Evaluation of Novel Human Immunodeficiency Virus Type 1 Gag DNA Vaccines for Protein Expression in Mammalian Cells and Induction of Immune Responses (original) (raw)
Abstract
Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocytes (CTL) are an important parameter of host defenses that limit viral replication after infection. Induction of effective CTL against conserved viral proteins such as Gag may be essential to the development of a safe and effective HIV type 1 (HIV-1) vaccine. DNA vaccination represents a novel strategy for inducing potent CD8+ CTL responses in vivo. However, expression of HIV-1 structural proteins by DNA vectors has been hampered by a stringent requirement for coexpression with other viral components, such as Rev and RRE. Furthermore, even with Rev and RRE present, the level of expression of HIV-1 Gag, Pol, or Env is very low in murine cells. These problems have limited our ability to address the key issue of how to generate effective CTL responses to Gag in a mouse model. To overcome this problem, we compared several novel DNA expression vectors for HIV-1 Gag protein expression in primate and mouse cells and for generating immune responses in mice after DNA vaccination. A DNA vector containing wild type HIV-1 gag coding sequences did not induce detectable Gag expression in any of the cells tested. Attempts to increase nuclear export of Gag expression RNA by adding the constitutive transport element yielded only a moderate increase in Gag expression in monkey-derived COS cells and an even lower increase in Gag expression in HeLa cells or several mouse cell lines. In contrast, silent-site mutations in the HIV-1 gag coding sequences significantly increased Gag expression levels in all cells tested. Furthermore, this construct induced both Gag-specific antibody and CTL responses in mice after DNA vaccination. Using this construct, we achieved stable expression of HIV-1 Gag in the mouse cell line p815, which can now be used as a target cell for measuring HIV-1 Gag-specific CTL responses in immunized mice. The DNA vectors described in this study should make it possible to systematically evaluate the approaches for maximizing the induction of CTL responses against HIV-1 Gag in mouse and other animal systems.
There is increasing evidence that CD8+ cytotoxic T lymphocytes (CTL) may play an important role in controlling human immunodeficiency virus type 1 (HIV-1) infection. Containment of primary HIV-1 infection in infected individuals correlates with the emergence of virus-specific CTL responses (3, 12, 22). In chronically infected individuals, a high-frequency CTL response against HIV-1 is also correlated with low viral load and slow disease progression (19, 20). An HIV-1-specific CTL response has also been demonstrated in certain highly exposed seronegative individuals (2, 13, 28). Broad, cross-clade CTL responses recognizing conserved epitopes in HIV-1 Gag have been detected in HIV-1-infected individuals (7, 18). It is therefore reasonable to hypothesize that induction of an effective CTL response against conserved internal virion proteins of HIV-1 such as Gag is essential for the development of a safe and effective HIV-1 vaccine.
In order to generate an efficient major histocompatibility complex (MHC) class I-restricted cellular immune response to a vaccine, viral proteins must be synthesized endogenously. Efficient production of CTL responses requires endogenous antigen synthesis, usually achieved by using a live, attenuated virus or recombinant virus vectors. Concerns about using a live, attenuated virus vaccine for HIV-1 include potential pathogenic replication and disease development over a longer period of time as well as potential adverse effects of integrated viral DNA. Using recombinant virus-based vectors, it is difficult to achieve repeated boosting because of the strong immune response generated against the viral proteins of the virus vector. Certain virus vectors, such as vaccinia virus, may also inhibit class I MHC-restricted CTL responses (32).
Recently, a new approach (DNA vaccination) has been used to express antigens in vivo for the generation of both humoral and cellular immune responses (6). Several groups have used the DNA vaccination approach against HIV-1 (10, 17, 21, 34). Unfortunately, expression of HIV-1 Gag, Pol, and Env proteins by DNA vectors has been hampered by the presence of multiple inhibitory sequences (INS) in the structural genes encoding Gag, Pol, and Env proteins of HIV-1. This makes expression of the structural HIV-1 proteins dependent on the viral regulatory protein Rev, which is responsible for the nuclear export and efficient expression of unspliced HIV-1 mRNAs (5, 8, 23, 24). Rev binds specifically to an RNA site within HIV-1 mRNA named RRE. In the absence of functional Rev/RRE, mRNAs containing INS are either retained in the nucleus or degraded rapidly; therefore, little protein can be expressed from these mRNAs. Furthermore, even with Rev and RRE, expression of HIV-1 Gag, Pol, or Env is very low in certain murine cell lines (10, 33), limiting our ability to study the DNA vaccine-induced immune response against Gag or Pol using a mouse model.
It has been reported that type D retroviruses contain a _cis_-acting posttranscriptional control element (CTE) that can replace the Rev and RRE regulation of HIV-1 (4, 36). When a CTE is inserted into an HIV-1 Rev−/RRE− clone, replication of the CTE-containing clone can approach that of the wild-type HIV-1 clone in cell lines (4, 36). The CTEs of different type D retroviruses are approximately 200 nucleotides in length and form extensive RNA stem-loop structures. They contain binding sites for cellular factors that contain nuclear export signals (9). CTEs function in a wide spectrum of cell types and species (31) and have been identified in both type D simian retroviruses (4, 36) and murine intracisternal-A particles (31).
