Domain Structure of Salmonella FlhB, a Flagellar Export Component Responsible for Substrate Specificity Switching (original) (raw)
Abstract
We have investigated the properties of the cytoplasmic domain (FlhBC) of the 383-amino-acid Salmonella membrane protein FlhB, a component of the type III flagellar export apparatus. FlhB, along with the hook-length control protein FliK, mediates the switching of export specificity from rod- and hook-type substrates to filament-type substrates during flagellar morphogenesis. Wild-type FlhBC was unstable (half-life, ca. 5 min), being specifically cleaved at Pro-270 into two polypeptides, FlhBCN and FlhBCC, which retained the ability to interact with each other after cleavage. Full-length wild-type FlhB was also subject to cleavage. Coproduction of the cleavage products, FlhBΔCC (i.e., the N-terminal transmembrane domain FlhBTM plus FlhBCN) and FlhBCC, resulted in restoration of both motility and flagellar protein export to an flhB mutant host, indicating that the two polypeptides were capable of productive association. Mutant FlhB proteins that can undergo switching of substrate specificity even in the absence of FliK were much more resistant to cleavage (half-lives, 20 to 60 min). The cleavage products of wild-type FlhBC, existing as a FlhBCN–FlhBCC complex on an affinity blot membrane, bound the rod- and hook-type substrate FlgD more strongly than the filament-type substrate FliC. In contrast, the intact form of FlhBC (mutant or wild type) or the FlhBCC polypeptide alone bound FlgD and FliC to about the same extent. FlhBCN by itself did not bind substrates appreciably. We propose that FlhBC has two substrate specificity states and that a conformational change, mediated by the interaction between FlhBCN and FlhBCC, is responsible for the specificity switching process. FliK itself is an export substrate; its binding properties for FlhBC resemble those of FlgD and do not provide any evidence for a physical interaction beyond that of the export process.
Salmonella secretes large amounts of proteins into the culture media. The major secreted proteins are either flagellar proteins or virulence factors (7). They penetrate the cytoplasmic and outer membranes through either flagellar or needle-like structures, which look fairly similar to each other (8). Both systems are called type III export pathways, and their components share substantial sequence similarities (12).
The flagellum, which works as a rotary motor, consists of a basal body, a hook, and a filament. Flagellar assembly begins with the basal body, proceeds with the hook, and finishes with the filament (12). Most of the flagellar proteins do not undergo signal peptide cleavage during export. Following their translocation across the cytoplasmic membrane by an ATP-driven mechanism, they diffuse down a central channel in the nascent flagellar structure and assemble at its distal end (11).
The flagellar export apparatus consists of six membrane proteins, FlhA, FlhB, FliO, FliP, FliQ, and FliR, and three cytoplasmic proteins, FliH, FliI, and FliJ (17, 18). These proteins are called general export components because they are required for rod-type (FlgB, FlgC, FlgF, and FlgG), hook-type (FlgD and FlgE), and filament-type (FlgM, FlgK, FlgL, FliC, and FliD) export substrates (17, 18). The membrane components are believed to be located in a central pore within the membrane-supramembrane ring (MS ring) (2, 6, 21). FliI is an ATPase whose enzymatic activity is necessary for flagellar assembly, suggesting that it may provide the energy for translocation of export substrates across the cytoplasmic membrane (1). FliJ functions as a flagellum-specific chaperone to prevent its export substrates from premature aggregation in the cytoplasm (12a). In addition to these export components, other cytoplasmic proteins—FliS, FlgN, and FliT—are proposed to be substrate-specific chaperones which facilitate the export of their filament-type export substrates (3, 24).
The flagellar export apparatus switches substrate specificity during flagellar morphogenesis. Two of its components, FlhB, which is a 39-kDa cytoplasmic membrane protein with a substantial C-terminal cytoplasmic domain (15), and the hook length control protein FliK, are proposed to be involved in this process (10, 19, 22). During hook assembly, the flagellar export apparatus preferentially exports the rod-type and hook-type export substrates, including FliK itself (14). Upon completion of the hook, the C-terminal domain of FliK (FliKC) somehow communicates (though perhaps not directly) with the C-terminal cytoplasmic domain of FlhB (FlhBC), resulting in substrate specificity switching from rod- and hook-type substrates to filament-type substrates. This means that FlhBC may exist in two substrate specificity states.
flhB mutants that support filament assembly even in the absence of FliK have been isolated (4, 10, 22), demonstrating that these mutant proteins can switch export specificity autonomously. In this study, we have analyzed the properties of the cytoplasmic domain of both wild-type and mutant FlhB proteins. In a previous study (18), N-terminally His-tagged FlhBC (N-His-FlhBC) had an anomalously low apparent molecular mass; however, since this protein had been purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and migrated as a single band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), we assumed it was a single species. In this study we show that FlhBC is fairly unstable, being specifically cleaved into two polypeptides, which we call FlhBCN and FlhBCC. The FlhBCC domain appears to be predominantly responsible for substrate binding. Full-length FlhB (including the transmembrane domain) exhibits the same susceptibility to cleavage; the cleavage products are capable of associating with each other and support both export and assembly of flagellar proteins. All mutant FlhBC versions tested are much less susceptible to cleavage than the wild type, indicating that the suppressor mutations affect the state of the putative hinge region between the FlhBCN and FlhBCC subdomains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Strains and plasmids used in this study are listed in Table 1. Luria-Bertani broth, motility agar plates, and M9 medium were used as described previously (18).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Escherichia coli | ||
| NovaBlue | Recipient for cloning experiments | Novagen |
| BL21 (DE3) pLysS | Overproduction of proteins from pET-based plasmids | Novagen |
| Salmonella | ||
| JR501 | Conversion of _E. coli_-derived plasmids to compatibility with Salmonella | 20 |
| SJW1103 | Wild-type strain for motility and chemotaxis | 23 |
| SJW1383 | flhB | 17 |
| MY2701 | flhB extragenic suppressor of fliK polyhook mutant | 22 |
| MY2714 | flhB extragenic suppressor of fliK polyhook mutant | 22 |
| MY3018 | flhB extragenic suppressor of fliK polyhook mutant | 22 |
| MY3201 | flhB extragenic suppressor of fliK polyhook mutant | 22 |
| MY3501 | flhB extragenic suppressor of fliK polyhook mutant | 22 |
| MY3519 | flhB extragenic suppressor of fliK polyhook mutant | 22 |
| Plasmids | ||
| pMM1 | pET19b/N-His-FlhBC | 18 |
| pMM7 | pET19b/N-His-FLAG-FlhB | This study |
| pMM12 | pET19b/N-His-FLAG-FlhBTM | This study |
| pMM14 | pET22b/C-His-FlhBC | This study |
| pMM15 | pET22b/untagged FlhBC | This study |
| pMM20 | pET19b/N-His-FlhBCC | This study |
| pMM23 | pTrc99A/untagged FlhBΔCC | This study |
| pMM24 | pET19b/N-His-FLAG-FlhBΔCC | This study |
| pMM25 | pTrc99A/untagged FlhBΔCC + N-His-FlhBCC | This study |
| pMM26 | pTrc99A/untagged FlhB | This study |
| pMM28 | pTrc99A/N-His-FlhBCC | This study |
| pMM29 | pTrc99A/untagged FlhBTM + N-His-FlhBCC | This study |
| pMMBc2701NH | pET19b/N-His-FlhBC of MY2701 | This study |
| pMMBc2714NH | pET19b/N-His-FlhBC of MY2714 | This study |
| pMMBc3018NH | pET19b/N-His-FlhBC of MY3018 | This study |
| pMMBc3201NH | pET19b/N-His-FlhBC of MY3201 | This study |
| pMMBc3501NH | pET19b/N-His-FlhBC of MY3501 | This study |
| pMMBc3519NH | pET19b/N-His-FlhBC of MY3519 | This study |
PCR and cloning.
