The Role of Alpha/Beta and Gamma Interferons in Development of Immunity to Influenza A Virus in Mice (original) (raw)
Abstract
During influenza virus infection innate and adaptive immune defenses are activated to eliminate the virus and thereby bring about recovery from illness. Both arms of the adaptive immune system, antibody neutralization of free virus and termination of intracellular virus replication by antiviral cytotoxic T cells (CTLs), play pivotal roles in virus elimination and protection from disease. Innate cytokine responses, such as alpha/beta interferon (IFN-α/β) or IFN-γ, can have roles in determining the rate of virus replication in the initial stages of infection and in shaping the initial inflammatory and downstream adaptive immune responses. The effect of these cytokines on the replication of pneumotropic influenza A virus in the respiratory tract and in the regulation of adaptive antiviral immunity was examined after intranasal infection of mice with null mutations in receptors for IFN-α/β, IFN-γ, and both IFNs. Virus titers in the lungs of mice unable to respond to IFNs were not significantly different from congenic controls for both primary and secondary infection. Likewise the mice were comparably susceptible to X31 (H3N2) influenza virus infection. No significant disruption to the development of normal antiviral CTL or antibody responses was observed. In contrast, mice bearing the disrupted IFN-α/β receptor exhibited accelerated kinetics and significantly higher levels of neutralizing antibody activity during primary or secondary heterosubtypic influenza virus infection. Thus, these observations reveal no significant contribution for IFN-controlled pathways in shaping acute or memory T-cell responses to pneumotropic influenza virus infection but do indicate some role for IFN-α/β in the regulation of antibody responses. Recognizing the pivotal role of CTLs and antibody in virus clearance, it is reasonable to assume a redundancy in IFN-mediated antiviral effects in pulmonary influenza. However, IFN-α/β seems to be a valid factor in determining tissue tropism and replicative rates of highly virulent influenza virus strains as reported previously by others, and this aspect is discussed here.
Influenza virus is a major cause of morbidity and mortality worldwide, making the understanding of disease mechanisms and immunity to this pathogen of great interest (47). While events occurring comparatively late in the course of infection, such as development of cytotoxic T lymphocytes (CTLs) and specific antibodies, are known to contribute to viral clearance and recovery (8, 34), comparatively little is known about the initial stages of the immune response to influenza virus infection prior to the engagement of specific antiviral effector mechanisms. During the initial phase of infection, influenza virus interacts with cells on the luminal side of the airways to induce the release of immunoactive mediators, which attract infiltrating cells to the site of infection and/or exert antiviral activities, providing an early defense against viral infection. Induction of pulmonary inflammation appears to be particularly important in the translocation of antigen from the lung to lymphatic tissue and has an intricate role in the recruitment, immigration, and activation of virus-specific lymphocytes. A variety of cytokines and chemotactic factors are likely involved in the initiation of the inflammatory response in addition to the later recruitment and activation of specific lymphocytes (14).
It has been long recognized that interferons (IFNs) are an essential part of the innate cytokine response to viral infection, indeed, IFN-α/β and IFN-γ were originally identified as antiviral (31) but also have many other important functions in the immune system. In other RNA virus models, such as lymphocytic choriomeningitis virus (LCMV), Venezuelan equine encephalitis virus (VEE), or vesicular stomatitis virus (VSV) infections, the IFN system is prominently associated with antiviral immunity (23, 44). It is well known that IFNs are induced by many stimuli and that several viruses, notably vaccinia virus and adenovirus, have specific mechanisms for counteracting IFN-dependent host defenses (33). Such defenses include de novo transcription of a number of host genes, including cytokine genes, and induction of cellular antiviral mechanisms such as the Mx proteins, 2′-5′ oligoadenylate synthetase and the IFN-induced double-stranded RNA activated protein kinase (16, 32, 50, 55). These systems act to promote a cellular antiviral state, resulting in the inhibition of viral gene transcription and expression and, in certain cases, apoptosis of infected cells (10). In addition to inducing an antiviral state in susceptible cells, IFNs are also noted for their immunomodulatory effects (2, 4, 48). Thus, both types of IFNs upregulate the expression of major histocompatibility complex (MHC) class I and II molecules and are major activators of natural killer cells (62). In addition, IFN-α/β has recently been reported to be of importance in the augmentation of dendritic cell responses (6) and in promoting the survival of activated lymphocytes (39, 60), whereas IFN-γ exerts stimulatory effects on macrophage function and regulates the balance of cytokine production during immune responses (43). Cellular sources of IFNs vary, with IFN-α being produced by cells of the lymphoid lineage, IFN-β being produced by epithelial and fibroblast cells (28), and IFN-γ being produced by T cells and large granular lymphocytes but also by macrophages and B cells (64).
In humans and mice infected with influenza virus, a close correlation is observed between IFN levels and virus titers in secretions and lung fluids (21, 27, 40, 63). Thus, both IFN-α/β and IFN-γ are induced early in the airways of mice infected with different strains of influenza virus (27, 40, 58). More detailed information on the role of IFNs in influenza has been obtained from studies on infected mice depleted of IFN-α/β or IFN-γ either by treatment with antibodies to selectively inhibit extracellular interferon and/or by using mice unable to respond to IFN-α/β or IFN-γ due to gene disruption. These studies show that IFN-γ is nonessential for CD8+ T-cell-mediated recovery from primary influenza virus infection but exerts a protective effect during the response to heterotypic challenge independent from the generation and local recruitment of effector CTL (5). Studies on the role of IFN-α/β in protection from acute influenza have led to various conclusions. Administration of neutralizing antibodies led to enhanced mortality of infected mice expressing the Mx protein (24), while in contrast no significant effects on virus replication were found in another study in which mice infected with influenza virus were treated with anti-IFN globulin (22). Finally, a more recent report suggests that IFN-α/β plays an important role in determining the replicative rate of the A/WSN/33 strain in extrapulmonary tissues (17). However, in the same experimental setting, no significant antiviral effects were observed in the lungs after intranasal (i.n.) infection. Thus, further studies are required to better define the role of IFN-α/β in antiviral protection and in particular in the shaping and regulation of downstream adaptive immunity to influenza virus infection.
