Identification of SopE2, a Salmonella Secreted Protein Which Is Highly Homologous to SopE and Involved in Bacterial Invasion of Epithelial Cells (original) (raw)
Abstract
Type III secreted Sop protein effectors are delivered into target eukaryotic cells and elicit cellular responses underlying Salmonella pathogenicity. In this work, we have identified another secreted protein, SopE2, and showed that SopE2 is an important invasion-associated effector. SopE2 is encoded by the sopE2 gene which is present and conserved in pathogenic strains of Salmonella. SopE2 is highly homologous to SopE, a protein encoded by a gene within a temperate bacteriophage and present in only some pathogenic strains.
A variety of gram-negative pathogenic bacteria possess a dedicated protein secretion system, denoted type III, which is of primary importance for the successful engagement of eukaryotic host cells (for a recent review, see reference 8). Type III secretion systems play a central role in virulence, directing secretion and translocation of several bacterial effector proteins into the cytoplasm of host cells. It is becoming apparent that the mechanism of type III secretion and translocation is conserved in a variety of bacteria, although each pathogen appears to have a specific set of secreted protein effectors. Upon translocation, the bacterial protein effectors affect different host cell functions and elicit a variety of responses. This, in turn, has a major impact on the development and progress of infection.
Recent work by our group and others has identified a number of protein effectors (3, 4, 5, 9, 12, 17) secreted and translocated into eukaryotic cells by the Salmonella Inv/Spa type III secretion system, which is one of two type III systems present in salmonellae (for a review, see reference 2). The Inv/Spa system is encoded within a pathogenicity island, SPI-1 (13), and is required for eliciting host cell responses which in turn result in host cell cytoskeleton rearrangements which assist bacterial entry into nonphagocytic epithelial cells and aid in the production of proinflammatory signals (for a recent review, see reference 1). Four of the key proteins in these processes are Sip A, B, C, and D. All these proteins are encoded by a single polycistronic operon located adjacent to the inv/spa loci (7, 10, 11, 16). At least some of the Sip proteins appear to have multiple activities and perform several functions, but it is believed that the major function of Sip B, C, and D is to execute the translocation of a set of specific effector proteins into eukaryotic cells. One such translocated effector is SopE (5, 17). SopE contributes to the expression of Salmonella invasion by stimulating membrane ruffling via guanidine nucleotide exchange on Rho GTPases CDC42 and Rac (6). The sopE gene is located outside SPI-1 and is encoded within a temperate bacteriophage (5, 15). Interestingly, not all strains of Salmonella carry the sopE gene (15), suggesting either that the SopE function is redundant or that there is another protein with functions similar to those of SopE, in at least some Salmonella strains. We have investigated this suggestion.
SopE was initially identified as a component of protein aggregates accumulated in the culture media of Salmonella enterica serovar Dublin B1 (sipB mutant) (17). This suggested that sipB mutants of different Salmonella strains may have different profiles of Sop proteins deposited into filamentous aggregates. In order to assess the Sop protein profiles from Salmonella strains with different host specificities, we first transduced the polar sipB mutation from serovar Dublin B1 into a variety of other Salmonella strains (virulent field isolates from the IAH strain collection) by using P22 transduction. The correct mutations in Cmr transfectants (one for each strain) were confirmed by PCR (data not shown). Different sipB mutant strains were then grown overnight at 25°C in Luria-Bertani medium, were diluted 10-fold in fresh LB medium, and were incubated for 5 h at 37°C. All Salmonella strains tested produced filaments similar to those produced by serovar Dublin B1. The protein filament aggregates accumulated in each flask were collected and washed in phosphate-buffered saline, essentially as described earlier (17). Filaments were dissolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and proteins were separated by SDS–12% PAGE. The gel was stained with Coomassie brilliant blue (Fig. 1A) or was blotted onto a nitrocellulose membrane and probed with an anti-SopE monoclonal antibody (Fig. 1B). The analysis of the Coomassie-stained gel revealed that filament aggregates isolated from each Salmonella strain were composed of a similar set of proteins, migrating in each case with a profile closely resembling that of serovar Dublin (Fig. 1A). The monoclonal anti-SopE antibodies (17), however, did not recognize the ∼30-kDa protein from Salmonella enterica serovar Typhimurium F98 isolated from a chicken and serovar Typhimurium ST5306 isolated from a pig (Fig. 1B). In both cases, however, this protein appeared to migrate with the same apparent molecular weight as SopE from serovar Dublin. We then attempted to amplify a _sopE_-specific DNA fragment by PCR by using genomic DNA from different Salmonella strains as a template with SEup (5′-ACCGAGAAAAGATCTTTAGCAAAAA-3′) and SEdown (5′-CAAGATCAGCTCACACT-3′) primers, based on the known DNA sequence of sopE from serovars Dublin and Typhimurium. No DNA fragment was amplified when DNA from serovar Typhimurium F98 or ST5306 was used as a template, but a _sopE_-specific DNA fragment of the expected size was amplified in all other samples (data not shown). This further suggested that serovar Typhimurium F98 and ST5306 did not possess sopE and that the ∼30-kDa protein from these two strains may be a different protein. To investigate this protein further, we extracted the 30-kDa protein produced by serovar Typhimurium F98, subjected it to N-terminal amino acid sequencing, and used it for immunization of mice to raise monoclonal antibodies. The sequence of 15 amino acids was determined to be X-N-I-T-L-S-T-Q-H-Y-R-I-H-R-S. This sequence was then analyzed by using the BLAST search program against a partial genomic sequence of serovar Typhimurium and Salmonella enterica serovar Paratyphi available from the Genome Sequencing Center at Washington University School of Medicine, St. Louis, Mo. (http://genome.wustl.edu/gsc/bacterial/salmonella.shtml) and against that of serovar Typhi available from Sanger Center (http://www.sanger.ac.uk/Projects/S_typhi/blast_server.shtml). This analysis revealed a match with the product of an open reading frame (ORF) present in all three Salmonella serotypes. (In the available nucleotide sequence from serovar Typhi there is a deletion of one nucleotide at position 73 within this ORF leading to a frame shift. It is unclear at this moment if this is due to a posisble sequencing mistake.) The deduced amino acid sequence of the protein product of this ORF was highly homologous to SopE (see below). We therefore designated this protein SopE2, and the ORF was designated sopE2. DNA fragments containing a complete sopE2 gene and upstream areas from serovars Dublin 2229 and Typhimurium F98 were amplified by PCR by using an upstream primer SE3 (5′-TTAAGCATAAGCTTAATTCCATTTGTT-3′) and a downstream primer SE4 (5′-CTGATAAATGAATTCAGGCCGCATC-3′) based on the available sequence from serovar Typhi and modified to include restriction sites. Chromosomal DNA from serovars Dublin 2229 and Typhimurium F98 were used as templates. The resulting DNA fragments were cloned into pBluescript to yield pSopE2sd and pSopE2st, respectively. The sequence of the cloned DNA in both plasmids was identical to that of an ORF obtained from the serovar Typhimurium genome database. The deduced amino acid sequence of the SopE2 protein was 64% identical to that of SopE (Fig. 2). The SopE2 sequence was found to be remarkably conserved in different Salmonella serotypes, with only six amino acid substitutions in SopE2 in serovar Paratyphi compared to that in serovars Typhimurium and Dublin. We next designed two primers, SE2up (5′-TCGTGGGAGCGGATCCGAGGGTAGGGCAGTAT-3′) and SE2down (5′-TGCGCAGCCTCGAGTATCTCTTTCAGAAA-3′), from an internal part of sopE2 and used them in PCR with DNA from different Salmonella strains as template. The _sopE2_-specific DNA was present in all Salmonella strains analyzed, including strains with different host specificities (data not shown). Together, these data suggest that the sopE2 sequence is present and conserved in different S. enterica strains.
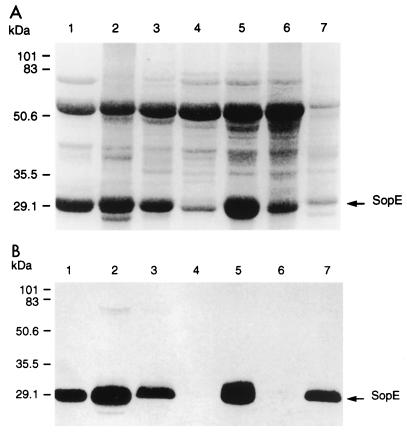
FIG. 1.
