. Thus, colonization factors are expressed only when they are beneficial to the bacterial cell (e.g., when they are associated with the host), avoiding the execution of costly processes in nonpermissive environments (11, 47). In gram-negative bacteria the most commonly used signals are either (i) acylated homoserine lactones (acyl-HSLs) synthesized by a LuxI or LuxM type of enzyme or (ii) autoinducer-2 (AI-2), a furanosyl borate diester synthesized by the LuxS enzyme (5, 47). While acyl-HSLs are generally species specific due to differences in the acyl side chain, the LuxS signal appears to be essentially ubiquitous; i.e., many gram-negative and gram-positive species produce and respond to this signal molecule (2).
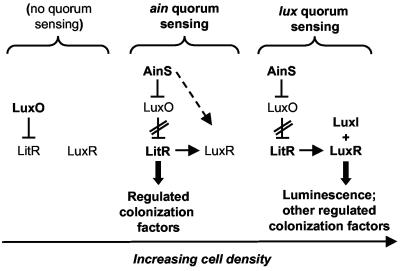
Quorum sensing based on the lux system was first described in V. fischeri as the principal process regulating luminescence expression (7). The expression of the lux genes, luxICDABEG, in V. fischeri is regulated in a cell density-dependent fashion through the LuxI-directed synthesis of _N-_3-oxo-hexanoyl homoserine lactone (3-oxo-C6-HSL) and its subsequent binding to the transcriptional activator protein LuxR (8, 25). The LuxR-acyl-HSL complex interacts with the lux promoter and induces the transcription of the luxICDABEG locus (12). The V. fischeri lux system is required for both luminescence expression in vivo and persistence of the bacterial symbiont in the squid light organ (41). V. fischeri possesses a second quorum-sensing system based on AinS, an enzyme belonging to the LuxM type of acyl-HSL synthases, which synthesizes an _N_-octanoyl homoserine lactone signal (C8-HSL) (13). We previously proposed a model of luminescence gene regulation by the two quorum-sensing systems ain and lux (22) (Fig. 1). At low cell densities, luminescence gene regulation is repressed through LuxO (22), a negative modulator of the transcriptional regulator LitR (29), (Fig. 1). At the intermediate cell densities achieved in laboratory culture, the AinS-synthesized signal has two effects, i.e., induction of luminescence gene expression through direct interaction with LuxR and inactivation of LuxO (22); the second effect results in increased transcription of litR (29) (Fig. 1). LitR positively regulates transcription of V. fischeri luxR (9), thereby connecting the two quorum-sensing systems ain and lux and allowing the sequential induction of luminescence gene expression with increasing cell density (22) (Fig. 1). Inactivation of the V. fischeri ain system results in a luminescence defect and affects colonization competence (22). The V. fischeri LuxS signal AI-2 was shown to be involved in the regulation of luminescence expression and colonization competence as well; however, compared to that of ain and lux quorum sensing, the impact of AI-2 was quantitatively small (21).
FIG. 1.
Model illustrating gene regulation by the two major V. fischeri quorum-sensing systems, ain and lux. See the introduction for explanation.
We asked here whether the stepwise activation of the ain and lux quorum-sensing systems with increasing cell density is functional in the symbiosis. We found that (i) ain, but not lux, quorum sensing is required for efficient initiation of colonization; (ii) the lux quorum-sensing system is not maximally induced until the later stages of colonization; and (iii) ain quorum sensing differentially regulates several factors, such as flagellar motility, with potential importance for colonization initiation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. V. fischeri strains were grown at 28°C either in a seawater-based nutrient medium (SWT) (3) or in Luria-Bertani salt medium (LBS) (14). E. coli strains were grown at 37°C in Luria-Bertani medium (36). Media were solidified with 1.5% (wt/vol) agar as needed. Antibiotics were added to the media at the following concentrations when appropriate: chloramphenicol (Cam), 2.5 μg/ml for V. fischeri and 20 μg/ml for Escherichia coli; erythromycin, 5 μg/ml for V. fischeri and 150 μg/ml for E. coli; and kanamycin (Kan), 100 μg/ml for V. fischeri and E. coli. 3-Oxo-hexanoyl-l-homoserine lactone (3-oxo-C6-HSL) was obtained from Sigma-Aldrich Corp. (St. Louis, MO); octanoyl-l-homoserine lactone (C8-HSL) was obtained from Aurora Biosciences (Coralville, IA). Other medium reagents were purchased from Difco Laboratories (Sparks, MD) and Sigma-Aldrich.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| CC118λpir | Cloning strain | 18 |
| DH5α | Cloning strain | 17 |
| V. fischeri strains | ||
| Wild-type ES114 | Isolate from an E. scolopes light organ | 3 |
| ainS mutant CL21 | ainS gene partially deleted and replaced by a chloramphenicol resistance (cat) marker | 22 |
| litR mutant PMF8 | litR gene inactivated by insertion of a kanamycin resistance marker (kanR) | 9 |
| luxI mutant VCW2G7 | luxI gene inactivated by a frameshift mutation | 22 |
| luxO mutant CL42 | luxO gene inactivated by insertion of a kanamycin resistance marker (kanR) | 22 |
| luxR mutant CL53 | luxR gene inactivated by insertion of a erythromycin resistance marker (ermR) | 21 |
| luxS mutant CL39 | luxS gene inactivated by insertion of a kanamycin resistance marker (kanR) | 21 |
| ainS luxI mutant CL24 | Double mutant carrying mutations as described above | 22 |
| ainS luxO mutant CL64 | Double mutant carrying mutations as described above | 22 |
| ainS luxS mutant CL41 | Double mutant carrying mutations as described above | 21 |
| LuxO(D47E) mutant CL59 | LuxO protein carrying a T-to-G point mutation resulting in an aspartate-to-glutamate substitution at position 47 | This study |
| Plasmids | ||
| pCL145 | pEVS79 carrying a 1.8-kb V. fischeri ES114 DNA fragment with the luxO gene | 22 |
| pCL150 | pCL145 with LuxO mutated by an aspartate-to-glutamate substitution at position 47 | This study |
| pDM104-6 | pVO8 carrying a flaA::lacZ transcriptional fusion | 48 |
| pEVS79 | Allelic exchange vector | 40 |
| pKV112 | pVO8 carrying the gfp gene under Plac control | K. Visick |
| pKV31 | pVO8 carrying a 1-kb SalI-PvuII fragment with the intact luxR gene | K. Visick |
| pVO8 | V. fischeri cloning vector, camR and ermR | 43 |
Colonization assays.