The mouse provides an excellent model of influenza pneumonia, and murine gene targeting technologies provide a means to study individual components of the immune system, including cytokines. Due to the large number of IFN-α/β genes, strategies to render mice genetically deficient in IFN-α/β genes are currently not feasible. However, in both humans and mice the same receptor complex is used for both IFN-α and IFN-β (9). The IFN-α/β receptor is composed of two distinct chains, IFNAR1 and IFNAR2, which are encoded by separate genes (36, 45), with both IFNAR1 and IFNAR2 chains required for the induction of an antiviral response in cells treated with IFN-α/β (38, 45). Similarly, the IFN-γ receptor, which is distinct from the IFN-α/β receptor, also contains two chains: IFNγR1, which is responsible for ligand binding, and IFNγR2, which is required for signal transduction (46, 52). Genetically modified mice bearing disruptions in IFNAR1 (44, 61) and IFNγR1 knockout mice (30) have been available for some time. Such knockout mice show dramatically increased susceptibilities to a range of viruses. Clearly, there is considerable interest to further understand the roles played by cytokines and IFNs in the response to influenza virus infection. It is of particular importance to determine the impact of such cytokines on virus dissemination within the respiratory tract during the onset of infection and to understand their role in the initiation and regulation of the inflammatory response and, therefore, the outcome of viral infection. Although some studies on IFN-γ have been conducted, little information regarding the role of IFN-α/β in the development and regulation of the adaptive immune response to acute and secondary heterosubtypic infection with influenza A virus is available. This, along with gaining a better understanding of the functional differences between IFN-α/β and IFN-γ, calls for a systematic reassessment of the roles of the various IFNs during influenza virus infection. In this report the kinetics of virus replication, the antibody response, and the development of specific cellular immune responses in both primary and challenge infections of mice deficient in functional receptors for IFN-α/β, IFN-γ, or both IFN receptors have been analyzed in the context of pneumotropic influenza virus infection.
MATERIALS AND METHODS
Mice.
Mice deficient in IFN-α/β receptor (IFNα/βR−/−), IFN-γ receptor (IFNγR−/−), or both IFN-α/β and IFN-γ receptors (IFNα/β-γR−/−) on the 129/SvEv background (30, 44, 61), originally obtained from B&K Universal Limited (Hull, United Kingdom), were bred and maintained under specific-pathogen-free conditions. Age-matched 129/SvEv control mice were purchased from the Jackson Laboratories (Bar Harbor, Maine). All mice used in this study had the H-2b MHC, and animals were kept and experiments were performed in accordance with institutional animal welfare guidelines.
Viruses.
Stocks of influenza virus strains A/PR/8/34 (H1N1) and X31 (H3N2) were grown in the allantoic cavity of 10-day-embryonated hen's eggs and were free of bacterial, mycoplasma, and endotoxin contamination. X31 was originally obtained from John Skehel (National Institute of Medical Research, London, United Kingdom), while A/PR/8/34 virus was a kind gift of Peter Doherty (St. Jude Children's Research Hospital, Memphis, Tenn.). Viruses were titrated on MDCK cells by plaque assay as described previously (1). Mice were anesthetized with methoxyflurane (Metofane; Pitman-Moore, Mundelein, Ill.), and infected i.n. with 50 μl of the indicated virus doses diluted in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA).
Virus titers in lung tissue.
Tissues from infected mice were homogenized in 1 ml of cold PBS and 50 μl of log dilutions of clarified homogenates were adsorbed for 1 h at 37°C onto confluent monolayers of MDCK cells in 96-well plates. Infected monolayers were then overlaid with a solution of minimal essential medium supplemented with 0.5% BSA and 25 μg/ml of TPCK (tosylamido-phenylethyl chloromethyl ketone)-trypsin (Sigma, St. Louis, Mo.) and incubated for 72 h at 37°C and 5% CO2. Virus growth was assessed by hemagglutination with 1% chicken erythrocytes. The 50% tissue culture infective dose (TCID50) was determined by the method of moving averages (59), and virus titers are expressed as the TCID50/gram of tissue. The threshold of virus detection in the MDCK assay is ∼102 TCID50/g of lung tissue.
Influenza virus antigen.
X31 virus was harvested from MDCK cell supernatant fluid at 48 h postinfection, clarified by centrifugation (1,000 × g, 30 min) and concentrated by use of polyethylene glycol (PEG 8000, 5% [wt/vol]) precipitation. Virus was sedimented at 3,750 × g for 3 h and resuspended in a small volume of PBS. This was layered onto a discontinuous sucrose gradient (60 to 30% [wt/vol] sucrose in PBS) and spun at 100,000 × g for 90 min. Viral bands were collected by side puncture, diluted in PBS, and sedimented at 100,000 × g for 2 h. Virus was further purified by centrifugation on a 40 to 15% continuous sucrose gradient for 90 min at 100,000 × g; virus bands were then again collected by side puncture and pelleted for 2 h at 100,000 × g. Finally, virus was resuspended in PBS and disrupted by ultrasonication. Protein concentration was determined with a Coomassie assay kit (Pierce, Rockford, Ill.).
HI assay.
Specific antibody titers in sera from infected mice were determined by hemagglutination inhibition (HI) assay as follows. Sera were diluted 1:10 in receptor-destroying enzyme (cholera filtrate; Sigma) and incubated at 37°C overnight to destroy nonspecific serum inhibitor activity. Receptor-destroying enzyme activity was eliminated by incubation at 56°C for 2 h. Doubling dilutions of treated sera were made in PBS in a U-bottom 96-well plate, and an equal volume (50 μl) of the appropriate virus suspension (8 hemagglutinating units) was added. Virus and antibody were incubated for 60 min at room temperature, and then 100 μl of 1% chicken erythrocytes was added. HI titers were assessed after 45 min and expressed as the reciprocal of the final dilution of serum inhibiting hemagglutination.
Detection of X31 specific antibody levels in sera of infected mice.
Virus-specific antibodies in serum were assayed by enzyme-linked immunosorbent assay (ELISA) as described previously (42). Briefly, 96-well plates (Microtest III; Falcon, Oxnard, Calif.) were coated with 0.5 μg of purified X31 antigen overnight at 4°C and blocked with 1% BSA in PBS for 2 h at room temperature. Serial dilutions of serum samples in PBS were added to the wells and allowed to incubate for 2 h at 37°C. Specific antibody isotypes were detected with horseradish peroxidase-conjugated polyclonal antibody specific to mouse immunoglobulin isotypes (immunoglobulin G [IgG], IgM, or IgA [Sigma]; IgG1, IgG2a, IgG2b, or IgG3 [Zymed, San Francisco, Calif.]). The reaction was developed with _o_-phenylenediamine dihydrocholoride substrate (Sigma), and the absorbance was read at 492 nm.
Intracellular staining for IFN-γ or TNF-α following peptide stimulation.