A current hypothesis about the function of INS is that they are binding sites for cellular factors that contribute to mRNA instability (1). Recently, it has been demonstrated that substitutions of AT-rich regions in HIV-1 gag, mostly in third-site positions, without changing the amino acid sequence of the produced Gag protein can result in efficient expression of Gag in a Rev- and RRE-independent fashion (29, 30). In the present study, we have constructed a series of HIV-1 Gag DNA expression vectors and compared the influence of the constitutive transport element (CTE) or of silent-site mutations within HIV-1 gag on Gag expression in a variety of cells of human, simian, and mouse origin. We found that silent-site mutations in HIV-1 gag allowed efficient Gag expression in a species-independent fashion and induced both humoral and CTL responses after naked DNA vaccination in mice. On the other hand, CTE only increased Gag expression moderately in monkey-derived COS cells and negligibly in HeLa (human) or several mouse cell lines. A DNA vaccine containing wild-type gag coding sequence plus CTE did not induce any detectable humoral or CTL response in mice. In contrast, mutated gag not containing INS induced detectable humoral and CTL responses. We have also achieved stable expression of HIV-1 Gag protein in mouse p815 cells and shown that these cells can be used as CTL target cells. Reagents described in this study should facilitate the development and evaluation of HIV-1 DNA vaccine strategies against conserved viral Gag protein in animal models.
MATERIALS AND METHODS
Construction of HIV-1 Gag expression vectors.
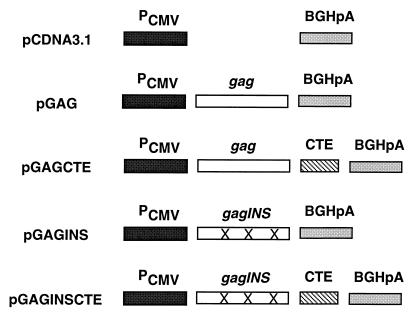
The HIV-1 gag coding region from HXB2 (25) was amplified by PCR, using primers Gag-for (5′GCTAGAAGGTCTAGAATGGGTGCGAGAGCG3′, with the _Xba_I site underlined) and Gag-rev (5′AGTTGCCCCCGAATTCTTATTGTGACGAGG3′, with the _Eco_RI site underlined). The PCR products were digested with _Xba_I and _Eco_RI and cloned into pcDNA3.1(−) (Invitrogen, San Diego, Calif.) downstream from the human cytomegalovirus (CMV) immediate-early promoter (Pcmv) sequence, using _Xba_I and _Eco_RI sites to generate pGAG (Fig. 1). To generate pGAGCTE, the CTE from simian retrovirus type 1 (SRV-1) (36) was obtained as a _Bam_HI-_Kpn_I restriction fragment from plasmid pS12 (31) and inserted into pGAG downstream from the gag coding sequences and upstream from the bovine growth hormone polyadenylation (BGHpA) signal (Fig. 1). pGAGINS was generated by PCR amplification of INS mutant gag coding sequences from p55M1-10 (29), using primers Gag-for and Gag-rev. The PCR products were digested with _Xba_I and _Eco_RI and cloned into pcDNA3.1(−) to generate pGAGINS (Fig. 1). The CTE from SRV-1 as a _Bam_HI-_Kpn_I restriction fragment from plasmid pS12 was inserted into pGAGINS downstream from the gag coding sequences but upstream from the BGHpA signal by using the _Bam_HI and _Kpn_I sites to generate pGAGINSCTE (Fig. 1). These plasmids were transformed into Dh5α bacteria, cultured in Luria broth, and purified with the QIAGEN Maxiprep kit (QIAGEN, Chatsworth, Calif.).
FIG. 1.
Schematic representation of HIV-1 Gag expression vectors. The CMV promoter and BGHpA signal flank wild-type (25) or INS mutant (29) HIV-1 gag coding sequences. The CTE from SRV-1 (36) was inserted downstream from either the wild-type HIV-1 gag coding sequences or the INS mutant HIV-1 gag coding sequences.
Antibodies and synthetic peptide.
An HIV-1-positive human serum was obtained from an HIV-1-infected subject in Baltimore, Md. Rabbit anti-p24 antibody and sheep anti-p17 antibody were obtained from the AIDS Research and Reference Reagents Program, NIH, Bethesda, Md. The anti-p6Gag antiserum has been described previously (35). The anti-p7 monoclonal antibody was obtained from Larry Arthur. Alkaline phosphatase (AP)-conjugated goat anti-human immunoglobulin G (IgG) and AP-conjugated goat anti-mouse IgG were purchased from Jackson ImmunoResearch Laboratories, Inc. HIV-1 Gag p24 peptide Gag199-207 (AMQMLKETI) was synthesized on an Applied Biosystems model 433A peptide synthesizer (Berkley, Calif.). Peptide stock solutions were prepared in phosphate-buffered saline (PBS) and diluted in culture medium to the appropriate concentration.
Cell lines and transfections.