Colonization levels were determined as the number of CFU per squid light organ present at 12 h postinoculation (34). For each experiment, 12 to 20 newly hatched squids were placed into 50 ml of filter-sterilized seawater containing about 3,000 CFU of the indicated strain per ml. Another group of animals was placed into filter-sterilized seawater without added bacteria. After incubation for 12 h in the dark to prevent the usual expulsion of bacteria at sunrise (15, 19), the animals were homogenized, and the homogenate was diluted and spread onto SWT agar. The colony number was counted after overnight incubation, and the mean number of CFU per squid was calculated.
To determine the relative competitive index of the mutant strains, 10 to 15 newly hatched squids were placed into 50 ml of filter-sterilized seawater containing approximately 4,000 CFU of each of two competing strains and were incubated for 12 h. An aliquot of the inoculated seawater was spread onto LBS agar to determine the number and exact ratio of the two strains in the inoculum. At 48 h postinoculation, squids were homogenized and a dilution of the homogenate spread onto LBS agar. Approximately 100 CFU from the inoculum, as well as from each of the homogenates, was patched onto both antibiotic-containing and antibiotic-free LBS agar to determine the ratio of V. fischeri wild-type (Cam and Kan sensitive) to luxO mutant (Kan resistant) or ainS mutant (Cam resistant) cells. The relative competitive index was calculated as the ratio of the two strains in the symbiosis divided by the ratio in the inoculum.
To observe aggregation behavior, newly hatched squids were incubated with an inoculum of 500,000 CFU per ml of either wild-type V. fischeri or the ainS mutant, each carrying pKV112, a green fluorescent protein expression plasmid (Table 1). Every 30 min, two squids per treatment were dissected and the formation of aggregates was observed using fluorescence microscopy as previously described (30).
Mutant construction.
Genetic techniques (DNA extraction, PCR, ligation, conjugation, and DNA sequencing) were carried out as previously described (22).
The LuxO(D47E) mutant was constructed using the QuikChange site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene Inc., La Jolla, CA). Plasmid pCL145 (Table 1), carrying the intact luxO gene in the allelic exchange vector pEVS79 (Table 1), served as a template to create the site-directed mutation. The asparagine GAT codon in position 47 of the LuxO protein was changed into a glutamine GAG codon by introducing a T→G point mutation. The resulting plasmid, pCL150, was transferred into the V. fischeri luxO mutant CL42 by triparental mating, and double recombinants that had lost kanamycin resistance were selected as previously described (39), generating the V. fischeri LuxO(D47E) mutant strain CL59. The introduction of the point mutation into the genome was confirmed by sequence analysis of the mutant's luxO gene. Luminescence and growth characteristics were determined as previously described (22).
Microarray comparison.
Wild-type V. fischeri and the ainS, luxO, and LuxO(D47E) mutants were grown in SWT medium to an optical density (OD) at 600 nm of 2.5, a point at which V. fischeri cells still grow exponentially and, based on luminescence expression data (22), ain quorum sensing is maximally induced. RNA extractions were performed as previously described (37). cDNA synthesis was carried out using 10 to 12 μg of purified RNA, 15 μg random decamer primers (IDT, Coralville, IA), a deoxynucleoside triphosphate mixture (Amersham Biosciences, Piscataway, NJ) containing a 3:2 ratio of aminoallyl-dUTP (Sigma-Aldrich) to dTTP, and 500 units of Superscript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA). RNA was removed by alkaline hydrolysis, and cDNA was purified and concentrated using Microcon-30 filters (Millipore Corp., Billerica, MA). Labeling of cDNA was carried out using monoreactive Cy3 and Cy5 dyes according to the manufacturer's protocol (Amersham Biosciences), and labeled cDNA was desalted with a PCR purification kit (QIAGEN Inc., Valencia, CA). Nucleotide and label dye concentrations were determined by measuring absorption at 260 nm (nucleotide), 550 nm (Cy3), and 650 nm (Cy5). Microarray glass slides spotted three times with PCR products representing approximately 95% of all V. fischeri open reading frames (Integrated Genomics Inc., Chicago, IL) were incubated for 45 min at 42°C in prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] [Sigma-Aldrich], 0.001% sodium dodecyl sulfate [SDS], 0.5% bovine serum albumin). The two differentially labeled samples were mixed at a Cy3:Cy5 dye ratio of approximately 1:1 and concentrated to a final volume of 10 μl using Microcon-30 columns (Millipore Corp.). To each sample, 3.5 μl of 1-mg/ml salmon sperm DNA, 3.5 μl of 1-mg/ml yeast tRNA, and 18 μl PerfectHyb Plus hybridization buffer (Sigma Chemical Co.) were added, and the mixture was denatured at 99°C for 1.5 min. Each sample was added to the microarray glass slides and hybridized for 14 to 18 h at 42°C. The slides were washed at approximately 24°C for 5 min each in (i) 1× SSC, 0.001% SDS; (ii) 0.1× SSC, 0.001% SDS; (iii) 0.001% SDS; and (iv) water. After drying, fluorescence intensities were determined using a 428 Array Scanner (Affymetrix Inc., Santa Clara, CA). For each hybridization spot, background intensities were subtracted from spot intensities. The data were then log transformed and normalized by equalizing 25% and 75% tiles. Ratios of the normalized Cy3 and Cy5 dye intensities were determined for each spot and averaged for each microarray. For statistical analysis, a two-sided, paired Student t test was performed.
Transcriptional activity of flaA.
Plasmid pDM104-6 (Table 1) was introduced into V. fischeri wild-type and ainS mutant strains by triparental mating as previously described (39). Both strains were cultured in SWT plus erythromycin, and flaA transcriptional activity was estimated by assaying beta-galactosidase activity in six samples from two independent cultures at different time points during growth, using a standard method (36).
Motility assays.