Cell populations recovered by bronchioalveolar lavage (BAL) or from spleen were cultured in 96-well U-bottom plates at 4 × 106 cells/well in 200 μl of RPMI 1640 (Gibco) supplemented with 10% fetal calf serum, plus 10 U of murine interleukin-2 (IL-2) and 1 μg of brefeldin A (Pharmingen, San Diego, Calif.) per well in the presence or absence of CTL epitope peptide at a concentration of 1 μg/ml. Viral peptides were the NP366–374 (ASNENMETM) which binds H-2Db or NS2114–121 (RTFSFQLI) which binds H-2Kb. After 6 h of culture, cells were harvested, washed once in fluorescence-activated cell sorter buffer (PBS with 1% BSA and 0.2% sodium azide), and surface stained with phycoerythrin-conjugated monoclonal rat antibody specific to mouse CD8α (clone 53-6-72). After being washed, cells were stained for intracellular cytokines using the Cytofix/Cytoperm kit (Pharmingen) according to the manufacturer's instructions. Fluorescein isothiocyanate-conjugated monoclonal rat antibodies specific to murine IFN-γ or tumor necrosis factor alpha (TNF-α [Caltag, Burlingame, Calif.]; clones XMG1.2 and MP6-XT22, respectively) and its isotype control antibody (rat IgG1 and IgG2a, respectively) were used to identify cytokine-positive cells. Stained cells were washed a further time and fixed in PBS containing 0.1% paraformaldehyde. Samples were acquired on a FACSCalibur flow cytometer (Becton-Dickinson, San Jose, Calif.), and data were analyzed using CellQuest software.
Histology.
Histology was performed on lung tissues fixed in 10% buffered formalin, paraffin embedded, and sectioned. Each lung specimen was stained with hematoxylin and eosin and then subjected to gross and microscopic pathologic analysis.
RESULTS
Susceptibility of mice lacking receptors for IFN-α/β, IFN-γ, or both IFN-α/β and IFN-γ to influenza virus infection.
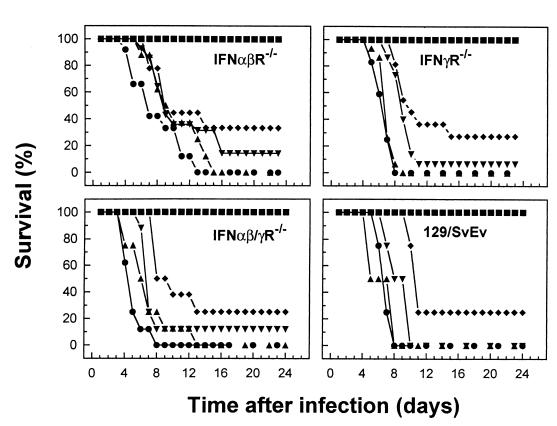
To assess the impact of IFN-γ or IFN-α/β on susceptibility to infection with influenza virus, groups of age-matched IFNα/βR−/−, IFNγR−/−, or IFNα/β-γR−/− IFNγR−/− mice or 129/SvEv mice as a control were infected with various doses of X31 virus (107, 106, 105, 104, or 102 PFU), and the rate of survival of the animals was observed over a period of 25 days (Fig. 1). At the highest dose (107 PFU), the mean survival time for control mice was 6 days, with similar survival kinetics in IFNγR−/−, IFNα/βR−/−, or IFNα/β-γR−/− mice. A progressive delay in the time of death and increased survival rate was observed when the viral inoculum was decreased, with complete protection observed at a dose of ≤103 PFU. Comparable patterns of survival were observed when mice were infected with the less-virulent A/Memphis/102/72 (H3N2) virus (data not shown). Overall, these data indicate that mice unable to respond to IFN-α/β, IFN-γ, or both IFNs did not significantly differ from congenic controls with regard to the outcome of influenza virus infection.
FIG. 1.
Susceptibility to influenza virus infection of mice lacking receptors for IFN-α/β, IFN-γ, or both IFNs. 129/SvEv, IFNα/βR−/−, IFNγR−/−, and IFNα/β-γR−/− mice were infected with X31, and the survival of infected mice was observed over a period of 25 days. The percent survival is shown for groups of 10 to 15 mice. Virus was administered i.n. at doses of 107 PFU (●), 106 PFU (▴), 105 PFU (▾), 104 PFU (⧫) or 102 PFU (■).
IFN-α/β or IFN-γ is not essential for clearance of infectious virus from the lungs during acute or secondary pulmonary influenza.
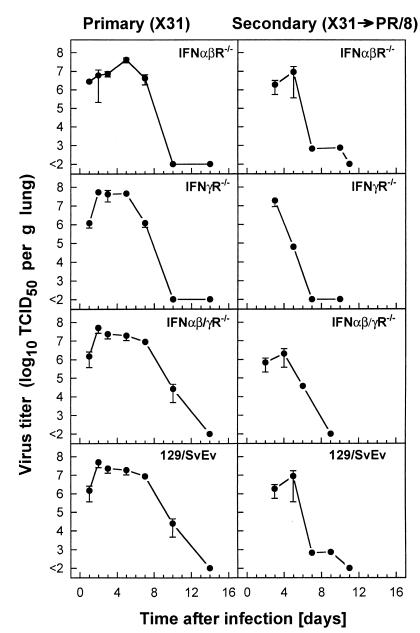
The lack of responsiveness to IFN-γ, IFN-α/β, or both IFN-γ and IFN-α/β did not result in a reduced ability of mice to recover from primary infection with X31 influenza virus. The i.n. administration of a sublethal dose (500 PFU) of X31 to IFN receptor-deficient mice or their congenic controls resulted in a pulmonary infection with viral replication peaking between days 2 and 5 (Fig. 2, left panels), followed by a rapid decline in virus lung titers by day 10. There was no significant difference in the peak lung virus titers between the controls and the mice with disrupted IFN receptor genes, and the virus was cleared by day 14 after infection in all groups of mice. Further studies examined dissemination to tissues outside the respiratory tract in mice lacking IFN responsiveness. No virus was detectable in extrapulmonary tissues (heart, liver, kidney, spleen, and brain) taken at the preterminal stages of a lethal infection with 107 PFU of X31 (data not shown).
FIG. 2.
Kinetics of lung virus replication after primary (X31) or challenge infection with A/PR/8/34 influenza A virus in mice lacking receptors for IFN-α/β, IFN-γ, or both IFNs compared to their 129/SvEv congenic controls. Lung virus titers were measured following infection of naive (primary) IFNα/βR−/−, IFNγR−/−, IFNα/β-γR−/−, or 129/SvEv control mice with 500 PFU of X31 (left panels) or following challenge of mice which had been primed with 500 PFU of X31 30 days previously with the heterologous A/PR/8/34 influenza (500 PFU i.n.) (right panels). Lung virus titers are expressed as the mean ± the standard error of the mean (SEM) log10 TCID50/gram of lung tissue of three to five mice.