COS-7, NIH 3T3, and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum–100 U of penicillin/ml–100 μg of streptomycin/ml and passaged upon confluence. p815 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 15% fetal bovine serum–100 U of penicillin/ml–100 μg of streptomycin/ml (complete medium). HeLa cells are human cervical epithelial carcinoma cells. NIH 3T3 cells are mouse fibroblasts. p815 cells were derived from a mouse mastocytoma with an H-2d MHC background. Cell culture reagents were obtained from Life Technologies, Gaithersburg, Md. COS-7 cells were transiently transfected by the DEAE-dextran method as previously described (15). NIH 3T3, p815, and HeLa cells were transfected by the lipofectin method as suggested by the manufacturer (Life Technologies). At 24 h after transfection, the transfected p815 cells were washed once with complete medium and selected with the antibiotic G418 (1.2 mg/ml) for 2 to 3 weeks. The established p815 cell lines were maintained in complete medium and 0.4 mg of G418/ml.
Immunoblotting and p24 assay.
Three days after transfection, cell lysates were prepared as previously described (14). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by using standard methods. Proteins were transferred to nitrocellulose (Schleicher & Schuell) by washing the gel and nitrocellulose in transfer buffer (50 mM NaCl, 1.8 mM EDTA, 10 mM Tris, pH 7.0) and making a sandwich of nitrocellulose sheets with the gel in the middle, wrapping with plastic wrap, and incubating overnight between two glass plates with a 2-kg weight on top, at 4°C (2a, 14–16, 35). The blots were stained with HIV-1-positive human serum in a PBS solution with 3% nonfat dried milk. Secondary antibodies were AP-conjugated anti-human (Jackson Immunoresearch, Inc., West Grove, Pa.) and staining was with BCIP and NBT solutions prepared from chemicals obtained from Sigma. Cell culture supernatants were collected 3 days after transfection and clarified by centrifugation at 3,000 × g for 10 min, and p24 antigen capture enzyme-linked immunosorbent assays (ELISAs) were performed as specified by the manufacturer (DuPont, Wilmington, Del.).
DNA vaccination of mice.
Plasmid DNAs for vaccinations were prepared using QIAGEN plasmid purification kits. DNA was dissolved in saline at a concentration of 1 mg/ml. Female BALB/c mice, 6 to 8 weeks old, purchased from Charles River Laboratories (Wilmington, Mass.), were immunized by intramuscular injection with 0.05 ml of plasmid DNA to a separate site on each side of the quadriceps muscle, followed by two booster vaccinations at 2-week intervals as previously described (6). All animals used in this study were maintained at the Johns Hopkins University, Baltimore, Md., under the supervision of University Laboratory Animal Resources.
Assay for anti-Gag antibodies.
BALB/c mice were intramuscularly injected three times with 100 μg of plasmid DNA each at 0, 2, and 4 weeks. Anti-Gag antibodies were measured at week 6. Sera were collected from each mouse group injected with the various DNA constructs, pooled, and analyzed by immunoblotting, using purified immature viral particles containing unprocessed Gag p55 or mature HIV-1 virions from H9 cells (16).
Cytotoxic T-cell assays.
At week 6 (2 weeks after the last inoculation), animals were sacrificed. Spleens were removed from immunized mice and naive mice and compressed through sterile nylon mesh with a rubber stopper, then washed twice with RPMI 1640. Splenic mononuclear cells were isolated by centrifugation through Ficoll-Hypaque discontinuous density gradients, and T cell enrichment was performed with sterilized nylon wool columns. The harvested cells were centrifuged for 10 min in a Sorval H-1000B rotor at 1,000 rpm (200 × g) and resuspended for the CTL assay. Cell viability was determined by trypan blue exclusion.
The stimulator cells were splenocytes harvested from naive mice and purified by Ficoll-Hypaque gradient. The stimulator cells (5 × 106 cells/ml) were incubated for 20 min in 10 μg of psoralen/ml and then transferred to a T-25 culture plate and exposed to UV light (365 nm) for 5 min. The cells were pelleted, washed 4 to 5 times with RPMI medium, and pulsed with 10 μg of MHC class I-restricted p24 peptide (199 AMQMLKETI 207)/ml for 1 h at 37°C before use. The effector cells in 24-well plates were incubated with stimulator cells at an effector-to-stimulus (E/S) ratio of 5/1 for 6 days in culture medium containing 15 U of interleukin-2/ml. The target cells (p815; 107 cells/ml) were incubated with 10 μg of MHC class I-restricted p24 peptide (199 AMQMLKETI 207)/ml for 30 min at 37°C and then labeled with 100 to 200 μCi of [51Cr]Na2O for 2 h and washed three times with RPMI 1640. Stimulated splenocytes were cocultivated at 37°C for 5 h with 104 radiolabeled target cells at varied effector-to-target ratios of 64/1, 16/1, 4/1, and 1/1. Target cell lysis was measured by liquid scintillation counting. The percent specific lysis of labeled target cells for a given effector cell sample was calculated as follows: specific lysis = (Cr release in sample − control Cr release)/(maximal Cr release − control Cr release) × 100. Maximal release was determined as the average release in wells to which 1% Nonidet P-40 was added in place of effector cells. Control release was determined as the averaged release in control wells in the absence of effector cells, which was 15 to 25% of maximal release. All determinations were performed in triplicate.
RESULTS
Construction of HIV-1 Gag expression vectors.