Methods were as described previously (18), except that Red Taq DNA polymerase (Sigma, St. Louis, Mo.) was used.
Radiolabeling experiment with [35S]methionine.
An overnight culture of BL21 (DE3) pLysS (125 μl) carrying pET-based plasmids was inoculated into 2.5 ml of Luria-Bertani broth containing ampicillin and was grown at 37°C with shaking to an optical density at 600 nm of 0.7 to 0.9. The cells were washed twice with M9 medium containing ampicillin. A constant number of cells was suspended in 350 μl of M9 medium containing ampicillin and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and was incubated at 37°C for 20 min. Then, 3.5 μl of 25-mg/ml rifampin was added, and incubation was continued for another 1 h. Two microliters of 1.5-μCi/ml [35S]methionine (Amersham Pharmacia, Piscataway, N.J.) was added, and the mixture was incubated at 37°C for 5 min. The cells were washed twice with 500 μl of 50 mM Tris-HCl (pH 8.0), resuspended in 100 μl of SDS loading buffer, and boiled for 3 min.
For pulse-chase experiments, 2 μl of 1.5-μCi/ml [35S]methionine was added and incubated for 1 min, and then 10 μl of 40-mg/ml l-methionine was added. After the start of the chase, samples were collected at 0, 5, 20, and 60 min, and trichloroacetic acid (TCA) was added to a final concentration of 10%. After centrifugation, cells were washed twice with 500 μl of acetone, resuspended in 100 μl of SDS loading buffer, and boiled for 3 min.
After SDS-PAGE, proteins were transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). After the membranes were dried, they were exposed on X-ray film.
Preparation of whole-cell proteins.
Whole-cell proteins were prepared from BL21 (DE3) pLysS carrying pET-based plasmids as described previously (18).
Immunoblotting and affinity blotting.
Immunoblotting with anti-FLAG (Sigma), anti-FlhBC (a gift of K. Kutsukake [15]), anti-FlgD, anti-FliC, and anti-FliK antibodies was performed as described previously (17). Affinity blotting was carried out as described previously (18).
Purification of C-terminally His-tagged FlhBC and N-terminally His-tagged FlgD, FliC, and FliK.
C-terminally His-tagged FlhBC and N-terminally His-tagged FlgD, FliC, and FliK were purified with a Ni-NTA agarose column (Qiagen, Valencia, Calif.) as described previously (18).
Swarming assay.
SJW1383 (flhB) was transformed with pTrc99A-based plasmids, and the resulting transformants were inoculated onto motility agar plates containing ampicillin and incubated at 30°C for 8 h.
Preparation of soluble cellular fractions and culture supernatants.
Soluble cellular fractions and culture supernatants were prepared from SJW1383 (flhB) carrying pTrc99A-based plasmids as described previously (17). The proteins in the supernatants were precipitated by 10% TCA, suspended in Tris-saturated SDS loading buffer, and boiled for 3 min.
Amino acid sequencing.
N-terminal amino acid sequencing was performed by the Keck Foundation Biotechnology Resource Laboratory, Yale University.
RESULTS
Cytoplasmic domains of mutant FlhB proteins.
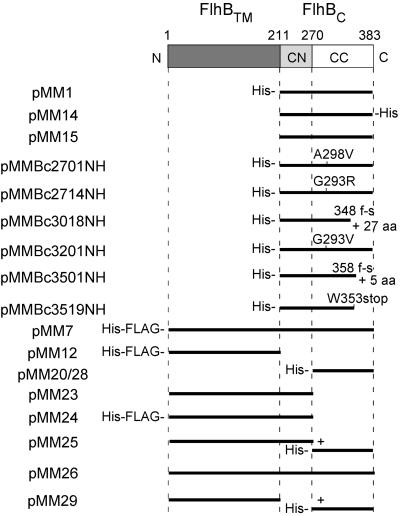
We had previously cloned the wild-type cytoplasmic domain of FlhB (FlhBC) into pET19b to give plasmid pMM1 (18). To better understand the role of FlhBC in substrate specificity switching of the flagellar export apparatus, we cloned the cytoplasmic domains of various flhB alleles from fliK extragenic suppressor mutants to produce plasmids pMMBc2701NH through pMMBc3519NH (Table 1 and Fig. 1). All of these FlhBC domains have a His tag at their N terminus, which starts at Phe-211 (18). The plasmids were introduced into BL21 (DE3) pLysS, and the resulting transformants were labeled with [35S]methionine and subjected to SDS-PAGE and autoradiography.
FIG. 1.