Analysis of the proteins secreted by different Salmonella strains (sipB mutants). The secreted proteins deposited by each strain into proteinaceous filaments were isolated as described earlier (17). The protein samples from serovar Dublin B1 (lane 1), Salmonella enterica serovar Gallinarum S9.B1 (lane 2), Salmonella enterica serovar Enteritidis S13.B1 (lane 3), serovar Typhimurium F98.B1 (lane 4), serovar Typhimurium ST4/74.B1 (lane 5), serovar Typhimurium ST5306.B1 (lane 6), and Salmonella enterica serovar Choleraesuis A57.B1 (lane 7) were separated by SDS–12% PAGE and were stained with Coomassie brilliant blue (A) or were transferred onto nitrocellulose membrane and probed with an anti-SopE monoclonal antibody (B). Molecular mass is shown on the left.
FIG. 2.
Sequence alignment of serovars Dublin SopE and Typhimurium F98 SopE2. Lines indicate identical amino acids, and colons and periods indicate conservative amino acid substitutions.
A DNA fragment amplified by using serovar Typhimurium DNA was cloned into the pDM4 suicide plasmid (14) to yield pSB1. sopE2 mutant serovar Typhimurium SE2.1 was constructed by crossing pSB1 from Escherichia coli S17.1 λpir into parental serovar Typhimurium F98, and correct insertion of the suicide plasmid was confirmed by PCR. The mutation was transcomplemented by introducing the pSopE2st plasmid into serovar Typhimurium SE2.1. The growth characteristics of the sopE2 mutant and the transcomplemented strains in LB medium at 37°C were undistinguishable from those of the wild-type strain (data not shown). The proteins produced and secreted by the different Salmonella strains in LB medium at 37°C were isolated as described earlier (17) and were analyzed by SDS-PAGE and Western blotting by using monoclonal anti-SopE2 antibodies. The SDS-PAGE analysis revealed that the sopE2 mutation did not cause any apparent effect on the expression and secretion of secreted proteins other than a 30-kDa protein (data not shown). The Western blot analysis revealed that a faint protein band at 30 kDa, present in the wild-type strain, was missing in the sopE2 mutant but reappeared in the transcomplemented strain (Fig. 3), suggesting that the observed 30-kDa protein is SopE2.
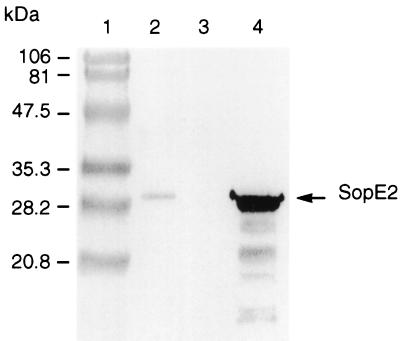
FIG. 3.
Western blot analysis of the secreted proteins of the wild-type serovar Typhimurium F98 (lane 2), serovar Typhimurium SE2.1 sopE2 mutant (lane 3), and serovar Typhimurium SE2.1/pSopE2st transcomplemented sopE2 mutant (lane 4) strains. Culture supernatant proteins from different Salmonella strains were prepared by precipitation with 10% trichloroacetic acid as previously described (17), were separated by SDS–PAGE, were transferred to nitrocellulose membrane, and were probed with anti-SopE2 monoclonal antibody.
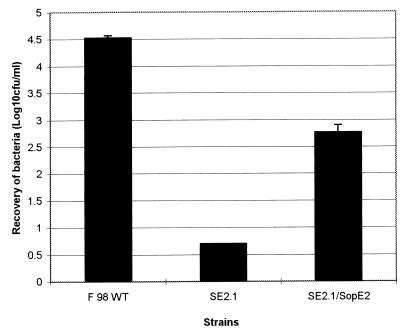
The wild-type, mutant, and transcomplemented strains were then assessed in a cultured cell invasion assay, essentially as described earlier (17). The results of this experiment clearly showed that serovar Typhimurium SE2.1 had a reduced ability to invade HeLa cells (Fig. 4). The invasion phenotype was partially restored by complementing the mutations in trans, demonstrating that the invasion defect in the mutant strain is due to inactivation of sopE2.
FIG. 4.