Motility and chemotaxis behaviors of V. fischeri were determined by growing wild-type and mutant strains in the appropriate liquid medium to an optical density of approximately 0.4. Three microliters of the culture was spotted on the surface of a fresh 0.25% agar medium plate, and the relative diameters of the colony and the expanding rings of migrating cells were determined after 6 to 24 h of incubation at 25°C. Assays were carried out on (i) SWT medium or (ii) defined medium (10 mM NH4Cl, 0.33 mM K2HPO4, 300 mM NaCl, 50 mM MgSO4, 10 mM CaCl2, 10 mM KCl, 0.01 mM FeSO4, and 50 mM Tris-HCl [pH 7.4]) containing either 0.3% (wt/vol) Casamino Acids (CAA) or 0.5 g/liter _N_-acetylglucosamine (NAG) as a carbon source.
RESULTS
Colonization initiation by V. fischeri quorum-sensing mutants.
To determine whether any of the V. fischeri quorum-sensing systems is important for the initial stages of light-organ colonization, we determined the number of CFU in the organ 12 h after inoculation with either wild-type V. fischeri or the luxI, ainS, or luxS mutant (Table 2). At this early point in colonization, the number of CFU in squid colonized with the luxI mutant was indistinguishable from wild-type levels (Table 2). The fact that luxI mutant-colonized animals do not produce detectable luminescence (i.e., <1% of the wild-type level) (22) suggests that even a severe defect in light production does not affect colonization efficiency during the initial stages of colonization. In contrast, the ainS mutant, while expressing low (only 10% of wild-type) luminescence levels (22), was able to colonize the light organ only to <50% of the wild-type number of bacterial cells. Thus, although light emission was affected by a mutation in ainS, this gene probably regulates additional factors beyond luminescence that are important for colonization initiation. Squid colonized by the luxS mutant displayed colonization levels indistinguishable from those of wild-type-colonized animals at 12 h postinoculation (Table 2), indicating that luxS is not required for normal initiation of colonization. A secondary mutation in either luxI or luxS did not enhance the colonization defect of the ainS mutant (Table 2), suggesting the absence of a synergistic effect of these mutations in colonization initiation.
TABLE 2.
Colonization levels of quorum-sensing mutants at 12 h postinoculation
| Strain | % of wild-type colonization levela |
|---|---|
| Wild type | 100 (17) |
| ainS | 45 (14) |
| luxI | 115 (19) |
| ainS luxI | 79 (15) |
| luxS | 95 (30) |
| ainS luxS | 48 (7) |
| luxO | 37 (12) |
| ainS luxO | 36 (12) |
| litR | 51 (12) |
| luxR | 119 (17) |
One of the early events in colonization initiation is the formation of bacterial aggregates outside the squid light organ (30). Because a defect in this activity could explain the observed initiation phenotype, we examined aggregation behavior of the ainS mutant in comparison to wild-type V. fischeri. However, we could not detect any difference between the two strains, either in the timing of aggregate formation or in the size of the bacterial aggregate formed (data not shown). These results suggest that the ainS mutant is not defective in attaching to the squid mucus and that the colonization process(es) regulated by ainS becomes important after bacterial aggregation.
Regulation cascade of the initiation defect.
In our previous study (22) we demonstrated that the ainS mutant displays a persistence defect; i.e., the number of CFU is similar to that of the wild type at 24 h after colonization (the earliest time point examined) but becomes significantly lower by 48 and 72 h. This persistence defect could be reversed by introducing a luxO mutation into the ainS mutant background, suggesting that AinS regulates gene expression through the inactivation of LuxO (22). LuxO, in turn, down-regulates transcription of litR, a gene encoding a positive regulator of luxR (9, 29). To investigate whether the colonization factors underlying the initiation defect of the ainS mutant are regulated through this AinS-LuxO-LitR pathway (Fig. 1), we compared light-organ colonization levels at 12 h postinoculation with either the V. fischeri wild-type, luxO, ainS-luxO, litR, or luxR strain (Table 2).
Previous work (22) has shown that a luxO mutation does not interfere with the ability of V. fischeri to persist in the squid light organ after 24 h of colonization. However, the results presented here demonstrate that luxO, like ainS and litR but not luxR, is required for a normal level of colonization by 12 h (Table 2). Furthermore, the luxO mutant displayed a competition defect that was comparable to that of the ainS mutant. Specifically, when juvenile squids were colonized with a 1:1 mixed inoculum of either luxO mutant and wild-type cells or ainS mutant and wild-type cells, the average relative competitive index of both mutants was 0.4 (data not shown). Thus, mutations in both positive (AinS and LitR) and negative (LuxO) regulators in the pathway (Fig. 1) resulted in a similar defect in the level of colonization at 12 h (Table 2), which might also lead to a competitive defect. One interpretation of this surprising outcome is that, rather than a simple on-off switch, the AinS-LuxO-LitR pathway serves to maintain the colonization factor(s) it regulates at a proper level of expression. Because inactivation of luxR had no effect on the initiation of symbiosis (Table 2), the lux system is apparently not important for early stages of colonization, and thus the initiation defect of the ainS mutant does not operate through LuxR (Fig. 1).
Growth, luminescence, and colonization phenotypes of the V. fischeri LuxO(D47E) mutant.
While inactivation of V. fischeri ainS results in both initiation and persistence defects, a mutation in litR, operating downstream of ainS, affected only colonization initiation (Table 2) (9, 22), suggesting that the effect of ainS on colonization initiation may be distinct from that on colonization persistence. We therefore hypothesized that the AinS-LuxO-LitR cascade might directly activate genes important for colonization initiation, while the absence of _ain_-dependent induction of lux quorum sensing results in a defect in colonization persistence (Fig. 1).