The primary structure of the FlhB protein and schematic representations of the FlhB products encoded by the plasmids listed in Table 1. FlhB consists of an N-terminal transmembrane region (FlhBTM; dark gray) imbedded in the cytoplasmic membrane and a C-terminal cytoplasmic domain (FlhBC), which is responsible for the switching of export substrate specificity. The present study establishes that FlhBC can be divided into two subdomains, FlhBCN (light gray) and FlhBCC (white). Residue numbers defining the boundaries of these domains are indicated. For the plasmids, the extent of sequence present is indicated by solid bars. Plasmids pMM20 and pMM28 encode the same protein, but on different vectors (see Table 1). Plasmid pMM25 encodes two proteins, the first consisting of FlhBTM plus FlhBCN (i.e., FlhBΔCC) and the second consisting of FlhBCC; similarly, plasmid pMM29 encodes both FlhBTM and FlhBCC. Missense mutations are indicated as, e.g., A298V. Frameshift mutations are indicated by f-s, along with the number of additional (frame-shifted) residues generated. His-, N-terminal His tag; -His, C-terminal His tag; His-FLAG-, N-terminal His and FLAG tags.
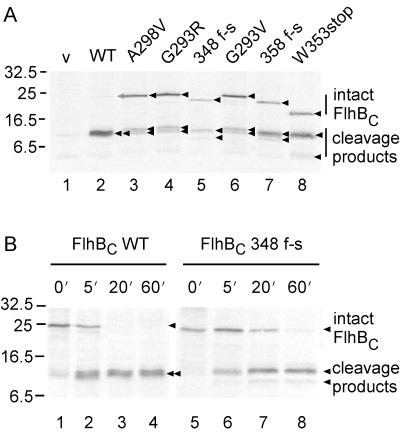
With pMM1 (N-His-FlhBC [wild-type]), a single major product with an apparent molecular mass of ca. 11.5 kDa was detected (Fig. 2A, lane 2), as was reported previously (18). With the plasmids encoding mutant versions of FlhBC, in addition to a band or bands at about 11.5 kDa, there was another major band, with an apparent molecular mass of about 22 kDa in the case of the missense mutants (lanes 3, 4, and 6) and with slightly smaller masses in the cases of the frameshift and nonsense mutants (lanes 5, 7, and 8). The apparent molecular masses of these upper bands correspond closely to their deduced molecular masses. With FlhBC A298V (lane 3), G293R (lane 4), and G293V (lane 6), the 11.5-kDa band was resolved into a closely spaced doublet. With FlhBC 348 f-s (lane 5), 358 f-s (lane 7), and W353stop (lane 8), the 11.5-kDa band appeared as a singlet, but there was another band with a lower apparent molecular mass. These results suggested that the bands of 11.5 kDa and smaller might result from cleavage of FlhBC.
FIG. 2.
(A) Autoradiogram of wild-type and mutant cytoplasmic domains (FlhBC) of FlhB. BL21 (DE3) pLysS cells carrying pET19-based plasmids were radiolabeled with [35S]methionine and subjected to SDS-PAGE. Lane 1, pET19b (vector, v); lane 2, pMM1 (wild-type FlhBC, WT); lane 3, pMMBc2701NH (FlhBC A298V); lane 4, pMMBc2714NH (FlhBC G293R); lane 5, pMMBc3018NH (FlhBC frame-shifted, f-s, at residue 348); lane 6, pMMBc3201NH (FlhBC G293V); lane 7, pMMBc3501NH (FlhBC 358 f-s); lane 8, pMMBc3519NH (FlhBC W353stop). The proteins in lanes 2 to 8 all carry an N-terminal His tag. The various intact forms and specific cleavage products are indicated by arrows. The intact form of wild-type FlhBC is barely visible and is indicated by a gray arrow. Molecular mass markers are shown to the left. (B) Pulse-chase experiments with wild-type and mutant versions of N-terminally His-tagged FlhBC. BL21 (DE3) pLysS cells carrying pET19B-based plasmids were radiolabeled with [35S]methionine for 1 min. After addition of excess unlabeled l-methionine, the cells were incubated for the times indicated and subjected to SDS-PAGE and autoradiography. Lanes 1 to 4, pMM1 (wild-type FlhBC, WT); lanes 5 to 8, pMMBc3018NH (FlhBC 348 f-s). The various intact forms and specific cleavage products are indicated by arrows. Molecular mass markers are shown to the left.
Stability of wild-type and mutant FlhBC proteins.
To examine whether wild-type FlhBC is cleaved, we carried out pulse-chase experiments (Fig. 2B). Intact wild-type FlhBC was detected at the zero time point but was rapidly cleaved (half-life, about 5 min; lanes 1 to 4). After prolonged incubation (more than 20 min) in the presence of excess unlabeled methionine, no intact FlhBC could be detected.
We had found that the intact forms of the mutant FlhBC proteins could be readily detected by autoradiography (Fig. 2A), indicating that they were more stable. This was confirmed by the pulse-chase experiments. In all cases, as is illustrated for FlhBC 348 f-s (Fig. 2B, lanes 5 to 8), the intact forms could be detected even after an extended chase (half-lives, about 20 to 60 min).
Identification of C-terminally His-tagged FlhBC and untagged FlhBC products.
As described above, wild-type N-terminally His-tagged FlhBC revealed only a single band, at 11.5 kDa. We suspected that this band must be a fortuitous superposition of two cleavage products. To test this, we constructed two plasmids, pMM14 and pMM15, which encode C-terminally His-tagged and untagged wild-type FlhBC, respectively.