The HIV-1 gag coding region from HXB2 (25) was amplified by PCR and cloned into pcDNA3.1(−) downstream from the human CMV immediate-early promoter (Pcmv) sequence, using _Xba_I and _Eco_RI sites to generate pGAG (Fig. 1). To generate pGAGCTE, the CTE from SRV-1 (36) was inserted downstream from the gag coding sequence but upstream from the BGHpA signal, using the _Bam_HI and _Kpn_I sites (Fig. 1). The CMV promoter provides a high level of constitutive expression in a range of mammalian cells. The BGHpA signal provides efficient transcription termination and polyadenylation of mRNA. pGAGINS was generated by PCR amplification of mutant gag coding sequence contained in plasmid p55M1-10 (29) and cloned into pcDNA3.1(−) downstream from the human CMV immediate-early promoter (Pcmv) sequence, using the _Xba_I and _Eco_RI sites (Fig. 1). The CTE from SRV-1 was inserted downstream from the gag coding sequence in pGAGINS to generate pGAGINSCTE (Fig. 1).
Comparison of HIV-1 Gag expression by the various constructs in primate cells.
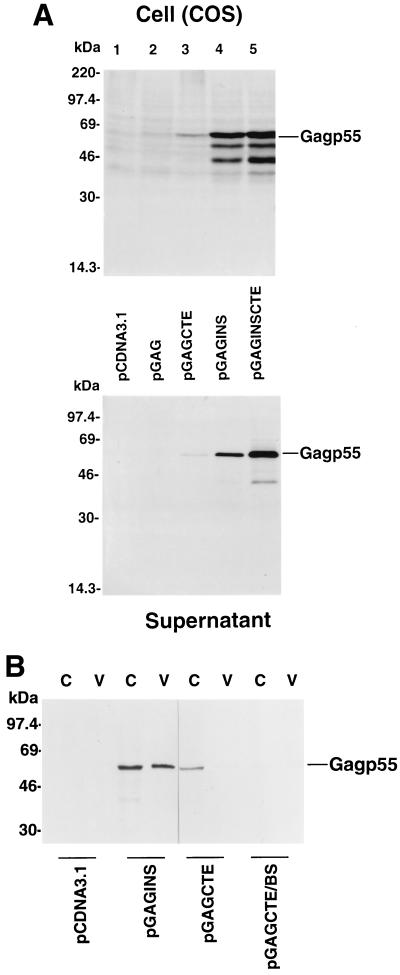
We initially tested the protein expression by various HIV-1 Gag expression vectors in transfected COS-7 cells, an African green monkey kidney-derived cell line. When cell lysates from transfected COS-7 cells were analyzed by immunoblotting with an HIV-1-positive human serum, Gag p55 precursor protein was not detected in pcDNA3.1-transfected COS-7 cells, as expected (Fig. 2A, lane 1, top panel). Also, Gag p55 precursor protein was not detected in pGAG-transfected COS-7 cells (Fig. 2A, lane 2, top panel). Gag p55 precursor protein was detected in transfected COS-7 cells with the pGAGCTE, pGAGINS, and pGAGINSCTE constructs (Fig. 2A, top panel). However, expression of Gag p55 by the pGAGINS (lane 4) construct was much more efficient than that with the pGAGCTE (lane 3) construct (Fig. 2A, top panel). The expression of Gag p55 by the pGAGINSCTE (lane 5) construct was approximately twofold higher than that of the pGAGINS (lane 4) construct (Fig. 2A, top panel).
FIG. 2.
HIV-1 Gag expression in transfected COS cells with various DNA constructs. (A) Cell lysates from pcDNA3.1-, pGAG-, pGAGCTE-, pGAGINS-, and pGAGINSCTE-transfected cells were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum (top panels). Virus lysates from transfected COS-7 cells were prepared as described in Materials and Methods, separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum (bottom panels). (B) Cell lysates from pcDNA3.1-, pGAGINS-, pGAGCTE-, and pGAGCTE/BS-transfected cells were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum (lanes C). Virus lysates were analyzed by immunoblotting with an HIV-1-positive human serum (lanes V).
Gag p55 in particle form was also detected in culture supernatants of pGAGCTE (lane 3)-, pGAGINS (lane 4)-, and pGAGINSCTE (lane 5)-transfected COS-7 cells by the HIV-1-positive human serum (Fig. 2A, bottom panel). As expected, no particulate form of Gag p55 was detected in culture supernatants of pcDNA3.1 (lane 1)- or pGAG (lane 2)-transfected COS-7 cells (Fig. 2A, bottom panel). Again, the pGAGINS-transfected COS-7 cells produced much more Gag p55 in particle form than the pGAGCTE-transfected COS-7 cells (Fig. 2A, bottom panel). These results suggest that the mutations eliminating the INS sequences in the HIV-1 gag coding sequence increase Gag expression in COS-7 cells to a greater extent than insertion of a CTE element at the 3′ untranslated region of the wild-type gag coding sequence.