To investigate this possibility further, we constructed a site-directed LuxO(D47E) mutant encoding an aspartate-47-to-glutamate substitution that has been shown to mimic the phosphorylated, and thus active (i.e., repressive), state of the Vibrio harveyi LuxO protein (10). According to the model of quorum sensing (Fig. 1), we predicted that this mutation would simulate the effects of an ainS mutation on the LuxO-LitR regulatory cascade but would allow the induction of lux quorum sensing (22). When we compared the growth, luminescence, and colonization phenotypes of the LuxO(D47E) mutant with those of the ainS, litR, and luxO mutants (Table 3), we found that their behavior was consistent with our hypothesis. Specifically, the growth yield of the LuxO(D47E) mutant in culture was reduced, while its doubling time was indistinguishable from that of the wild-type strain. However, the growth yield defect of the LuxO(D47E) mutant was smaller than those of the ainS and litR mutants, suggesting that LuxO(D47E) might not repress downstream genes to the same extent as the native phosphorylated LuxO protein. As expected, the growth defect could not be eliminated by exogenous addition of C8-HSL (data not shown), because the mutated LuxO(D47E) protein only mimics the phosphorylated LuxO state and cannot be dephosphorylated (and thus deactivated) by ain signaling. Luminescence of the LuxO(D47E) and litR mutants in culture was decreased compared to wild-type levels but was not eliminated (Table 3) (9). This result is consistent with the model of regulation (Fig. 1) in which AinS has two effects on luminescence regulation: the inactivation of LuxO repression and the activation of the lux operon through a C8-HSL-LuxR interaction (22). Thus, inactivation of AinS is expected to result in a more severe luminescence defect than a constitutively active LuxO protein or a mutation in litR. The luminescence levels of the LuxO(D47E) and litR mutants did not change in response to exogenous C8-HSL addition, implying that both mutant strains synthesize sufficient amounts of C8-HSL to induce luminescence through C8-HSL-LuxR interaction. When 3-oxo-C6-HSL was provided, luminescence reached nearly wild-type levels, indicative of proper lux quorum-sensing induction. As previously shown, the ainS mutant displayed essentially opposite effects, i.e., an increase in luminescence to wild-type levels when C8-HSL was added but a reduced response to 3-oxo-C6-HSL (22) (Table 3). This effect may be due to the absence of C8-HSL-LuxR interaction, which is necessary for proper lux quorum-sensing induction (22).
TABLE 3.
Comparison of the V. fischeri ainS and luxO deletion mutants with the constitutively active LuxO(D47E) mutant
| Phenotype | Value for V. fischeri strain | ||||
|---|---|---|---|---|---|
| Wild type | LuxO(D47E) | ainS | litR | luxO | |
| Doubling timea | 48 (7) | 46 (4) | 47 (6) | 51 (6) | 45 (6) |
| Growth yielda | 7.8 (0.7) | 7.4 (0.1) | 6.1 (0.3) | 5.9 (0.1) | 7.8 (0.5) |
| Luminescenceb | 3.2 (0.4) | 0.6 (0.1) | BD | 0.1 (0.0) | 3.5 (0.8) |
| Luminescence with C8-HSLb | 4.3 (0.9) | 0.7 (0.1) | 3.3 (0.3) | 0.2 (0.1) | 7.8 (2.0) |
| Luminescence with 3-oxo-C6-HSLb | 842 (70) | 724 (87) | 2.9 (0.6) | 659 (168) | 1036 (73) |
| Colonization level 12 h postinoculationc | 100 (17)d | 52 (10) | 45 (14)d | 51 (12)d | 37 (12)d |
| Colonization level 72 h postinoculationc | 100 (11) | 97 (7) | 29 (15)e | ND | 95 (12)e |
Finally, while the colonization levels of the LuxO(D47E), litR, and ainS mutants were comparable at 12 h postinoculation, only the ainS mutant displayed a persistence defect at 72 h (Table 3). Thus, the ainS mutant is compromised in both the initiation and persistence of symbiosis; however, the underlying causes of these two colonization defects appear to be distinct.
Onset of lux quorum sensing.
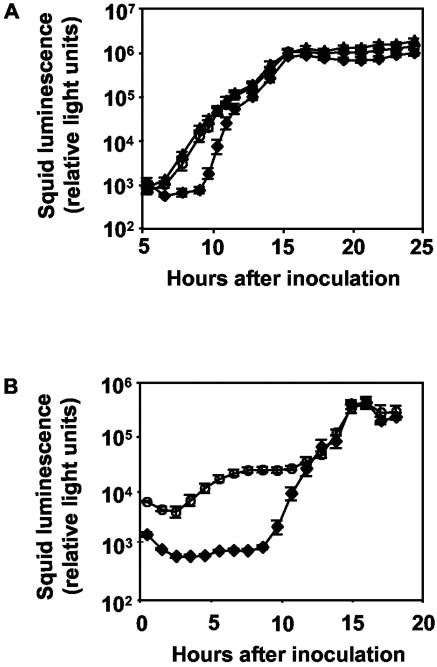
Having established that lux quorum sensing does not affect the ability of V. fischeri to initiate colonization, we asked whether this system is fully active during the early stages of colonization. Because induction of the lux system is required for light expression in the host (i.e., luxI mutant-colonized animals are dark [22]), lux activity can be easily monitored by measuring symbiotic light production. In culture, where the lux system is inactive, the exogenous addition of saturating concentrations of the LuxI-synthesized 3-oxo-C6-HSL stimulates light expression more than 1,000-fold (22). We used a similar approach to determine the level of lux activity during the initiation of symbiosis by comparing luminescence expression by wild-type-colonized juvenile squid exposed to seawater containing (i) no addition, (ii) 120 nM 3-oxo-C6-HSL, or (iii) 1,200 nM 3-oxo-C6-HSL during the first 24 h of colonization (Fig. 2A). To minimize any possible effects of 3-oxo-C6-HSL on colonization phenotypes other than luminescence expression, juvenile squids were incubated with wild-type V. fischeri cells for 6 h before being transferred into seawater containing 3-oxo-C6-HSL. Almost immediately after exposure to 3-oxo-C6-HSL, and 3 h earlier than the no-addition control, squid became detectably luminous (Fig. 2A). This positive effect of 3-oxo-C6-HSL on luminescence expression continued until approximately 12 h postinoculation, after which luminescence levels were indistinguishable between exposed and control animals (Fig. 2A). In contrast, exposure of wild-type-colonized squid to 1,200 nM of the AinS-synthesized signal C8-HSL had no significant effect on the development of luminescence expression (data not shown), suggesting that this signal is produced in saturating concentrations by 6 h postinoculation. Because acyl-HSLs are susceptible to hydrolysis, it was a concern that the absence of any positive effect on luminescence at time points beyond 12 h was due to degradation of the compound. However, because luminescence expression by animals exposed to 10 times the saturating concentration (1,200 nM 3-oxo-C6-HSL) was indistinguishable from that by animals exposed to 120 nM 3-oxo-C6-HSL (Fig. 2A), it seemed unlikely that degradation of 3-oxo-C6-HSL affected the outcome of this experiment.