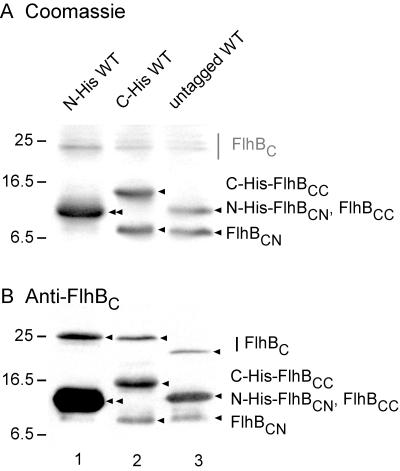
After SDS-PAGE of whole cells transformed with these plasmids, we carried out Coomassie staining (Fig. 3A). Unlike the case with N-His-FlhBC (lane 1), the cleavage products of both C-His-FlhBC and untagged FlhBC were cleanly resolved into two bands (lanes 2 and 3). In the case of C-His-FlhBC (lane 2), one band was at a higher position and the other was at a lower position than that of the single band seen with N-His-FlhBC (lane 1). With the untagged protein (lane 3), the upper band coincided with the single band of N-His-FlhBC, while the lower band coincided with the lower band of C-His-FlhBC. All bands (including the uncleaved forms at around 22 to 25 kDa) were recognized by anti-FlhBC antibody (Fig. 3B). We identified the upper of the two lower bands shown in lanes 2 and 3 as C-terminally His-tagged and untagged FlhBCC, respectively, and the lower of the two lower bands in both cases as untagged FlhBCN; the polyclonal antibody appears to recognize FlhBCC more strongly than FlhBCN. We conclude that the single band in lane 1 is indeed a superposition of N-His-FlhBCN and FlhBCC.
FIG. 3.
Cleavage products from variously tagged versions of wild-type FlhBC. BL21 (DE3) pLysS cells transformed with pET-based plasmids were grown, subjected to SDS-PAGE, and stained with Coomassie blue (A) or immunoblotted using polyclonal anti-FlhBC antibody (B). Lane 1, pMM1 (N-terminally His-tagged FlhBC, N-His WT); lane 2, pMM14 (C-terminally His-tagged FlhBC, C-His WT); lane 3, pMM15 (untagged FlhBC, untagged WT). Intact forms of FlhBC are not clearly evident in panel A, and so their approximate positions are indicated by a gray bar; in panel B, their positions are indicated by arrows. In both A and B, the positions (arrows) and identities of cleavage products are identified (see the text for nomenclature). In lane 1, N-His-FlhBCN and untagged FlhBCC comigrate and are indicated by a double arrow. Molecular mass markers are shown to the left.
N-terminal amino acid sequencing of cleaved C-His-FlhBC.
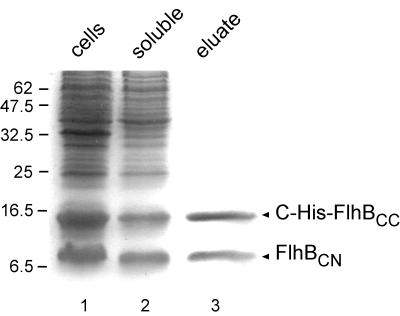
The sharpness of the bands shown in Fig. 3, lanes 2 and 3, suggested there was a unique cleavage site. We therefore purified the cleavage products of C-His-FlhBC using Ni-NTA affinity chromatography. Following elution by 200 mM imidazole, two major bands were seen by SDS-PAGE (Fig. 4, lane 3) at the same positions seen in Fig. 3, lane 2. Since FlhBCN did not carry a His tag, coelution from the Ni-NTA column established that FlhBCN and FlhBCC maintain a stable association with each other even after cleavage. (With untagged FlhBC, all proteins were eluted in the flowthrough [data not shown]). The two bands were transferred onto polyvinylidene difluoride membranes, and their N-terminal sequences were determined. For the lower band, the sequence was (M)FQIFSXLXXLXMSRQDIRDEF, where X is unknown, which (after a methionine deriving from the vector) corresponds to amino acids Phe-211 to Phe-231 of FlhB (15) and is in agreement with the deduced N-terminal sequence of the cloned fragment. For the upper band, the sequence was PTXYXVALQYDENKMSAPKVVAK, corresponding to the sequence of FlhB from Pro-270 to Lys-292 (15). This confirms the identification of the lower and upper bands as FlhBCN and C-His-FlhBCC, respectively, and establishes that there is a unique cleavage site between Asn-269 and Pro-270 of the FlhB sequence.
FIG. 4.
Copurification by Ni-NTA chromatography of the cleavage products (C-terminally His-tagged FlhBCC and untagged FlhBCN) of C-terminally His-tagged FlhBC. Coomassie blue-stained SDS-PAGE gel of samples during purification. Lane 1, whole-cell lysates of BL21 (DE3) pLysS carrying pMM14 (C-His-FlhBC); lane 2, soluble fraction (supernatant after ultracentrifugation; load to Ni-NTA column); lane 3, eluate from Ni-NTA column at an imidazole concentration of ca. 200 mM. Molecular mass markers are shown to the left.
Affinity blotting with export substrates and wild-type and mutant FlhBC proteins.
Our previous study had shown that on affinity blots, N-His-FlhBC binds more strongly to rod- and hook-type substrates than to filament-type substrates (18). In this study, we have shown that the 11.5-kDa band seen with N-His-FlhBC is a superposition of two cleavage fragments, N-His-FlhBCN and FlhBCC (Fig. 3). Thus, it was FlhBC, cleaved, refolded, and presumably reassociated (cf. Fig. 4) on the nitrocellulose membrane, that was predominantly in the rod- and hook-type specificity state.
The mutant FlhB proteins used in this study were isolated as second-site suppressors of FliK mutants and can switch substrate specificity from the rod and hook type to the filament type even in the absence of FliK (10, 22), suggesting that unlike wild-type FlhBC, they might bind to both rod- and hook-type substrates and filament-type substrates. To examine this hypothesis, we performed affinity blotting with mutant versions of FlhBC as targets and purified N-terminally His-tagged export substrates as probes.
The intact forms of the mutant FlhBC proteins could in all cases be detected on Coomassie blue-stained gels, as is illustrated in Fig. 5A for FlhBC A298V (lane 3) and FlhBC 348 f-s (lane 4) (cf. Fig. 2A). The cleavage products from wild-type FlhBC (lanes 1 and 2), FlhBC 348 f-s (lane 4), and FlhBC 358 f-s and FlhBC W353stop (data not shown) were also detected, while those from FlhBC A298V (lane 3) and FlhBC G293R and FlhBC G293V (data not shown) were not detected, even by immunoblotting; however, they had been detected by radiolabeling (Fig. 2A).