A previous study suggested that CTE can significantly increase simian immunodeficiency virus SIVmac Gag expression when placed after the wild-type SIVmac gag coding sequences (11). A difference between the expression vectors in the two studies is the presence of an intron between the CTE and the polyadenylation signal in the SIVmac Gag expression vector. We have cloned the wild-type HIV-1 gag coding sequences plus CTE into the same location in the same DNA vector, pBK-CMV (11) to generate pGAGCTE/BS. HIV-1 Gag expression was again analyzed in transfected COS-7 cells. Immunoblotting analyses using cell lysates (Fig. 2B, lanes C) and supernatant lysates (Fig. 2B, lanes V) indicated that pGAGCTE/BS did not induce better HIV-1 Gag expression than did the pGAGCTE vector (Fig. 2B). Both pGAGCTE and pGAGCTE/BS induced less HIV-1 Gag expression than did pGAGINS (Fig. 2B). These results suggest that CTE could not significantly stimulate HIV-1 Gag expression, irrespective of the DNA vectors used.
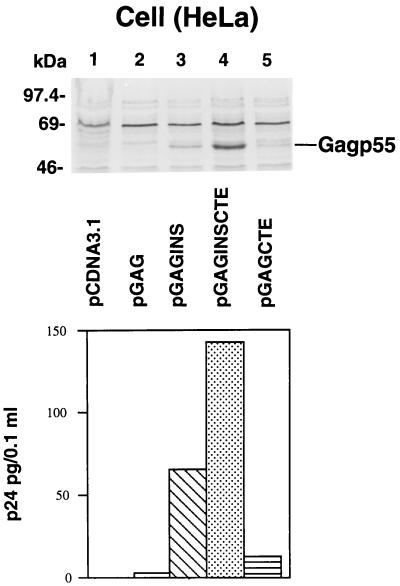
Gag p55 expression was also evaluated with the various constructs in transfected HeLa cells. Immunoblot analysis of cell lysates prepared from transfected HeLa cells is shown in Fig. 3 (top panel). Transfection of the HeLa cells with pcDNA3.1, pGAG, and pGAGCTE did not produce detectable levels of Gag protein by immunoblot analysis (Fig. 3, top panel, lanes 1, 2, and 5). On the other hand, pGAGINS- and pGAGINSCTE-transfected HeLa cells produced detectable Gag p55 (Fig. 3). Gag p55 was also detected in the supernatants of pGAGINS- and pGAGINSCTE-transfected HeLa cells (Fig. 3, bottom panel) but not to any appreciable extent from those of pcDNA3.1-, pGAG-, and pGAGCTE-transfected HeLa cells (Fig. 3, bottom panel).
FIG. 3.
HIV-1 Gag expression in transfected HeLa cells. HeLa cells were transfected with various DNA constructs as described in Materials and Methods. Cell lysates were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum (top panels). Soluble Gag released from transfected HeLa cells was monitored by p24 ELISA (bottom panel).
Comparison of HIV-1 Gag expression by the various constructs in mouse cells.
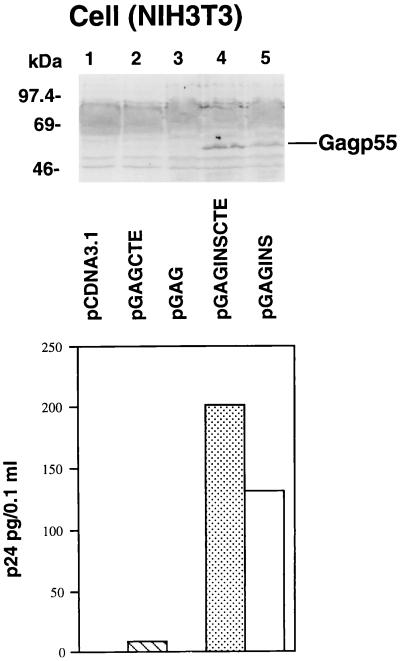
To compare levels of Gag expression in mouse cells by the various HIV-1 Gag constructs, we performed transient transfection experiments using mouse NIH 3T3 cells. Three days after transfection, cell lysates from transfected NIH 3T3 cells were analyzed by immunoblotting for the presence of cell-associated Gag p55 molecules (Fig. 4, top panel). Gag p55 was detected in the cell lysates of NIH 3T3 cells transfected by pGAGINS (lane 5) and pGAGINSCTE (lane 4) (Fig. 4, top panel). No Gag p55 was detected in the cell lysates of pcDNA3.1-, pGAG-, and pGAGCTE-transfected NIH 3T3 cells by HIV-1-positive serum (Fig. 4, top panel, lanes 1 to 3). Gag p55 was also detected in the supernatants of pGAGINS- and pGAGINSCTE-transfected NIH 3T3 cells (Fig. 4, bottom panel). No Gag p55 was detected in the supernatants of pcDNA3.1-, pGAG-, and pGAGCTE-transfected NIH 3T3 cells (Fig. 4, bottom panel).
FIG. 4.
HIV-1 Gag expression in transfected NIH 3T3 cells. NIH 3T3 cells were transfected with various DNA constructs as described in Materials and Methods. Cell lysates from transfected cells were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum (top panel). Soluble Gag released from transfected NIH 3T3 cells was monitored by p24 ELISA (bottom panel).
Anti-Gag antibody responses in mice immunized with naked DNA vaccine.