FIG. 2.
Symbiotic lux quorum-sensing activity during initial stages of colonization. Presented are the mean luminescence levels of 24 animals for each treatment, with standard errors. A. Effect of 3-oxo-C6-HSL addition on luminescence of _V. fischeri_-colonized juvenile squid. Animals were inoculated with V. fischeri wild-type cells for 6 h and then transferred to seawater only (diamonds) or seawater containing either 120 nM (circles) or 1,200 nM (triangles) 3-oxo-C6-HSL. B. Effect of luxR gene overexpression on luminescence of _V. fischeri_-colonized juvenile squid. Animals were inoculated with V. fischeri wild-type cells carrying either pKV31 (circles) or pVO8 (diamonds), a vector control.
Finally, we tested our hypothesis that LuxR, a target of LitR induction (Fig. 1), is limiting during the early stages of colonization. When juvenile squids were colonized with wild-type V. fischeri overexpressing the luxR gene in trans, luminescence levels were higher than those of animals colonized with V. fischeri carrying the vector control in trans (Fig. 2B). Again, the differences in the levels of light expression were observed only during the initiation of colonization. These data suggest that, because of the limited expression of LuxR, the lux system is not maximally induced until approximately 12 h postinoculation, coincident with the time at which the squid light-organ crypts become densely colonized with V. fischeri cells (30).
Microarray comparison of wild-type V. fischeri and quorum-sensing mutants.
In an effort to identify factors regulated by V. fischeri ain quorum sensing that could be responsible for the observed colonization defects, we performed three independent microarray experiments using culture-grown cells. Two microarray experiments compared the transcriptomes of the ainS mutant and the wild type. To ensure that the regulation was ain quorum-sensing specific and not a result of gene regulation by the C8-HSL-LuxR interaction (Fig. 1), a third experiment compared the transcriptomes of the LuxO(D47E) and luxO mutants. We used the two selection criteria of (i) greater than twofold differential regulation in each of the three experiments and (ii) a P value of <0.05 to identify genes that were differentially regulated by ain quorum sensing (Table 4).
TABLE 4.
Transcriptome analysis of V. fischeri quorum-sensing mutants
| Category and ORFb | Annotation (putative function) | Ratioa | |
|---|---|---|---|
| Wild type/ainSc | luxO/LuxO(D47E)d | ||
| Motility | |||
| VF0777 | Methyl-accepting chemotaxis protein | −3.0 (0.6) | −2.9 (6.5) |
| VF1836 | Flagellar biosynthesis protein, FlhF | −2.3 (0.9) | −3.3 (0.7) |
| VF1846e | Flagellar protein, FliL | −1.8 (6) | −3.1 (0.3) |
| VF1853e | Flagellar hook-basal body protein, FliE | −1.9 (0.4) | −4.2 (0.9) |
| VF1862 | Flagellin, FlaE | −5.0 (0.4) | −15.5 (0.2) |
| VF1863 | Flagellin, FlaD | −4.0 (0.3) | −8.3 (0.7) |
| VF1864e | Flagellin, FlaC | −1.7 (18) | −4.2 (0.1) |
| VF1865e | Flagellin, FlaB | −1.6 (12) | −3.0 (6.4) |
| VF1866e | Flagellin, FlaA | −1.8 (7) | −7.2 (4.1) |
| VF1874e | Flagellar hook protein, FlgE | −1.4 (28) | −3.8 (0.9) |
| VF1875 | Basal-body rod modification protein, FlgD | −5.6 (0.2) | −5.1 (0.1) |
| VF1876 | Flagellar basal-body rod protein, FlgC | −11.8 (0.1) | −11.1 (0.1) |
| VF1877 | Flagellar basal-body rod protein, FlgB | −3.3 (0.3) | −8.6 (0.7) |
| VF1881 | Negative regulator of flagellin synthesis, FlgM | −3.7 (0.7) | −7.2 (32.3) |
| VF2079 | Flagellin, FlaF | −7.1 (0.1) | −8.0 (0.1) |
| Transcriptional regulators | |||
| VF1573 | DNA-binding protein | −3.1 (0.1) | −2.7 (2.5) |
| VF2177 | Transcriptional regulator, LitR | 10.3 (4.2) | 5.9 (<0.1) |
| VFA1014 | Hypothetical protein (sensory transduction) | 2.5 (10.6) | 2.2 (17.5) |
| VFA1015 | Sigma factor, RpoS like | 5.5 (0.1) | 2.3 (2.9) |
| VFA1017 | Possible response regulator | 7.0 (4.0) | 4.9 (0.3) |
| Metabolism | |||
| VF0008 | Cystine-binding protein transporter | −3.8 (<0.1) | −2.5 (25) |
| VF0316 | Drug/metabolite exporter | −3.2 (3.8) | −5.2 (5.5) |
| VF2247 | Dihydroneopterin aldolase (folate biosynthesis) | −2.5 (19) | −3.1 (2.0) |
| VFA0452 | Formiminoglutamase (histidine metabolism) | 2.3 (0.8) | 2.6 (0.5) |
| VFA0806 | Glucose-1-phosphate adenylyltransferase (polysaccharide biosynthesis) | 3.9 (0.4) | 3.1 (1.6) |
| VFA0995 | High-affinity ferrous iron permease | −5.6 (0.3) | −2.3 (0.6) |
| EPS biosynthesis | |||
| VF0171 | Hypothetical protein | 4.2 (0.4) | 2.4 (17.5) |
| VF0172 | _O_-Acetyltransferase | 3.6 (0.1) | 3.8 (0.5) |
| Hypothetical proteins | |||
| VF1355 | Hypothetical protein | 2.0 (3.5) | 2.8 (1.1) |
| VF2255 | Hypothetical protein | 2.3 (0.3) | 3.2 (0.2) |
The most striking finding of the microarray study was that genes encoding several flagellins and flagellar basal-body proteins, appeared to be negatively regulated by the V. fischeri ain quorum-sensing system (Table 4). In addition, one of the 31 putative methyl-accepting chemotaxis protein genes (http://www.ergo-light.com/ERGO/) appeared to be differentially repressed by ainS, suggesting a possible role of quorum sensing in the regulation of a behavioral response to a specific environmental condition (Table 4). Finally, the gene for the V. fischeri FlgM homolog, a putative sigma-28 antagonist, appeared to be down-regulated as well, indicating that the repression of sigma-28-dependent motility genes by ain quorum sensing is unlikely to function through FlgM (Table 4). The majority of the V. fischeri motility genes are located in a distinct chromosomal region (VF1826 to VF1881 [http://www.ergo-light.com/ERGO/]). When we lowered the selection criteria somewhat to include genes that were on average greater than twofold differentially regulated (yet still with a P value of <5%), we found an additional six motility genes that were down-regulated by ain quorum sensing (Table 4).