The next experiment (Fig. 2, right panels) explored the role of interferons in control of heterosubtypic challenge of primed mice. The kinetics of virus replication and elimination in the lung following A/PR/8/34 (500 PFU/mouse) challenge of mice primed 30 days previously by i.n. inoculation of 500 PFU of X31 was compared between mice with disrupted IFN receptor genes and their congenic controls. X31 is a reassortant virus which expresses the surface hemagglutinin (HA) and neuraminidase (NA) proteins of A/Aichi/2/68 (H3N2) and the internal components of A/PR/8/34 (H1N1) (35). Thus, the neutralizing antibody response to HA and NA of these two viruses is not cross-reactive. Both IFN receptor-deficient mice and controls cleared the heterologous A/PR/8/34 virus from their lungs with comparable kinetics (Fig. 2, right panels), with virus titers falling from their peak (ca. 107 TCID50/g) at days 3 through 7 and diminishing to below the limit of detection by 10 days postinfection. This is compatible with the hypothesis of accelerated virus clearance due to an anamnestic CTL response against epitopes conserved between X31 and A/PR/8/34. No overall difference in the pulmonary virus elimination kinetics was seen between the different cohorts of mice. Note that the entire population of primed mice were protected against challenge with A/PR/8/34 (500 PFU), while naive C57BL/6 mice infected with the same virus inoculum succumbed to influenza pneumonia, between days 9 and 14 after infection (unpublished results).
Role of IFN-α/β or IFN-γ in generation of CTLs in primary and secondary influenza pneumonia.
Besides a direct effect on virus replication, IFNs are generally believed to play a pivotal role in the maturation of virus-specific immune responses in viral infection (3). Previous studies have demonstrated no effect or redundancy for IFN-γ in the development of an efficient CTL response during influenza virus infection (5, 20, 51). However, the role of IFN-α/β in the proliferation and recruitment of virus-specific CTLs to the site of pathology in the lung is unknown.
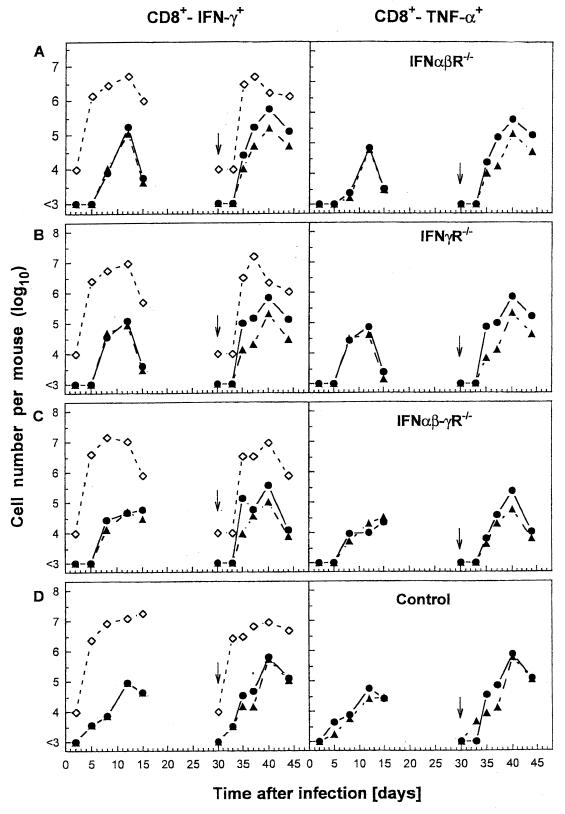
This was evaluated by studying the kinetics and magnitude of leukocyte and CD8+ CTL responses recovered by BAL from mice during primary and heterosubtypic challenge (Fig. 3). The inflammatory responses did not differ greatly between controls and receptor-deficient mice, and comparable kinetic profiles for virus-specific CD8+ T-cell localization to the BAL, as analyzed by staining CD8+ T cells for intracellular IFN-γ (Fig. 3, left panels) or TNF-α (Fig. 3, right panels) following NP366–374 or NS2114–121 peptide stimulation, were obtained. A similar pattern of virus-specific CD8+ T-cell responses were obtained in the spleen (data not shown). It is noteworthy that the numbers of IFN-γ or TNF-α secreting cells detectable by intracellular staining were in close agreement, even in mice bearing disrupted IFN-γ receptors. Thus, the absence of IFN-γ receptor does not appear to affect IFN-γ production by individual CD8+ CTLs in response to an antigenic stimulus. In a further set of experiments, lung tissues from virus-infected mice were analyzed histologically because it is likely that cells obtained by BAL do not fully reflect the overall pulmonary inflammatory process. Gross and microscopic pathologic analysis of hematoxylin-and-eosin-stained paraffin sections of lung tissue from IFN receptor-deficient or control mice obtained 3, 5, or 9 days after infection with 500 PFU of X31 did not reveal major differences in the spectrum or magnitude of the inflammatory process between the experimental groups of mice (data not shown). However, slightly increased inflammation was observed on day 9 after infection in IFNα/β-γR−/− mice in comparison to control animals. The inflammatory pathology in the respiratory tract, consisting of a few foci of perivascular and peribronchial inflammation of mononuclear cells (macrophages/monocytes) and numerous lymphoblasts at later stages of the infection, resolved rapidly subsequent to viral clearance.
FIG. 3.
Virus-specific CD8+ T cells in primary and challenge influenza infection of mice lacking receptors for IFN-α/β, IFN-γ, or both IFNs compared to controls. Naive IFNα/βR−/− (A), IFNγR−/− (B), IFNα/β-γR−/− (C), or control (D) mice were infected with 500 PFU of X31, and the numbers of virus-specific CD8+ T cells in the BAL fluid were measured. BAL samples from each group of three to five mice were pooled, and the numbers of virus-specific CTLs were determined by staining CD8+ T cells for intracellular IFN-γ (left panels) or TNF-α (right panels) secretion, following stimulation of cells with NP366–374 (●) or NS2114–121 (▴) viral peptide. Alternatively, X31 primed mice (500 PFU i.n.) were challenged with A/PR/8/34 (500 PFU i.n.) 30 days later (as indicated by the arrow), and the numbers of virus-specific CD8+ T cells were determined as described above. The BAL cell counts per mouse (◊) were used, together with the flow cytometry data, to calculate the average numbers for the total CD8+ T cells specific to NP366–374 or NS2114–121 peptide epitope. BAL samples (total volume, 1 ml/lung) containing <104 cells/ml (the limit of detection of our hemocytometer counting assay) were estimated as 104 cells per lung.