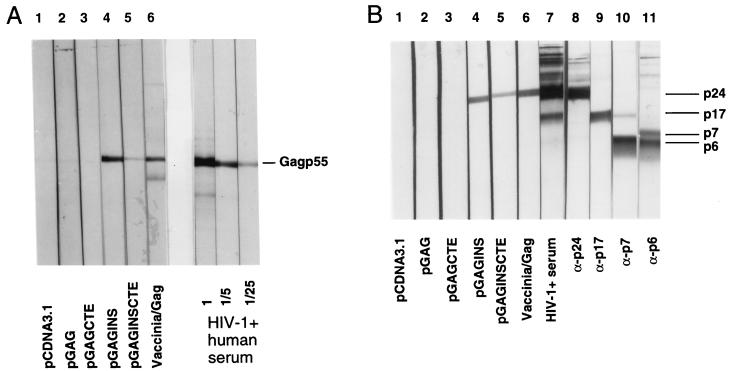
BALB/c mice were intramuscularly injected with 100 μg of plasmid DNA three times at 0, 2, and 4 weeks. Anti-Gag antibodies were measured at week 6. Sera were collected from the mice injected with the different DNA constructs and analyzed by immunoblot using purified particle-associated Gag p55 produced from H9 cells (16). As shown in Fig. 5A, anti-Gag antibodies were detected in mice vaccinated with either pGAGINS (lane 4) or pGAGINSCTE (lane 5) and in mice infected with recombinant vaccinia virus/Gag (lane 6). Serial dilutions of an HIV-1-positive human serum were used as a positive control (Fig. 5A). In contrast, no anti-Gag antibodies were detected in mice immunized with pcDNA3.1, pGAG, and pGAGCTE (Fig. 5A, lanes 1 to 3).
FIG. 5.
BALB/c mice were injected intramuscularly with 100 μg of plasmid DNA three times at 0, 2, and 4 weeks. Anti-Gag antibodies were measured at week 6. Sera were collected from each mice group injected with the different DNA constructs, pooled, and analyzed by immunoblot, using purified viral particle Gag p55 (A) or purified mature HIV-1 virions (B). Vaccinia/Gag, sera from mice infected with recombinant vaccinia virus/Gag. HIV-1-positive human serum was used as positive control.
The precursor Gag p55 is cleaved by viral protease into MAp17, CAp24, NCp7, and p6 during viral maturation. In HIV-1-infected individuals, CAp24 is the predominant region involved in the generation of an anti-Gag antibody response. To determine which region of Gag elicits the production of anti-Gag antibody in immunized mice, we performed an immunoblot analysis using purified mature virions as antigens. The positions of MAp17, CAp24, NCp7, and p6 in the blots were identified by specific antibodies (Fig. 5B). The anti-Gag antibodies generated in mice immunized with pGAGINS (lane 4) and pGAGINSCTE (lane 5) reacted predominantly with CAp24 (Fig. 5B). Interestingly, the pGAGINSCTE DNA construct did not induce a better anti-Gag antibody response than did the pGAGINS construct (Fig. 5A and B), although pGAGINSCTE seemed to induce a slightly higher level of protein expression in vitro than did pGAGINS (Fig. 2 to 4).
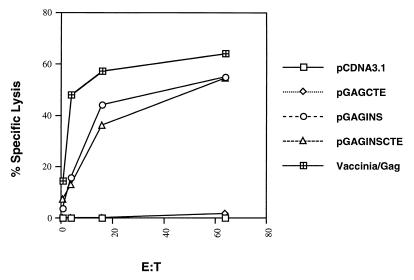
CTL responses in DNA-vaccinated mice.
Mice were immunized with the various DNA constructs as described above. At week 6, splenocytes from the mice in each group were harvested and pooled, and the CTL responses specific to HIV-1 Gag were measured following antigen restimulation. As a positive control for this experiment, mice were also vaccinated with recombinant vaccinia virus/Gag. As expected, those plasmids that did not express Gag p55 in vitro, such as pcDNA3.1 and pGAGCTE, did not elicit a CTL response against HIV-1 Gag (Fig. 6). Levels of specific lysis below 5% were considered not significant. The group of vaccinated mice that received pGAGINS developed a slightly higher level of HIV-1 Gag-specific lytic activity than those that received pGAGINSCTE. These results showed that intramuscular injection of Rev-independent expression plasmids can induce both humoral and cellular immune responses against HIV-1 Gag in a murine model.
FIG. 6.
BALB/c mice were intramuscularly injected three times with 100 μg of plasmid DNA at 0, 2, and 4 weeks. Anti-HIV-1 Gag CTL responses were measured at week 6. Splenocytes from the mice in each group were harvested and pooled, and the CTL responses specific to HIV-1 p24 peptide were measured following antigen restimulation in vitro. The target cells (p815) were pulsed with p24 peptide.