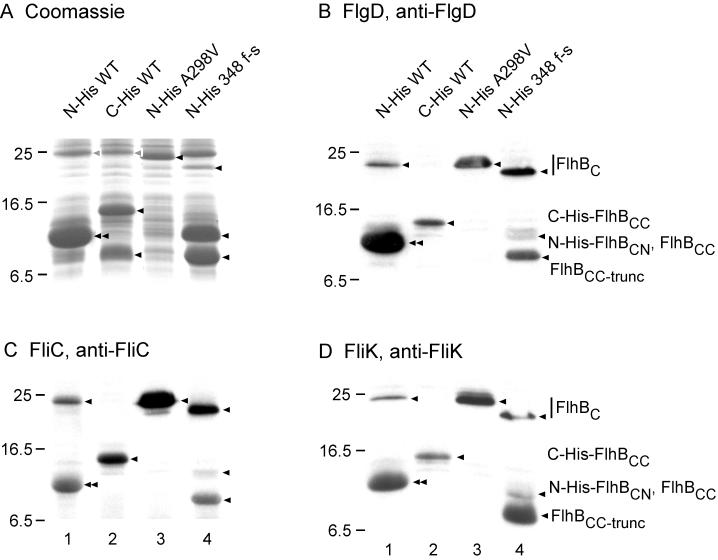
FIG. 5.
Affinity blotting of various forms of FlhBC and their cleavage products. (A) Coomassie blue-stained SDS-PAGE gels of whole-cell lysates from BL21 (DE3) pLysS carrying various pET19b- or pET22b-based plasmids. (B, C, and D) Affinity blotting of the same samples as shown in panel A, using purified N-His-FlgD, N-His-FliC, and N-His-FliK, respectively, as the probe and detection of the probe with the corresponding polyclonal antibody. Lanes 1, pMM1 (N-His-FlhBC WT); lanes 2, pMM14 (C-His-FlhBC WT); lanes 3, pMMBc2701NH (N-His-FlhBC A298V); lanes 4, pMMBc3018NH (N-His-FlhBC 348 f-s). The positions of the target proteins are indicated by arrowheads. The designation FlhBCC-trunc reflects the fact that FlhB 348 f-s is truncated near its C terminus because of the frameshift mutation. Molecular mass markers are shown to the left.
For affinity blotting, we used purified N-His-FlgD (a hook capping protein) and N-His-FliC (flagellin) as examples of rod- and hook-type substrates and filament-type substrates, respectively. Both FlgD and FliC bound strongly to intact mutant FlhBC (Fig. 5B and C, lanes 3 and 4). They also bound to intact N-terminally His-tagged wild-type FlhBC, although it was present only in small amounts (lane 1); intact C-terminally His-tagged wild-type FlhBC could not even be detected (lane 2). FlgD bound strongly to the (superposed) cleavage products of N-terminally His-tagged wild-type FlhBC (Fig. 5B, lane 1), whereas FliC bound more weakly (Fig. 5C, lane 1); this result is consistent with previous observations (18). Both FlgD and FliC bound to C-His-FlhBCC but not to FlhBCN (Fig. 5B and C, lane 2). N-His-FlhBCC 348 f-s interacted strongly with both FlgD and FliC (Fig. 5B and C, lane 4), whereas FlhBCN interacted only slightly if at all. This was also true of FlhBC 358 f-s and FlhBC W353stop (data not shown). Thus, the FlhBCN polypeptide does not appear to contribute directly to substrate binding, although it may enhance binding by the FlhBCC polypeptide (lane 1).
Affinity blotting with FliK as the probe.
Upon hook completion, FliK somehow communicates with FlhBC, resulting in a switch from rod- and hook-type substrates to filament-type substrates (10, 19, 22). However, whether FliK physically interacts with FlhBC has not been established. To address this issue, we performed affinity blotting with purified N-His-FliK as the probe and wild-type and mutant FlhBC as targets (Fig. 5D).
FliK bound to intact N-His-FlhBC (which was only present in small amounts) and bound strongly to its superposed cleavage products (lane 1). It bound to the C-His-FlhBCC cleavage product of C-His-FlhBC but not to the FlhBCN product (lane 2). It bound strongly to intact FlhBC A298V (lane 3). FliK bound to intact FlhBC 348 f-s (present in relatively small amounts) and strongly to its FlhBCC cleavage fragment (lane 4), but slightly if at all to its FlhBCN fragment. The overall pattern of binding resembles that of FlgD (Fig. 5B) and suggests that FliK is a rod and hook type of export substrate, as has also been concluded from its export characteristics (14).
Full-length wild-type FlhB also undergoes specific proteolytic cleavage within its cytoplasmic domain.
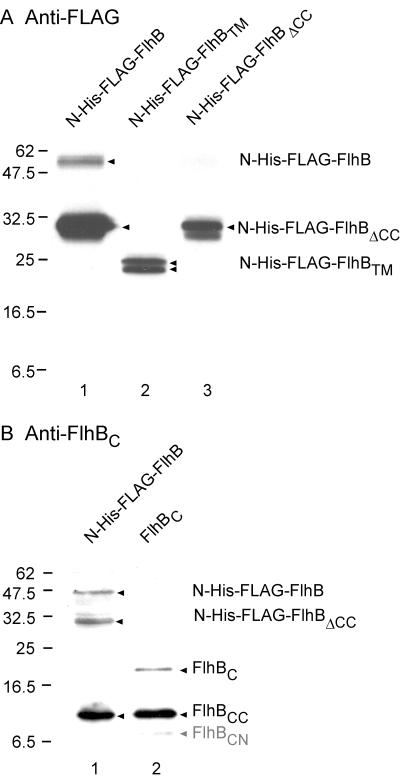
Is cleavage of FlhBC a consequence of its isolation from the transmembrane domain FlhBTM, or is the intact FlhB protein also subject to cleavage? We constructed plasmid pMM7, which encodes N-terminally His-FLAG-tagged intact wild-type FlhB (containing the transmembrane domain as well as the cytoplasmic domain) and demonstrated by immunoblotting with both anti-FLAG and anti-FlhBC antibodies that it too was cleaved into two polypeptides, N-His-FLAG-FlhBΔCC (Fig. 6A and B, lane 1) and FlhBCC (Fig. 6B, lane 1). The identity of N-His-FLAG-FlhBΔCC was confirmed by pMM24, which encodes this protein (Fig. 6A, lane 3). The identity of FlhBCC was confirmed by plasmid pMM15, which encodes FlhBC and yields the cleavage product FlhBCC (Fig. 6B, lane 2). Thus, the property of specific proteolytic cleavage is retained in intact FlhB. The instability of the intact molecule was further indicated by its weak detection by anti-FLAG antibody (Fig. 6A, lane 1) and anti-FlhBC antibody (Fig. 6B, lane 1); the identity of the full-size molecule was confirmed by its comigration with full-length N-His-FLAG-FlhB G293R (data not shown), which withstands cleavage to a considerable degree (cf. Fig. 2A, lane 4).