Virus-specific antibody response.
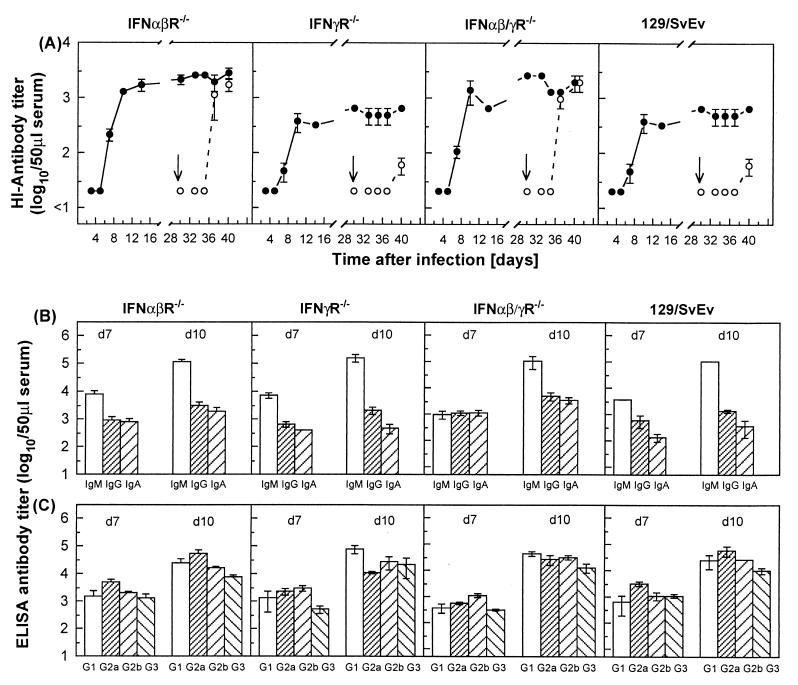
The role of IFNs in the generation and maintenance of primary or memory virus-specific B-cell-mediated responses was studied by the determination of HI antibody titers. IFN receptor-deficient mice or their controls produced significant levels of X31-specific HI antibodies (Fig. 4A). The rise in serum antibody activity detectable from day 7 after infection paralleled the kinetics of CD8+ T-cell localization at the site of pathology in the lung and was associated with the resolution of the viral infection. Surprisingly, mice bearing the disrupted IFN-α/β receptor (IFNα/βR−/− or IFNα/β-γR−/−) exhibited an accelerated development of antibody activity, and significantly higher maximal titers of HI antibodies were detectable compared to mice deficient in IFN-γ receptor or 129/SvEv controls. Likewise, mice primed i.n. with X31 (H3N2) and challenged 30 days later with the heterologous A/PR/8/34 (H1N1) virus (indicated as an arrow in the figure) developed primary responses in terms of HI antibodies specific to A/PR/8/34, with mice bearing the disrupted IFN-α/β receptor (IFNα/βR−/− or IFNα/β-γR−/−) exhibiting more rapid appearance of A/PR/8/34 specific antibodies and higher levels of HI antibodies on days 7 and 10 after virus challenge. It is of note that X31 primed mice challenged with A/PR/8/34 also mounted the expected anamnestic antibody response against X31, with the magnitude of the recall response heightened in IFNα/βR−/− and IFNα/β-γR−/− mice compared to IFNγR−/− mice and 129/SvEv controls.
FIG. 4.
Generation and maintenance of primary or memory virus-specific antibody responses of mice lacking receptors for IFN-α/β, IFN-γ, or both IFNs compared to controls. (A) The ability of IFNα/βR−/−, IFNγR−/−, IFNα/β-γR−/−, or 129/SvEv control mice to produce protective neutralizing antibodies was tested by measuring HI antibody titers in the sera of mice infected i.n. with 500 PFU of X31 (H3N2) (primary infection), or following their challenge on day 30 after primary infection (as indicated by the arrow), with 500 PFU of the heterologous A/PR/8/34 (H1N1) virus. The titers of X31 (●)- and A/PR/8/34 (○)-specific HI antibodies were estimated individually, and the results are expressed as the mean ± the SEM log10 HI antibody titers of groups of three to five mice. The isotype pattern of antibodies in the sera of IFNα/βR−/−, IFNγR−/−, IFNα/β-γR−/−, or 129/SvEv control mice following i.n. infection with 500 PFU of X31 was measured on days 7 and 10 after virus inoculation. (B and C) The results are shown as an ELISA titer of virus-specific antibody (mean ± the SEM log10 of three to five mice) of the IgM, IgG, or IgA isotype (B) or the IgG1, IgG2a, IgG2b, or IgG3 isotype (C).
In the course of the early immune response to influenza virus, interaction of CD4+ T cells with B cells regulates the activation of virus-specific IgG- and IgA-producing B cells mandatory for effective antiviral responses (13). In addition to its antiviral activity, IFN-γ is known to modulate the production of several cytokines and, in particular, to play a central role in the regulation of Th1-Th2 CD4+ T-cell subsets. Thus, the isotype pattern of virus-specific antibodies produced in the serum of IFN receptor deficient or congenic control mice infected with 500 PFU X31 was studied by ELISA (Fig. 4B and C). Generally, levels of each immunoglobulin isotype were similar between IFN receptor-deficient and control mice at the time points examined. No statistically significant differences were observable between mice lacking IFN-α/β or IFN-γ responsiveness and the control mice in terms of serum IgG, IgM, or IgA or in the IgG isotype pattern of virus-specific serum antibody at days 7 and 10, despite the documented role of IFN-γ in IgG isotype switching. It should be noted that the HI and ELISA titers are not directly comparable since HI distinguishes HA-specific antibody only, while ELISA detects all virus-specific antibodies. Hence, it is possible that the differences in HI titers between the various mice reflect qualitative rather than quantitative changes in the antibody response.
DISCUSSION
The natural history of influenza virus infection in humans follows a defined pattern with well-characterized features. However, the rates of development as well as the overall severity of disease vary widely in different individuals. Since factors determining the pathogenesis of influenza in humans are complex, involving epidemiological considerations as well as inherent viral properties (cytopathic phenotype, antigenic diversity, etc.), it is unlikely that a single virus gene dictates the virulence of a given virus strain; rather, a combination of viral genes and host susceptibility determines the outcome of infection (57). The fact that influenza is both an IFN-sensitive virus and an IFN-inducing virus led to the hypothesis that a very early line of antiviral defense by the IFN system prevents the virus from spreading efficiently, allowing the adaptive immune response enough time to develop and eliminate the virus.