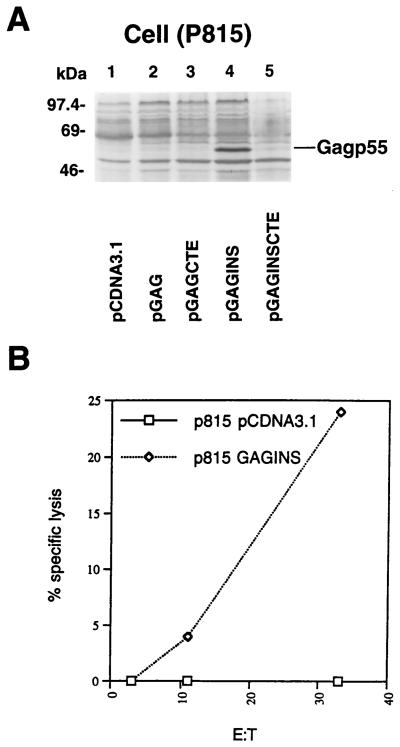
Stable expression of HIV-1 Gag in mouse p815 cells and as target cells for measuring CTL responses.
p815 cells were transfected and established as described in Materials and Methods. Cell lysates from stably transduced p815 cells were analyzed by immunoblotting for cell-associated Gag p55 molecules (Fig. 7A). Gag p55 was only detected in the cell lysates of pGAGINS-transduced p815 cells (Fig. 7A, lane 4). No Gag p55 was detected with the HIV-1-positive serum in the cell lysates of pcDNA3.1-, pGAG-, pGAGCTE-, or pGAGINSCTE-transduced cells (Fig. 7A). Stable expression of HIV-1 Gag in mouse cell lines has not been described previously. We can reproducibly transduce mouse p815 and obtain stable HIV-1 Gag expression with the pGAGINS construct.
FIG. 7.
p815 cells were transfected with various DNA constructs. Stable cell lines were established by selection with G418. (A) Cell lysates from transduced p815 cells were separated by SDS-PAGE, transferred to nitrocellulose filters, and analyzed by immunoblotting with an HIV-1-positive human serum. Gag expression was only detected in p815 cells transduced with the pGAGINS construct. (B) Transduced p815 cells as CTL target cells. CTL effector cells were obtained from splenocytes of BALB/c mice infected with recombinant vaccinia virus/Gag.
We have also examined whether HIV-1 Gag-transduced p815 cells can serve as HIV-1 Gag-specific CTL target cells (Fig. 7B). HIV-1 Gag-specific effector cells were obtained from mice infected with recombinant vaccinia virus/Gag. Two weeks after infection, splenocytes from mice infected with recombinant vaccinia virus/Gag were harvested, pooled, and stimulated with p24 peptide in vitro. CTL responses specific to HIV-1 Gag, using HIV-1 Gag-transduced p815 cells as target cells, were measured. As expected, p815 cells transduced with control vector pcDNA3.1 did not elicit a CTL response, whereas p815 cells transduced with pGAGINS did (Fig. 7B). These latter cells are useful reagents for evaluating HIV-1 Gag processing and MHC class I presentation in mouse cells and HIV-1 Gag-specific CTL responses elicited by various vaccine strategies in the mouse model.
DISCUSSION
In this report, we have compared several strategies using DNA vectors to induce efficient expression of HIV-1 Gag molecules in various cell types. The presence of multiple INS in the structural genes gag, pol, and env of HIV-1 make the expression of these structural proteins Rev and RRE dependent (24). Consistent with previous observations (29), we did not detect any expression of HIV-1 Gag proteins when a DNA expression vector containing wild-type gag coding sequences was transfected into human (HeLa), monkey (COS-7), or mouse (NIH 3T3) cells. Addition of the mRNA nuclear export signal CTE from SRV-1 increased Gag expression only slightly in COS cells and did not yield detectable levels of Gag in either HeLa or NIH 3T3 cells. In contrast, mutations of the INS in the wild-type gag coding sequences (29) significantly increased Gag protein expression in all the cell lines tested, including HeLa, COS, and NIH 3T3 cells.
The failure to achieve significant stimulation of Gag expression by adding the CTE could not be attributed to a suboptimal location of the CTE (26). The CTE in our DNA constructs is located just 5′ to the polyadenylation signal, a location considered to be the most efficient (26). A previous study has suggested that the CTE can significantly increase SIVmac Gag expression when placed after the wild-type SIVmac gag coding sequences (11). Those investigators used a vector containing an intron between the CTE and the polyadenylation signal. To exclude the possibility that the presence of the intron is responsible for the different results, we cloned the wild-type HIV-1 gag coding sequences plus the CTE into the same location as the DNA vector used in the previous study (pGAGCTE/BS). We did not observe any increased HIV-1 Gag expression relative to that with the pGAGCTE vector (Fig. 2B). These results suggest that the CTE is not able to significantly stimulate HIV-1 Gag expression, irrespective of the DNA vector used. It is possible that the wild-type HIV-1 gag coding sequences behave differently than do the wild-type SIVmac gag coding sequences when combined with the CTE. A side-by-side comparison of the CTE in the context of the wild-type SIVmac gag coding sequences and the INS mutant form of SIVmac gag coding sequences will be needed to determine which strategy is more efficient.
Although the CTE strategy appears to work efficiently to induce SIVmac Gag expression in simple DNA expression vectors (11, 27), the results obtained in the present study suggest that the most efficient strategy for expressing HIV-1 Gag via DNA vectors involves the elimination of the INS. More importantly, we found that this latter strategy apparently allowed efficient Gag protein expression in vivo after intramuscular DNA injection into mice and also induced efficient humoral as well as cellular immune responses against HIV-1 Gag. The precise function of the INS that limit HIV-1 Gag expression in the absence of Rev/RRE remains to be defined. It has been proposed that INS are binding sites for cellular factors which contribute to mRNA instability (1). Therefore, inactivation of INS in the gag coding region should increase mRNA stability and allow efficient Gag expression.