FIG. 6.
Cleavage properties of full-size FlhB (i.e., the transmembrane region FlhBTM plus the cytoplasmic domain FlhBC. (A) Immunoblot using monoclonal anti-FLAG antibody. Lane 1, pMM7 (N-His-FLAG-FlhB); lane 2, pMM12 (N-His-FLAG-FlhBTM); lane 3, pMM24 (N-His-FLAG-FlhBΔCC). (B) Immunoblot using polyclonal anti-FlhBC antibody. Lane 1, pMM7 (N-His-FLAG-FlhB); lane 2, pMM15 (untagged FlhBC). The proteins indicated at the right are marked by arrowheads. In panel B, the band corresponding to FlhBCN is very faint and is indicated in gray. Molecular mass markers are shown to the left.
We found no evidence for cleavage of full-size FlhB at around Phe-211, i.e., the approximate boundary between the transmembrane and cytoplasmic domains; pMM12, which encodes N-His-FLAG-FlhBTM, indicated the position of that protein (Fig. 6A, lane 2) as a doublet that may indicate a slight degree of proteolysis at its C terminus. The lack of cleavage at the membrane-cytoplasm interface of FlhB is not particularly surprising, since FlhBC was an engineered domain, not a naturally generated fragment.
Coproduction of FlhBΔCC and N-His-FlhBCC complements the motility and export defects of a flhB mutant.
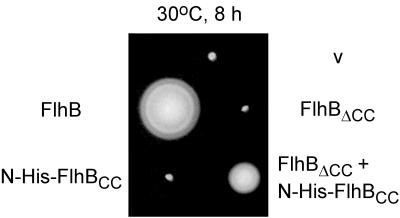
FlhBCN and FlhBCC interact with each other, as judged by Ni-NTA chromatography (Fig. 4). We wanted to know whether the interaction still existed with FlhBΔCC and FlhBCC or whether the transmembrane domain might disrupt it. Because we were dealing here with the entire protein sequence, we could address the issue directly by functional assays, namely, swarming and protein export. We constructed plasmid pMM25, which encodes both FlhBΔCC and N-His-FlhBCC on pTrc99A; as controls, we also constructed pMM23 (FlhBΔCC), pMM26 (wild-type FlhB), pMM28 (N-His-FlhBCC), and pMM29 (FlhBTM + N-His-FlhBCC). SJW1383 (flhB) was transformed with these plasmids, and the resulting transformants were inoculated on motility agar plates containing ampicillin (Fig. 7).
FIG. 7.
Swarming ability of SJW1383 (flhB) transformed with various pTrc99A-based plasmids: pTrc99a (v), pMM26 (untagged FlhB), pMM23 (untagged FlhBΔCC), pMM28 (N-His-FlhBCC), and pMM25 (untagged FlhBΔCC + N-His-tagged FlhBCC).
FlhBΔCC, lacking the essential FlhBCC subdomain, did not complement. N-His-FlhBCC also failed to complement, presumably because the transmembrane domain is essential for export. However, FlhBΔCC + N-His-FlhBCC restored motility to a considerable degree, although not to the wild-type level. Thus, FlhBΔCC and N-His-FlhBCC still have the ability to interact with each other to make a functional complex. The FlhBCN region is apparently necessary for this interaction, since FlhBTM + N-His-FlhBCC (plasmid pMM29) failed to complement (data not shown).
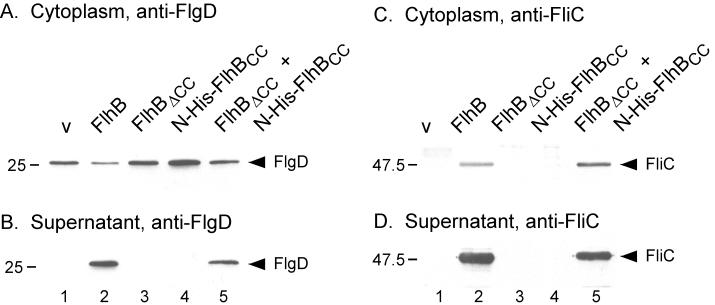
To examine whether restoration of motility caused by FlhBΔCC + N-His-FlhBCC results from restoration of flagellar protein export, we prepared the cytoplasmic contents and culture supernatants from SJW1383 (flhB) carrying pTrc99A, pMM26, pMM23, pMM28, or pMM25 and carried out immunoblotting with anti-FlgD (hook-capping protein) (Fig. 8A and B) and anti-FliC (flagellin) antibodies (Fig. 8C and D).
FIG. 8.
Immunoblotting, using polyclonal antibodies against the hook-capping protein (FlgD) (A and B) and flagellin (FliC) (C and D) of the cytoplasmic fraction (A and C) and culture supernatants (B and D) from SJW1383 (flhB) cells transformed with various pTrc99A-based plasmids. Lane 1, pTrc99a (v); lane 2, pMM26 (untagged FlhB); lane 3, pMM23 (untagged FlhBΔCC); lane 4, pMM28 (N-His-FlhBCC); and lane 5, pMM25 (untagged FlhBΔCC + N-His-tagged FlhBCC). The positions of FlgD and FliC are indicated. Molecular mass markers are shown to the left.
Cytoplasmic levels of FlgD were similar in all of the transformants (Fig. 8A), although those of FlhB (lane 2) and of FlhBΔCC + N-His-FlhBCC (lane 5) were somewhat lower. Consistent with the complementation data, FlgD was found in the culture supernatants when intact FlhB (Fig. 8B, lane 2) or FlhBΔCC and N-His-FlhBCC (lane 5) were being produced; the lower cytoplasmic levels of FlgD shown in Fig. 8A, lanes 2 and 5, presumably are a consequence of this export. No FlgD was found in the culture supernatants when either FlhBΔCC or N-His-FlhBCC was being produced individually (Fig. 8B, lanes 3 and 4).