In this report mice genetically deficient in receptor for IFN-α/β, IFN-γ, or both IFNs were utilized to study the role of IFNs during influenza virus pneumonia. While numerous in vitro studies demonstrate that influenza virus replication is sensitive to the effects of IFN-α/β, no major effect on overall lethality, virus replication, the kinetics of the cellular immune response, or the ability to maintain an effective recall CTL response to heterosubtypic challenge was detectable in IFN receptor-deficient mice. These findings are in general agreement with and extend other studies of mice deficient in IFN-γ or STAT-1 infected with pneumotropic influenza virus strains (15, 20). However, one striking observation in this study is that the disruption of IFN-α/β receptor responsiveness imparted an accelerated specific antibody response of increased magnitude compared to controls. This increase in antibody response occurred despite the fact that the virus replicated to equal peak titers in the lung over the same time course in control and IFNα/βR−/− mice. The underlying mechanism for this effect is unknown. However, it is possible that the lack of IFN-α/β responsiveness results in an enhanced infection of MHC-II-positive inflammatory cells (monocytes/macrophages and dendritic cells) not normally permissive for influenza virus. This may promote the induction of helper T cells, resulting in a more efficient antibody response. Indeed, recent reports suggest that autocrine production of IFN-α/β mediates the protection of human dendritic cells from influenza virus infection (6). Alternatively, the lack of IFN-α/β responsiveness may cause a shift toward a more Th2-like cytokine profile. This is supported by the observation that IFN-α/β has been reported to inhibit Th2-like responses by blocking IL-4 secretion by human CD4+ T lymphocytes (41). However, increased antibody responses as a result of skewing toward a Th2 phenotype is perhaps less likely due to the lack of effect following the disruption of IFN-γ responsiveness, a factor known to be critical for development of Th1 responses.
A recent study utilizing the virulent Wilson-Smith neurotropic (WSN [A/WSN/33]) (H1N1) influenza virus strain has demonstrated enhanced virus replication outside the respiratory tract in IFNα/βR−/− and STAT-1−/− mice, which are deficient in the IFN-α/β signaling pathway, implying a role for IFN-α/β in restricting viral replication to the respiratory mucosa (17). However, WSN has unusual virulence properties, including an unique mechanism of HA cleavage involving sequestration of host plasmin by the WSN NA (19). Due to the lack of viral recovery from extrapulmonary organs in our study, we feel that these earlier findings primarily reflect the ability of WSN to undergo HA cleavage and replicate in a range of tissues following disruption of the IFN-α/β response, as opposed to a broadly applicable role for IFN in limiting influenza virus replication to the respiratory tract, although this may be the case for viruses bearing highly cleavable HA molecules. Indeed, previous studies using polyclonal antibodies to neutralize the activity of IFN-α/β failed to show any effect on the course of pneumotropic A/PR/8/34 infection in mice, even when the sera were administered i.n. (22), which supports the observations reported here. In contrast, antibody neutralization of IFN-α/β in A2G mice infected with a virulent mouse-passaged influenza virus strain led to a 100-fold increase in lethality (24). It is noteworthy, however, that A2G mice have a functional Mx locus which correlates with their increased resistance to influenza virus infection. In mice the Mx1 gene product is a long-recognized antiviral protein induced by IFN-α/β, and the presence of Mx1 efficiently blocks early stages of influenza virus replication (29). Most inbred laboratory mouse strains, including those of the 129 genetic background as used in this study, have a functionally inactive Mx gene (53, 54). Thus, in the case of influenza virus infection, the antiviral effects of IFN-α/β in the lungs may be mediated predominantly via the Mx system, and thus disruption of IFN-α/β signaling in Mx-deficient mice has little direct effect on antiviral defense. By extension, it is possible that in extrapulmonary tissues, the antiviral effects of IFN-α/β may be mediated by factors other than Mx, which could explain the inability of IFN-deficient mice to control systemic infection with unusually virulent influenza virus strains. Our findings, and those of others, therefore support the hypothesis that the IFN system is involved in defense against systemic rather than localized viral infection. It is worthy of note that all previously reported studies utilizing IFN receptor-deficient mice have concentrated on such systemic infections (such as vaccinia virus, LCMV, VSV, and VEE), and many of the earlier studies of IFN in the context of influenza utilized virus strains of unusually high virulence.
It is presently unclear how the data in this report relates to observations that influenza virus has specific mechanisms, mediated via the NS1 gene product, to counteract IFN-induced responses (18). Indeed, A/PR/8/34 mutants lacking NS1 show reduced growth in MDCK cells but are less restricted in Vero cells, which are defective in their response to IFN-α/β. These NS1 deletants grow in STAT-1−/− mice at reduced titers compared to the parental strain but cannot grow in STAT-1+/+ mice (18). The apparent requirement for a viral gene product capable of inhibiting IFN-mediated pathways is thus puzzling in the light of the in vivo findings reported here. An alternative, and attractive, hypothesis is that the lack of IFN-α/β signaling is compensated for by overlapping pathways, such as IFN-γ in the absence of IFN-α/β or vice versa, or by other factors unrelated to the IFNs, as would be the case in IFNα/β-γR−/− mice. Such alternate pathways could conceivably also require STAT-1-mediated signaling, explaining the influenza virus-susceptible phenotype of STAT-1-deficient mice. STAT-1 is a rather pleiotropic transcription factor utilized in several signaling pathways in addition to the IFN pathway (15, 49). Thus, it is possible that NS1 is involved in disrupting other host cell responses in addition to the IFN pathway. Indeed, it has recently been shown that NS1 is capable of inhibiting double-stranded-RNA-mediated activation of PKR which would otherwise result in a translational block of viral protein synthesis (25).
Finally, it must be noted that it is possible, if not likely, that IFNs have a significant role in the expression of symptoms during influenza virus infection. In the case of human influenza, numerous systemic symptoms, particularly fever, occur in the early stages of infection, prior to the development of a specific immune response. It has long been hypothesized that IFNs are involved in these disease manifestations (11, 12, 26). While it would be possible to analyze the severity of some of the symptoms of influenza in IFN receptor-deficient mice, such as suppression of appetite and weight loss, the mouse does not represent the ideal model for the study of symptom expression in influenza as, in contrast to humans, there is a regulated and dramatic decline in body temperature following infection (7). Hence, it is possible that the mouse influenza pneumonia model is inherently unsuitable for extrapolating the effects of IFN in humans due to physiological differences in IFN responsiveness between species and, indeed, between mouse strains (37). If this were true, it would also imply that virus strategies, such as the NS1 gene product, act to counteract the effects of IFN in its natural hosts but are comparatively redundant in mice. It would be of great interest to examine the role of IFN in the development of symptoms in alternative animal models of influenza, such as the ferret (56).