We confirmed the microarray data for one of these genes (flaA) by comparing the beta-galactosidase activity levels of a flaA::lacZ fusion carried in trans by both wild-type V. fischeri and the ainS mutant. The beta-galactosidase activity was on average 3.3 times higher in the ainS mutant than in wild-type V. fischeri (data not shown), supporting a role of ainS in the expression of this important flagellin (26).
The genes for several regulatory proteins also appeared differentially regulated by ain quorum sensing (Table 4), suggesting that the V. fischeri quorum-sensing regulon is connected to other regulatory circuits. For instance, the previously proposed positive regulation of V. fischeri litR by AinS (22) was confirmed (Table 4). While the V. fischeri ainS mutant does not express detectable luminescence levels in culture and the specific luciferase activity is only 10% of that of the wild type (22), we could not detect differential luminescence (luxCDABE) or regulatory (luxR) gene expression by ain quorum sensing. We believe that this result may reflect how luciferase specific activity increases dramatically around an OD of approximately 1.0 and stays constant thereafter (22). Thus, at the OD of 2.5 at which the sample was taken for microarray analysis, positive regulation of lux transcription by ain quorum sensing may have already ceased.
One interesting regulatory locus contains the genes VFA1013 to VFA1017, some of which were positively regulated by ain quorum sensing (Table 4). The proteins encoded by both VFA1014/VFA1013 and VFA1016/VFA1017 have regions homologous to protein kinases and response regulator pairs, respectively (http://www.ergo-light.com/ERGO/). Located between these putative two-component regulatory systems is another gene (VFA1015) that was positively regulated by ain quorum sensing (Table 4). This gene is most closely related to the stationary-phase sigma factor gene V. fischeri rpoS (VF2067) (http://www.ergo-light.com/ERGO/). The functions of the genes in this regulatory locus remain to be determined; nevertheless, the ainS regulation of VFA1015, together with the growth yield defect of the ainS mutant (Table 3), led us to the hypothesis that the genes in the VFA1013-to-VFA1017 locus may encode important sensory and response systems used by V. fischeri to switch from the planktonic to the symbiotic lifestyle.
Motility phenotype of V. fischeri quorum-sensing mutants.
Because the microarray results indicated that flagellin and flagellar basal body genes are regulated by the ain system, we assessed the motility behaviors of several of the quorum-sensing mutants. As we reported previously (22), the swimming behaviors of the ainS and the luxI mutants on rich SWT medium containing either 0.4% or 0.7% agar were indistinguishable from that of the wild type. However, at a lower agar concentration (0.25%) on both SWT and a minimal salts medium supplemented with either CAA or NAG, differences in motility behavior between wild-type and mutant strains became apparent (Table 5).
TABLE 5.
Relative migration of V. fischeri quorum-sensing mutants in soft agar
| Strain | Migration distance ina: | ||
|---|---|---|---|
| SWT | CAA | NAG | |
| Wild type | 1 | 1 | 1 |
| ainS | 1.4 | 1.6 | 2.4 |
| luxO | 0.9 | BDb | BD |
| ainS luxO | 0.9 | BD | BD |
| luxS | 1.1 | 1 | 1 |
| ainS luxS | 1.4 | 1.6 | 2.4 |
| litR | 1.4 | 1.6 | 2.2 |
| luxI | 1 | 1 | 1 |
| luxR | 1 | 1 | 1 |
In general the effects of quorum-sensing gene mutations on motility were more pronounced in the defined media (Table 5), suggesting that nutrient availability affects V. fischeri motility by an unknown mechanism. While the ainS mutant displayed a hypermotile phenotype in all media, the luxO mutant produced slightly smaller migration ring diameters in SWT, and no migration could be observed in minimal medium containing either CAA or NAG (Table 5). The migration patterns of the ainS-luxO mutant in each medium were indistinguishable from those of the luxO mutant, suggesting that luxO has a dominant effect on the motility phenotype. The luxS mutant migrated slightly faster than the wild type in SWT, but the migration was not affected in minimal medium (Table 5). Furthermore, the introduction of the luxS mutation into the ainS mutant did not noticeably enhance the hypermotile phenotype of the ainS mutant (Table 5), suggesting the absence of a synergistic effect. The litR mutant displayed essentially the same phenotype as the ainS mutant (Table 5), consistent with the gene regulation model (Fig. 1) in which litR functions downstream of ainS (22). Neither mutation of luxI nor mutation of luxR resulted in an altered motility behavior under the conditions tested (Table 5). All of the strains grew with identical doubling times in each of the media, and only the hypermotile ainS, ainS luxS, and litR strains (22) (Table 3) displayed a growth yield defect (data not shown), indicating that the observed phenotypes are not due to a metabolic defect.
DISCUSSION
In this study we investigated the impact of the V. fischeri quorum-sensing systems ain and lux on the early stages of colonization. Our data demonstrate that (i) ain (but not lux) quorum sensing is required for the normal initiation of colonization, (ii) the lux system is not fully active during initiation of colonization, and (iii) the ain system regulates not only the lux system but also several putative early colonization genes, including those controlling motility. Taken together, these data demonstrate for the first time the differential activation of two bacterial quorum-sensing systems at distinct stages of a beneficial host tissue colonization.
A model of colonization factor regulation by V. fischeri quorum sensing.