In the case of FliC, production of intact FlhB or FlhBΔCC + N-His-FlhBCC resulted in similar cytoplasmic levels (Fig. 8C, lanes 2 and 5), and FliC was found in considerable quantities in the culture supernatants in both cases (Fig. 8D, lanes 2 and 5). Production of either no FlhB (Fig. 8C, lane 1) or of FlhBΔCC or N-His-FlhBCC alone (lanes 3 and 4) resulted in a failure to detect cytoplasmic FliC, presumably because of failure to export FlgM and activate class 3 gene expression (5, 9). As expected, FliC was not found in the culture supernatants in these cases either (Fig. 8D, lanes 1, 3, and 4).
DISCUSSION
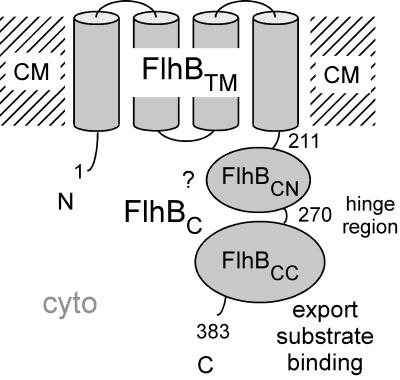
Salmonella FlhB, a component of the type III flagellar export apparatus (17), is a cytoplasmic membrane protein whose amino acid sequence indicates that it consists of two domains, a hydrophobic N-terminal domain (FlhBTM) that is predicted to be capable of crossing the membrane about four times and a hydrophilic C-terminal domain (FlhBC) that is predicted to be soluble and lie in the cytoplasm (Fig. 9).
FIG. 9.
Cartoon of the structure of FlhB, consisting of an N-terminal transmembrane domain, FlhBTM, and a C-terminal cytoplasmic domain, FlhBC. The boundary between FlhBTM and the cloned FlhBC protein is at Phe-211. FlhBC consists of two subdomains, FlhBCN and FlhBCC, linked by a proteolytically sensitive hinge centered at Pro-270. FlhBCC appears to be directly involved in binding export substrates. The role of FlhBCN is less clear (as indicated by the question mark), but since the two subdomains physically interact, its role may be to control the binding specificity state of FlhBCC. cyto, cytoplasmic compartment; CM, plane of the cytoplasmic membrane. FlhB and other membrane components of the export apparatus are believed to exist in a patch of membrane within the flagellar basal-body MS ring and deliver their substrates into the lumen of the nascent rod-hook-filament structure (cf. Fig. 6 of reference 17).
We previously cloned FlhBC and used it in a study of protein interactions among export apparatus components (18). Based on analyses of extragenic suppressors of the hook-length control gene fliK, FlhBC is likely to be responsible for substrate specificity switching of the flagellar export apparatus (10, 22). These flhB mutants can switch from export of rod- and hook-type substrates to export of filament-type substrates even in the total absence of FliK. All of the suppressor mutations lie within the C-terminal half of FlhBC. In this study, we have examined the properties of FlhBC, using the wild-type version and also versions from these extragenic fliK suppressor mutants.
FlhB can be divided into three domains, FlhBTM, FlhBCN, and FlhBCC.
N-terminally His-tagged FlhBC exhibited an apparent molecular mass of ca. 11.5 kDa, which is anomalously low compared to its deduced molecular mass of ca. 22 kDa (18). However, since this apparent mass was for a protein that had been purified by Ni-NTA affinity chromatography and that migrated as a single band on SDS-PAGE, we did not at the time suspect that it might consist of a superposition of two cleavage products. In this study, we have shown that FlhBC is fairly unstable (Fig. 2) and is specifically cleaved at Pro-270 into two polypeptides, FlhBCN and FlhBCC, which in the untagged case have apparent molecular masses of ca. 8.5 and 11.5 kDa, respectively (Fig. 3). Because these polypeptides are much more stable than the intact form of FlhBC, we conclude that they correspond to subdomains within the cytoplasmic domain structure. Therefore we conclude that FlhB consists of three domains, FlhBTM (the N-terminal transmembrane region), FlhBCN, and FlhBCC (Fig. 9).
On a Ni-NTA column, untagged FlhBCN was retained with C-His-FlhBCC and coeluted at an imidazole concentration of 200 mM (Fig. 4), indicating that the two fragments interact with each other. Coproduction of FlhBΔCC (FlhBTM + FlhBCN) with N-His-FlhBCC resulted in substantial complementation of a flhB mutant with respect to both swarming (Fig. 7) and protein export (Fig. 8), demonstrating that they too form a complex and that this complex is functional. Therefore, we feel confident in our conclusion that purified FlhBC exists in essentially its native state, even after it has been cleaved.
Although the existence of a highly specific proteolytically sensitive site within FlhBC has been useful for the analysis of this protein and probably indicates a hinge-like boundary between the FlhBCN and FlhBCC subdomains, we cannot conclude that cleavage plays any physiologically important role or even that it occurs in vivo to a significant extent. We suspect that it does not, since FlhBC domains from suppressor alleles can still undergo specificity switching yet are much less sensitive to cleavage (Fig. 2).
A conformation change of FlhBC, and specifically of FlhBCC, may be responsible for the substrate specificity switching of the flagellar export apparatus.
Based on a number of lines of evidence (7, 10, 13, 19, 22), it has been thought that FlhBC exists in two substrate specificity states: for export of rod- and hook-type substrates and of filament-type substrates. In this study, we have shown that intact forms of wild-type and mutant FlhBC bind to both FlgD (hook capping protein) and FliC in affinity blots (Fig. 5), suggesting that FlhBC can recognize both rod- and hook-type substrates and filament-type substrates. Cleaved N-terminally His-tagged wild-type FlhBC, where the N-His-FlhBCN and FlhBCC fragments remain associated, binds more strongly to FlgD than to FliC (18; this study). However, the wild-type or mutant FlhBCC fragment (not associated with FlhBCN) retains the ability to interact with both FlgD and FliC to a similar extent (Fig. 5). Therefore, we propose that it is the association of FlhBCN with FlhBCC that enables the latter to function as the export specificity switch. For reasons that we do not understand, proteolysis leaves the FlhBCN–FlhBCC complex with a bias towards the rod- and hook-type substrate specificity state.