Invasion of HeLa cells by the wild-type serovar Typhimurium F98, serovar Typhimurium SE2.1 sopE2 mutant, and serovar Typhimurium SE2.1/ pSopE2st transcomplemented sopE2 mutant strains. All strains were grown overnight at 26°C, then diluted 10-fold and grown at 37°C to an optical density at 600 nm of 0.5. The cells were infected with bacteria at an approximate ratio of 1:10. HeLa cells were infected for 1 h and then washed and exposed to gentamicin (100 μg/ml) treatment for 1 h. The cells were lysed with 1% Triton X-100, and the numbers of bacteria surviving gentamicin treatment were calculated by viable count. The experiments were carried out in triplicate. Each bar is derived from the mean of three samples per strain. The error bars indicate standard errors of the means calculated from triplicate plating.
Thus, we have identified SopE2, an invasion-associated secreted protein of Salmonella. Structurally, sopE2 is very similar to sopE, suggesting that these two genes have the same evolutionary origin and/or that one of these genes is a product of gene duplication. The high structural similarity between SopE2 and SopE and the similar phenotypic effects caused by inactivation of SopE- and SopE2-encoding genes suggest that these two proteins are likely to have similar mechanisms of action. Unlike sopE, sopE2 appears to be broadly distributed in salmonellae, suggesting that conservation of this gene may be more important than that of sopE. Thus, it appears that at least some Salmonella strains possess two functional genes, encoding virulence-associated type III secreted proteins highly similar to each other. This finding prompted us to compare sequences of other known secreted proteins with sequences deposited in the Salmonella genome database. No homologues were found for SopB (3), AvrA (4), or SptP (12). However, our analysis revealed that there is a gene encoding a SopD (9) homologue. The sequence of this protein is 41% identical to that of SopD (data not shown).
The type III secretion/translocation mechanism appears to be conserved in many gram-negative bacteria; however, each pathogen has a specific set of type III secreted effector proteins. An ability to expand the repertoire of such effectors without the risk of loosing essential functions may allow pathogens more flexibility in adapting within different host environments. It is interesting to note a remarkable conservation of the SopE2 sequence in different Salmonella serotypes, as this is in contrast to the conservation of SopE. SopE proteins from serovars Dublin and Typhimurium, for example, show only 90% overall identity, indicating a rapid evolution of the sopE gene in Salmonella. It is also worthy to note that the sopE gene is carried by a bacteriophage (5, 15). Together, these data suggest that Salmonella appears to possess a mechanism which enables the preservation of one copy of some genes encoding effector proteins while allowing simultaneous rapid evolution of the gene sequence. Those mutated gene variants that provide selective advantage can then be disseminated throughout the population via mobile genetic elements.
Nucleotide sequence accession number.
The sequence of the DNA fragment from serovar Typhimurium F98 identical to that from serovar Dublin 2229 was deposited in the EMBL database under accession no. AF200952.
Acknowledgments
The work presented here was supported by the Biotechnology and Biological Sciences Research Council of the United Kingdom; the United Kingdom Ministry of Agriculture, Fisheries and Food; the India/UK TOMBIT project financed by the United Kingdom Department for International Development; and the Indian Council for Agricultural Research.
C.S.B. and V.P.S. contributed equally to this work.
REFERENCES
- 1.Darwin K H, Miller V L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán J E. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 3.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 4.Hardt W-D, Galán J E. A secreted Salmonella protein with homology to a virulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardt W-D, Urlaub H, Galán J E. A target of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardt W-D, Chen L-M, Shuebel K E, Bustello X R, Galán J E. Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 7.Hermant D, Menard R, Arricau N, Parsot C, Popoff M Y. Functional conservation of the Salmonella and Shigella effectors of entry into epithelial cells. Mol Microbiol. 1995;17:781–789. doi: 10.1111/j.1365-2958.1995.mmi_17040781.x. [DOI] [PubMed] [Google Scholar]
- 8.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaniga K, Trollinger D, Galán J E. Identification of two targets of the type III secretion system encoded in the inv and spa loci of Salmonella typhimurium that share homology to IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaniga K, Tucker S G, Trollinger D, Galán J E. Homologues of the Shigella invasins IpaB and IpaC are required for Salmonella typhimurium entry into cultured cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaniga K, Uralil J, Bliska J B, Galán J E. A secreted tyrosine phosphatase with modular effector domains encoded by the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 13.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 14.Milton D L, O'Toole R, Hörstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirold S, Rabsch W, Rohde M, Stender S, Tschäpe H, Rūssmann H, Igwe E, Hardt W-D. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from epidemic Salmonella typhimurium strain. Proc Natl Acad Sci USA. 1999;96:9845–9850. doi: 10.1073/pnas.96.17.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 17.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]