In conclusion, the data reported here indicate no major contribution for IFN-α/β or IFN-γ pathways in protection or recovery from influenza virus infection in mice. However, increased antibody responses noted following the disruption of IFN-α/β receptor suggest a previously unappreciated role for these factors in the regulation of humoral immunity during viral respiratory infection. Further studies to dissect this mechanism may be relevant for vaccination strategies directed at producing heightened mucosal antibody responses.
REFERENCES
- 1.Barrett T, Inglis S C. Growth, purification and titration of influenza viruses. In: Mahy B W J, editor. Virology: a practical approach. Oxford, England: IRL Press; 1985. pp. 119–150. [Google Scholar]
- 2.Biron C A. Cytokines in the generation of immune responses to, and resolution of, virus infection. Curr Opin Immunol. 1994;6:530–538. doi: 10.1016/0952-7915(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 3.Biron C A. Initial and innate responses to viral infections—pattern setting in immunity or disease. Curr Opin Microbiol. 1999;2:374–381. doi: 10.1016/s1369-5274(99)80066-6. [DOI] [PubMed] [Google Scholar]
- 4.Biron C A. Role of early cytokines, including alpha and beta interferons (IFN-α/β), in innate and adaptive immune responses to viral infections. Semin Immunol. 1998;10:383–390. doi: 10.1006/smim.1998.0138. [DOI] [PubMed] [Google Scholar]
- 5.Bot A, Bot S, Bona C A. Protective role of gamma interferon during the recall response to influenza virus. J Virol. 1998;72:6637–45. doi: 10.1128/jvi.72.8.6637-6645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cella M, Salio M, Sakakibara Y, Langen H, Julkunen I, Lanzavecchia A. Maturation, activation, and protection of dendritic cells induced by double-stranded RNA. J Exp Med. 1999;189:821–829. doi: 10.1084/jem.189.5.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conn C A, McClellan J L, Maassab H F, Smitka C W, Majde J A, Kluger M J. Cytokines and the acute phase response to influenza virus in mice. Am J Physiol Reg Integ Comp Physiol. 1995;37:R78–R84. doi: 10.1152/ajpregu.1995.268.1.R78. [DOI] [PubMed] [Google Scholar]
- 8.Crowe J E., Jr The role of antibodies in respiratory viral immunity. Semin Virol. 1996;7:273–283. [Google Scholar]
- 9.Díaz M O. The human type I interferon gene cluster. Semin Virol. 1995;6:143–149. [Google Scholar]
- 10.Díaz-Guerra M, Rivas C, Esteban M. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology. 1997;236:354–363. doi: 10.1006/viro.1997.8719. [DOI] [PubMed] [Google Scholar]
- 11.Dinarello C A. Cytokines as endogenous pyrogens. J Infect Dis. 1999;179:S294–S304. doi: 10.1086/513856. [DOI] [PubMed] [Google Scholar]
- 12.Dinarello C A, Bernheim H A, Duff G W, Le H V, Nagabhushan T L, Hamilton N C, Coceani F. Mechanisms of fever induced by recombinant human interferon. J Clin Investig. 1984;74:906–913. doi: 10.1172/JCI111508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doherty P C, Allan W, Eichelberger M, Carding S R. Roles of αβ and γδ T cell subsets in viral immunity. Annu Rev Immunol. 1992;10:123–151. doi: 10.1146/annurev.iy.10.040192.001011. [DOI] [PubMed] [Google Scholar]
- 14.Doherty P C, Topham D J, Tripp R A, Cardin R D, Brooks J W, Stevenson P G. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol Rev. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 15.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 16.Gale M, Jr, Katze M G. What happens inside lentivirus or influenza virus infected cells: insights into regulation of cellular and viral protein synthesis. Methods. 1997;11:383–401. doi: 10.1006/meth.1996.0436. [DOI] [PubMed] [Google Scholar]
- 17.García-Sastre A, Durbin R K, Zheng H, Palese P, Gertner R, Levy D E, Durbin J E. The role of interferon in influenza virus tissue tropism. J Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 19.Goto H, Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Natl Acad Sci USA. 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham M B, Dalton D K, Giltinan D, Braciale V L, Stewart T A, Braciale T J. Response to influenza infection in mice with a targeted disruption in the interferon γ gene. J Exp Med. 1993;178:1725–1732. doi: 10.1084/jem.178.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green J A, Charette R P, Yeh T-J, Smith C B. Presence of interferon in acute- and convalescent-phase sera of humans with influenza or an influenza-like illness of undetermined etiology. J Infect Dis. 1982;145:837–841. doi: 10.1093/infdis/145.6.837. [DOI] [PubMed] [Google Scholar]
- 22.Gresser I, Tovey M G, Maury C, Bandu M-T. Role of interferon in the pathogenesis of virus diseases in mice as demonstrated by the use of anti-interferon serum. II. Studies with herpes simplex, Moloney sarcoma, vesicular stomatitis, Newcastle disease, and influenza viruses. J Exp Med. 1976;144:1316–1323. doi: 10.1084/jem.144.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grieder F B, Vogel S N. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology. 1999;257:106–118. doi: 10.1006/viro.1999.9662. [DOI] [PubMed] [Google Scholar]
- 24.Haller O, Arnheiter H, Gresser I, Lindenmann J. Genetically determined, interferon-dependent resistance to influenza virus in mice. J Exp Med. 1979;149:601–612. doi: 10.1084/jem.149.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatada E, Saito S, Fukuda R. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayden F G, Fritz R, Lobo M C, Alvord W, Strober W, Straus S E. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Investig. 1998;101:643–9. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennet T, Ziltener H J, Frei K, Peterhans E. A kinetic study of immune mediators in the lungs of mice infected with influenza A virus. J Immunol. 1992;149:932–939. [PubMed] [Google Scholar]
- 28.Hiscott J, Nguyen H, Lin R. Molecular mechanisms of interferon beta gene induction. Semin Virol. 1995;6:161–173. [Google Scholar]
- 29.Horisberger M A. Interferons, Mx genes, and resistance to influenza virus. Am J Respir Crit Care Med. 1995;152:S67–S71. doi: 10.1164/ajrccm/152.4_Pt_2.S67. [DOI] [PubMed] [Google Scholar]