Many gram-negative bacteria possess multiple quorum-sensing systems (47), a phenomenon that has led to an examination of how these systems may function together. Our results show that in the case of V. fischeri two systems are arranged to allow the induction or repression of colonization factors important during specific temporal phases of the symbiotic relationship. The fact that the ain system is functional and essential at early stages of colonization, whereas the lux system is neither fully induced nor required at this time (Fig. 2), suggests that the ain system is operative at a lower threshold cell density than the lux system and provides evidence for a stepwise pattern in their control during the onset of symbiosis. The sequential nature of quorum-sensing induction is further supported by our previous finding that in symbiotic strains of V. fischeri the effect of the ain system on luminescence is apparent at the relatively moderate bacterial concentrations occurring in culture (108 to 109 cells ml−1), whereas the lux system is the predominant inducer of luminescence expression at the very high cell densities (>1010 cells ml−1) achieved only within the squid light organ (22).
We proposed previously a model by which luminescence is regulated by ain and lux (22). The results of this present study expand this model to include the regulation of non-lux genes at specific phases in the colonization of E. scolopes by V. fischeri. At the very low cell densities found in seawater (<104 cells/ml), the absence of ain quorum sensing allows repression of colonization gene expression through the transcriptional regulator protein LuxO (Fig. 1). When V. fischeri initiates colonization of its host by aggregating and then migrating through the squid light-organ ducts (30), the cell density increases to the threshold cell density of ain quorum sensing. This threshold level is approximately 108 cells/ml, based on both _ain_-dependent luminescence induction and C8-HSL signal production in culture (21, 22), and correlates with the cell densities occurring during the initiation process (35). At this stage the AinS signal, C8-HSL, inactivates LuxO, allowing the appropriate level of expression of early colonization genes through LitR (Fig. 1). This model is consistent with two observations: the ainS, LuxO(D47E), and litR mutants are similarly defective in colonization initiation, and a double ainS luxO mutant displays an initiation defect comparable to those of the single ainS and luxO mutants (Table 2), suggesting that their effects are modulated through a common pathway. While our evidence supports the notion that both luminescence genes (22, 29) and motility genes (Tables 4 and 5) are regulated through the AinS-LuxO-LitR pathway (Fig. 1), it remains possible that LuxO regulates some colonization factors independently of LitR.
We have previously demonstrated that ain quorum sensing is required for appropriate expression of the lux quorum-sensing system (22). Thus, inactivation of ainS produces two effects in vivo: an initiation defect resulting from the loss of regulation of early colonization factors (Table 2) and a persistence defect caused by the absence of appropriate lux quorum-sensing induction (22). As predicted, both the LuxO(D47E) and the litR mutants display an initiation defect (Table 2) but not a persistence defect (Table 3) (9).
Finally, once V. fischeri has densely colonized (>1010 cells/ml) the deeper crypt regions of the squid light organ, and after initial induction by the ain system, lux quorum sensing induces expression of late colonization genes, such as those encoding luminescence (Fig. 1). A previous study demonstrated that mutations in either luxI, luxR, or luxA (luxA encodes one of the subunits of luciferase), produced similar defects in colonization persistence (41), suggesting that luminescence might be the only colonization factor regulated by the lux quorum-sensing system. However, a separate study of the fish light-organ symbiont V. fischeri MJ1 showed that lux quorum sensing regulated the expression of five non-lux proteins (4). Inactivation of one of the corresponding genes, qsrP, in the squid light-organ symbiont V. fischeri ESR1 resulted in a competitive defect, suggesting that at least this lux quorum-sensing-regulated protein is a colonization factor.
Quorum sensing affects V. fischeri motility.
When planktonic V. fischeri cells colonize juvenile E. scolopes, both the animal host and the bacterial symbiont undergo dramatic developmental changes (24). Two of the most apparent phenotypes of symbiotic V. fischeri cells are luminescence induction and an apparent loss of flagella (35). The ability to express luminescence during colonization is principally regulated by lux quorum sensing (42). Based on the data presented here (Tables 4 and 5), we hypothesize that, while flagellated planktonic V. fischeri initiate the colonization process, as they reach the deeper crypt tissues where proliferation occurs, flagellar elaboration is repressed as a consequence of ainS signaling.
There are several lines of evidence that the altered (either decreased or increased) motility behavior of mutants in the ain quorum-sensing pathway is the cause of the colonization initiation defect (Fig. 1; Table 2). Proper motility behavior has been shown previously to be essential for colonization initiation; i.e., both nonmotile and hypermotile mutants are significantly impaired in establishing a light-organ colonization (14, 27). However, at later stages of colonization the majority of bacterial cells have lost their flagella, implying that proper swimming motility is not required for maintenance of the symbiosis (35). The results presented here are consistent with this paradigm: both the hypermotile ainS and litR mutants and the motility-deficient luxO mutants (Table 5) display a delay in colonization initiation (Table 2). The presence of this delay in mutants predicted to have either higher (luxO) or lower (ainS or litR) expression of LitR suggests that targets of this regulator, like motility genes, must be carefully modulated.
Whole-genome transcription analysis showed that ain quorum sensing regulates both flagellar basal body and flagellin genes (Table 4). Motility gene regulation follows a hierarchical regulation cascade allowing the assembly of the flagellar apparatus in a sequential manner. The different classes of motility genes include early genes encoding regulatory proteins that control the entire motility regulon, middle genes encoding structural components of the hook-basal body and the type III secretion apparatus, and late genes encoding the flagellar filament, chemotaxis, and motor force generator proteins (1). The motility regulatory cascade in V. fischeri has not been completely elucidated; however, the analysis of regulatory proteins and the promoter regions of motility genes shows similarities to the regulatory cascade of Vibrio cholerae (26, 28, 31, 48). For example, inactivation of the early sigma-54-dependent master regulator (flrA), as well as sigma-54 (rpoN) itself, abolishes motility, suggesting that, as in V. cholerae, flrA is the master regulator of the V. fischeri motility regulon (28, 31, 48). Because ain quorum sensing regulates both sigma-54-dependent middle genes (i.e., flgBCDEM, flhF, fliEL, and flaA) (28) and sigma-28-dependent late genes (i.e., flaBCDEF) (26), it is not clear at which regulatory level quorum sensing affects motility gene regulation. Interestingly, while V. cholerae quorum sensing also negatively regulates motility, only chemotaxis genes were shown to be differentially regulated (49), suggesting that the regulatory level at which quorum sensing affects the motility regulon is different in these two Vibrio species.
ain quorum-sensing regulon.