The key role of the FlhBCC peptide in substrate binding is supported by data from the present study (Fig. 5). Its key role in switching is supported by the finding that all known extragenic fliK suppressor mutations lie within this peptide; none have been found within the FlhBCN peptide (10, 22). The suppressor mutations do not have to lie close to the subdomain boundary; indeed, some lie quite close to the C terminus of the protein (Fig. 1).
fliK suppressor activity and resistance to proteolysis are correlated.
Many flhB mutants have been isolated which have the ability to initiate export of filament-type export substrates even in the absence of FliK (4, 10, 22), i.e., they can be thought of as autonomous switchers of the specificity state. Pulse-chase experiments in this study show that the mutant FlhBC proteins are much more stable than wild-type FlhBC (half-life, about 20 to 60 min versus 5 min for the wild type) (Fig. 2B), indicating that the proteolytically sensitive site at Pro-270 is somehow protected in these proteins and therefore that their conformation must be different from that of the wild type. It is striking that these mutations all lie downstream of Pro-270 (ranging from residue 275 to the C terminus at residue 383), i.e., they are all located within FlhBCC. We therefore suggest that a conformational change of FlhBCC, FliK driven for the wild type but spontaneous for the mutants, is responsible for the substrate specificity switching of the flagellar export apparatus.
In one temperature-sensitive suppressor mutant, flhB4265, with the mutation S274P, both flagellin (FliC) and hook protein (FlgE) are exported at the permissive temperature of 30°C even in the absence of FliK. However, at the restrictive temperature of 42°C, only FliC export still occurs (16). Thus, at the restrictive temperature it is locked in the filament-type specificity state, further supporting our idea of conformational switching within FlhBCC.
The hook length control protein FliK binds to FlhBC.
Although it has been proposed that FliK interacts with FlhBC upon hook completion to effect the switch in substrate specificity (10, 19, 22), it has not been proven that the interaction involves actual binding of FliK to FlhBC. In this study, we have shown that FliK does in fact bind, both to FlhBC and to its C-terminal subdomain (Fig. 5D). However, it is now known that FliK is also an export substrate recognized by the flagellar export apparatus and that it belongs to the rod- and hook-type export class (14). Its binding properties for FlhB do not appear to be different from those of other export substrates, and in particular of substrates of the rod and hook type. Our data at this point do not permit us to say whether FlhBC also recognizes FliK in connection with its role in the hook length control process. Indeed, it is difficult to reconcile a location of FliK following its export with an interaction with a cytoplasmic domain of FlhB.
The role of FlhBCN.
The results of this study suggest that in contrast to intact FlhB and FlhBCC, FlhBCN does not play a major or direct role in substrate binding, regardless of substrate class. However, failure of complementation when FlhBTM and FlhBCC are coproduced demonstrates that FlhBCN is important for the overall process of export. It cannot simply be a passive linker between FlhBTM and FlhBCC, since it specifically associates with FlhBCC. Also, the fact that the binding specificity of FlhBCC is biased towards the rod- and hook-type state when it is interacting with FlhBCN argues for an active role for the latter.
The agreement between the proteolytically sensitive site at N269-P270 and the N-terminal boundary (S274) of the considerable spectrum of FlhB mutations that have been isolated (10, 22) seems too precise to be a coincidence. What is the reason for the phenotypic silence of the FlhBCN domain? To understand this, it must be remembered that these mutations were all isolated in a specific context, namely, acquisition of the ability to switch export substrate specificity even in the absence of instructions from FliK. To further explore the role of FlhBCN, it will be necessary to apply a different selection or screening strategy or to make a systematic scan of the region.
ACKNOWLEDGMENTS
We thank May Kihara for technical assistance and Kazuhiro Kutsukake for the gift of polyclonal anti-FlhBC antibody.
This work has been supported by USPHS grants AI12202 and GM40335.
REFERENCES
- 1.Fan F, Macnab R M. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- 2.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 3.Fraser G M, Bennett J C Q, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 4.Hirano T, Yamaguchi S, Oosawa K, Aizawa S-I. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 6.Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa S-I. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J Mol Biol. 1996;255:458–475. doi: 10.1006/jmbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 7.Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa S-I. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol. 1999;34:767–779. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- 8.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 9.Kutsukake K. Excretion of the anti-sigma factor through a flagellar structure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 10.Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macnab R M. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 12a.Minamino T, Chu R, Tamaguchi S, Macnab R M. Role of FliJ in flagellar protein export in Salmonella. J Bacteriol. 2000;182:4207–4215. doi: 10.1128/jb.182.15.4207-4215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minamino T, Doi H, Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci Biotechnol Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- 14.Minamino T, González-Pedrajo B, Yamaguchi K, Aizawa S-I, Macnab R M. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 15.Minamino T, Iino T, Kutsukake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol. 1994;176:7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minamino T, Kutsukake K. Role of the Salmonella typhimurium FlhB protein in the flagellum-specific export pathway. Genes Genet Syst. 1996;71:420. [Google Scholar]
- 17.Minamino T, Macnab R M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minamino T, Macnab R M. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 19.Muramoto K, Makishima S, Aizawa S-I, Macnab R M. Effect of the cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J Mol Biol. 1998;277:871–882. doi: 10.1006/jmbi.1998.1659. [DOI] [PubMed] [Google Scholar]
- 20.Ryu J, Hartin R J. Quick transformation in Salmonella typhimurium LT2. BioTechniques. 1990;8:43–44. [PubMed] [Google Scholar]
- 21.Suzuki H, Yonekura K, Murata K, Hirai T, Oosawa K, Namba K. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in Type III protein export. J Struct Biol. 1998;124:104–114. doi: 10.1006/jsbi.1998.4048. [DOI] [PubMed] [Google Scholar]
- 22.Williams A W, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, Macnab R M. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 24.Yokoseki T, Kutsukake K, Ohnishi K, Iino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141:1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]