- 30.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Immune response in mice that lack the interferon-γ receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 31.Isaacs A. Interferon. Adv Virus Res. 1963;10:1–38. [PubMed] [Google Scholar]
- 32.Jacobs B L, Langland J O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 33.Kalvakolanu D V. Virus interception of cytokine-regulated pathways. Trends Microbiol. 1999;7:166–171. doi: 10.1016/s0966-842x(99)01476-6. [DOI] [PubMed] [Google Scholar]
- 34.Karzon D T. Cytotoxic T cells in influenza immunity. Semin Virol. 1996;7:265–271. [Google Scholar]
- 35.Kilbourne E D. Future influenza vaccines and the use of genetic recombinants. Bull W H O. 1969;41:643–645. [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S H, Cohen B, Novick D, Rubinstein M. Mammalian type I interferon receptors consists of two subunits: IFNaR1 and IFNaR2. Gene. 1997;196:279–286. doi: 10.1016/s0378-1119(97)00240-0. [DOI] [PubMed] [Google Scholar]
- 37.Kurokawa M, Imakita M, Kumeda C A, Shiraki K. Cascade of fever production in mice infected with influenza virus. J Med Virol. 1996;50:152–158. doi: 10.1002/(SICI)1096-9071(199610)50:2<152::AID-JMV8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 38.Lewerenz M, Mogensen K E, Uzé G. Shared receptor components but distinct complexes for α and β interferons. J Mol Biol. 1998;282:585–599. doi: 10.1006/jmbi.1998.2026. [DOI] [PubMed] [Google Scholar]
- 39.Marrack P, Kappler J, Mitchell T. Type I interferons keep activated T cells alive. J Exp Med. 1999;189:521–529. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLaren C, Potter C W. The relationship between interferon and virus virulence in influenza virus infections of the mouse. J Med Microbiol. 1973;6:21–32. doi: 10.1099/00222615-6-1-21. [DOI] [PubMed] [Google Scholar]
- 41.McRae B L, Picker L J, van Seventer G A. Human recombinant interferon-β influences T helper subset differentiation by regulating cytokine secretion pattern and expression of homing receptors. Eur J Immunol. 1997;27:2650–2656. doi: 10.1002/eji.1830271026. [DOI] [PubMed] [Google Scholar]
- 42.Moskophidis D, Lehmann-Grube F. The immune response of the mouse to lymphocytic choriomeningitis virus IV. Enumeration of antibody-producing cells in spleens during acute and persistent infection. J Immunol. 1984;133:3366–3370. [PubMed] [Google Scholar]
- 43.Mosmann T R, Coffman R L. Th1-cell and Th2-cell—different patterns of lymphokine secretion lead to different functional-properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 44.Müller U, Steinhoff U, Reis L F L, Hemmi S, Pavlovic J, Zinkernagel R M, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 45.Owczarek C M, Hwang S Y, Holland K A, Gulluyan L M, Tavaria M, Weaver B, Reich N C, Kola I, Hertzog P J. Cloning and characterization of soluble and transmembrane isoforms of a novel component of the murine type I interferon receptor, IFNAR 2. J Biol Chem. 1997;272:23865–23870. doi: 10.1074/jbc.272.38.23865. [DOI] [PubMed] [Google Scholar]
- 46.Pestka S. The interferon receptors. Semin Oncol. 1997;24:S9-18–S9-40. [PubMed] [Google Scholar]
- 47.Potter C W. Unique features of influenza viruses, and their implications. Semin Respir Infect. 1992;7:2–10. [PubMed] [Google Scholar]
- 48.Ramshaw I A, Ramsay A J, Karupiah G, Rolph M S, Mahalingam S, Ruby J C. Cytokines and immunity to viral infections. Immunol Rev. 1997;159:119–135. doi: 10.1111/j.1600-065x.1997.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 49.Ransohoff R M. Cellular responses to interferons and other cytokines: the JAK-STAT paradigm. N Eng J Med. 1998;338:616–618. doi: 10.1056/NEJM199802263380911. [DOI] [PubMed] [Google Scholar]
- 50.Samuel C E. Antiviral actions of interferon: interferon-regulated cellular proteins and their surprisingly selective antiviral actions. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 51.Sarawar S R, Carding S R, Allan W, McMickle A, Fujihashi K, Kiyono H, McGhee J R, Doherty P C. Cytokine profiles of bronchoalveolar lavage cells from mice with influenza pneumonia: consequences of CD4+ and CD8+ T cell depletion. Reg Immunol. 1993;5:142–150. [PubMed] [Google Scholar]
- 52.Schreiber R D, Aguet M. The interferon-γ receptor. In: Nicola N A, editor. Guidebook to cytokines and their receptors. 1st ed. Oxford, England: Sambrook and Tooze; 1994. pp. 120–123. [Google Scholar]
- 53.Staeheli P, Grob R, Meier E, Sutcliffe J G, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 55.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 56.Sweet C, Fenton R J, Price G E. The ferret as an animal model of influenza virus infection. In: Zak O, Sande M, editors. Handbook of animal models of infection: Experimental models in antimicrobial chemotherapy. London, England: Academic Press; 1999. pp. 989–998. [Google Scholar]
- 57.Sweet C, Smith H. Pathogenicity of influenza virus. Microbiol Rev. 1980;44:303–330. doi: 10.1128/mr.44.2.303-330.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor P M, Meager A, Askonas B A. Influenza virus-specific T cells lead to early interferon gamma in lungs of infected hosts: development of a sensitive radioimmunoassay. J Gen Virol. 1989;70:975–978. doi: 10.1099/0022-1317-70-4-975. [DOI] [PubMed] [Google Scholar]
- 59.Thompson W R. The use of moving averages and interpolation to estimate median effective dose. Bacteriol Rev. 1947;11:115–147. [PubMed] [Google Scholar]
- 60.Tough D F, Borrow P, Sprent J. Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- 61.van den Broek M F, Müller U, Huang S, Aguet M, Zinkernagel R M. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welsh R M, Tay C H, Varga S M, O'Donnell C L, Vergilis K L, Selin L K. Lymphocyte-dependent ‘natural’ immunity to virus infections mediated by both natural killer cells and memory T cells. Semin Virol. 1996;7:92–105. [Google Scholar]
- 63.Wyde P R, Wilson M R, Cate T R. Interferon production by leukocytes infiltrating the lungs of mice during primary influenza virus infection. Infect Immun. 1982;38:1249–1255. doi: 10.1128/iai.38.3.1249-1255.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young H A. Regulation of interferon-γ gene transcription. Semin Virol. 1995;6:175–179. [Google Scholar]