While our findings are consistent with the hypothesis that the altered motility behavior of the ain quorum-sensing mutants is, by itself, sufficient to explain their inability to efficiently initiate colonization, it remains possible that AinS and LuxO also regulate other, perhaps disparate, initiation factors. Our transcriptome analysis revealed several additional genes that are regulated by ain quorum sensing (Table 4) and might play a role in colonization initiation. For instance, V. fischeri may adapt to growth conditions in the light-organ crypts by using ain quorum sensing to regulate factors involved in nutrient uptake or metabolism (Table 4). Furthermore, the differential regulation of these genes provides a possible explanation for the growth yield defect exhibited by the ain quorum-sensing mutants in vitro (22) (Table 3).
Another result of ain quorum sensing appears to be the induction of genes in a putative exopolysaccharide (EPS) locus (Table 4). This locus contains many genes homologous to the Escherichia coli wzy locus, a system involved in O-antigen biosynthesis (VF0151 to VF0201 [http://www.ergo-light.com/ERGO/]). The consequences of EPS production in bacterium-host associations are diverse, but they can include enhanced adherence to tissue surfaces and/or protection against the challenges of complement-mediated phagocytosis and killing by the innate immune system (32). While the role(s) of EPS in this association remains to be determined, both of these challenges are faced by symbiotic V. fischeri (42). Mutations in homologs of LuxO or LitR in other Vibrio species affect colony morphology and biofilm formation, which in some cases correlates to changes in EPS production (6, 20, 23, 38, 49). Accordingly, the V. fischeri litR mutant was reported to display a more translucent colony morphology and to produce more biofilm than the wild type (9; P. Fidopiastis, personal communication). Not surprisingly, we found that the V. fischeri ainS mutant displayed similar phenotypes (data not shown); however, it remains to be determined whether these phenotypes are related to LPS and/or EPS production.
Our analysis indicates that ain quorum sensing has an effect on the transcription of a number of genes, including some unique ones (VF1355 and VF2255) with unknown functions (Table 4). While the functions of others can be inferred, it is not yet clear whether their regulation is direct or is an indirect effect of a multistep signaling pathway involving other transcriptional factors. In any case, the discovery of these regulated genes not only has given us insight into the way in which ainS signaling controls both early and late stages of symbiosis but also has provided candidate genes that may encode novel colonization determinants.
Acknowledgments
We thank Amy Schaefer for developing the protocol used in the microarray study and for help with the experiment, David Goode for performing normalization of the microarray data sets and advice on statistical analysis, and K. Visick for kindly providing plasmids. We are grateful to Debbie Millikan and Amy Schaefer for critically reading the manuscript. V. fischeri ES114 genomic sequence information was made available by Integrated Genomics Inc., Chicago, Ill.
This work was supported by National Institutes of Health grant RR12294 to E. G. Ruby and M. McFall-Ngai, by National Science Foundation grant IBN0211673 to M. McFall-Ngai and E. G. Ruby, and by a W. M. Keck Foundation grant to E. P. Greenberg, E. G. Ruby, M. McFall-Ngai, and others.
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5**:**160-165. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179**:**4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172**:**3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan, S. M., and P. V. Dunlap. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182**:**2811-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, X., S. Schauder, N. Potier, A. v. Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415**:**545-549. [DOI] [PubMed] [Google Scholar]
- 6.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Camara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184**:**1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20**:**2444-2449. [DOI] [PubMed] [Google Scholar]
- 8.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81**:**4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidopiastis, P. M., C. M. Miyamato, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a newly described transcriptional activator homologue in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45**:**131-143. [DOI] [PubMed] [Google Scholar]
- 10.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31**:**665-677. [DOI] [PubMed] [Google Scholar]
- 11.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35**:**439-468. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50**:**727-751. [DOI] [PubMed] [Google Scholar]
- 13.Gilson, L., A. Kuo, and P. V. Dunlap. 1995. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177**:**6946-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176**:**6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95**:**1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37**:**168-179. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166**:**557-580. [DOI] [PubMed] [Google Scholar]
- 18.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172**:**6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K. H., and E. G. Ruby. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60**:**1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36**:**940-954. [DOI] [PubMed] [Google Scholar]
- 21.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 86**:**3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50**:**319-331. [DOI] [PubMed] [Google Scholar]
- 23.McCarter, L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180**:**3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McFall-Ngai, M. J., and E. G. Ruby. 1998. Sepiolids and vibrios: when they first meet. BioScience 48**:**257-265. [Google Scholar]
- 25.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55**:**123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millikan, D. S., and E. G. Ruby. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186**:**4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68**:**2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millikan, D. S., and E. G. Ruby. 2003. FlrA, a σ54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J. Bacteriol. 185**:**3547-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto, C. M., P. V. Dunlap, E. G. Ruby, and E. A. Meighen. 2003. LuxO controls luxR expression in Vibrio harveyi: evidence for a common regulatory mechanism in Vibrio. Mol. Microbiol. 48**:**537-548. [DOI] [PubMed] [Google Scholar]
- 30.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97**:**10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39**:**1595-1609. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, I. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50**:**285-315. [DOI] [PubMed] [Google Scholar]
- 33.Ruby, E. G. 1999. Ecology of a benign “infection”: colonization of the squid luminous organ by Vibrio fischeri, p. 217-231. In E. Rosenberg (ed.), Microbial ecology and infectious disease. American Society for Microbiology, Washington, D.C.
- 34.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50**:**591-624. [DOI] [PubMed] [Google Scholar]
- 35.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159**:**160-167. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schuster, M., C. P. Lohstroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185**:**2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao, C.-P., and L.-I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183**:**1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J. Bacteriol. 183**:**309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stabb, E. V., and E. G. Ruby. 2002. New RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358**:**413-426. [DOI] [PubMed] [Google Scholar]
- 41.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182**:**4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive encounter: specificity in the _Vibrio fischeri_-Euprymna scolopes partnership. J. Bacteriol. 182**:**1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick, K. L. and Ruby, E. G. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence. Wiley and Sons, New York, N.Y.
- 44.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180**:**2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183**:**835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whistler, C. A., and E. G. Ruby. 2003. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 185**:**7202-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25**:**365-404. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe, A. J., D. S. Millikan, J. M. Campbell, and K. L. Visick. 2004. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70**:**2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99**:**3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]