Using the pGAGINS construct, we were also able to reproducibly achieve stable expression of HIV-1 Gag in the p815 mouse cell line. However, attempts to obtain stable HIV-1 Gag expression in another mouse cell line, NIH 3T3, with either the pGAGINS or the pGAGINSCTE construct were unsuccessful. Using HIV-1 Gag-transduced p815 cells, we also demonstrated that these cells can present HIV-1 Gag antigen via MHC class I molecules and can serve as HIV-1 Gag-specific CTL target cells. These cells can therefore serve as useful reagents for evaluating the HIV-1 Gag-specific CTL response to various vaccine strategies in a mouse model system.
Although the combination of CTE and INS mutations (pGAGINSCTE) was slightly more effective than INS mutations alone (pGAGINS) in increasing in vitro Gag expression after transient transfections, pGAGINSCTE did not induce a better anti-Gag antibody response or anti-Gag CTL response after DNA immunization in mice. Furthermore, pGAGINS induced stable Gag expression in mouse p815 cells, whereas pGAGINSCTE did not. It is possible that pGAGINS induced more sustained protein expression than pGAGINSCTE in mice after intramuscular injection and therefore induced slightly better immune responses. Alternatively, stronger transient Gag expression with pGAGINSCTE but not with pGAGINS may induce some immune tolerance. Further study will be required to determine which of these factors is more significant. In summary, our results suggest that a simple strategy of eliminating the INS elements by mutagenesis is sufficient to express HIV-1 Gag in a Rev/RRE-independent and species-independent fashion. Such a strategy may facilitate future systematic evaluation of approaches to maximizing the induction of cellular immune responses, such as CTL and T-helper responses against HIV-1 Gag in the mouse model.
ACKNOWLEDGMENTS
We thank Bindong Liu from The Johns Hopkins School of Hygiene and Public Health, Baltimore, Md., for technical assistance. We are grateful to David Schwartz and Richard Markham for helpful discussions. The following reagents were obtained through the AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, NIH: recombinant vaccinia virus/Gag vP1287 (catalog no. 3542), antiserum to HIV-1 p24 (catalog no. 2930), antiserum to HIV-1 p17 (catalog no. 286), and purified p24Gag (catalog no. 382). We gratefully acknowledge the generous gifts of anti-p7 antibodies from Larry Arthur.
This work was supported by National Institutes of Health grants AI-42624 and AI-46324 to X.-F.Y. M.D. was supported in part by a training grant from NIEHS (ES07141).
REFERENCES
- 1.Afonina E, Neumann M, Pavlakis G N. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J Biol Chem. 1997;272:2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 2.Aldhous M C, Watret K C, Mok J Y, Bird A G, Froebel K S. Cytotoxic T lymphocyte activity and CD8 subpopulations in children at risk of HIV infection. Clin Exp Immunol. 1994;97:61–67. doi: 10.1111/j.1365-2249.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Barin F, M’Boup S, Denis F, Kanki P, Allan J S, Lee T-H, Essex M. Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of west Africa. Lancet. 1985;ii:1387–1389. doi: 10.1016/s0140-6736(85)92556-5. [DOI] [PubMed] [Google Scholar]
- 3.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly J J, Ulmer J B, Shiver J W, Liu M A. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 7.Durali D, Morvan J, Letourneur F, Schmitt D, Guegan N, Dalod M, Saragosti S, Sicard D, Levy J P, Gomard E. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J Virol. 1998;72:3547–3553. doi: 10.1128/jvi.72.5.3547-3553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 10.Haynes J R, Fuller D H, Eisenbraun M D, Ford M J, Pertmer T M. Accell particle-mediated DNA immunization elicits humoral, cytotoxic, and protective immune responses. AIDS Res Hum Retroviruses. 1994;10:S43–S45. [PubMed] [Google Scholar]
- 11.Indraccolo S, Feroli F, Minuzzo S, Mion M, Rosato A, Zamarchi R, Titti F, Verani P, Amadori A, Chieco-Bianchi L. DNA immunization of mice against SIVmac239 Gag and Env using Rev-independent expression plasmids. AIDS Res Hum Retroviruses. 1998;14:83–90. doi: 10.1089/aid.1998.14.83. [DOI] [PubMed] [Google Scholar]
- 12.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langlade-Demoyen P, Ngo-Giang-Huong N, Ferchal F, Oksenhendler E. Human immunodeficiency virus (HIV) nef-specific cytotoxic T lymphocytes in noninfected heterosexual contact of HIV-infected patients. J Clin Invest. 1994;93:1293–1297. doi: 10.1172/JCI117085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y-M, Tian C-J, Yu X-F. A bipartite membrane-binding signal in the human immunodeficiency virus type 1 matrix protein is required for the proteolytic processing of Gag precursors in a cell type-dependent manner. J Virol. 1998;72:9061–9068. doi: 10.1128/jvi.72.11.9061-9068.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y M, Tang X B, Cimakasky L M, Hildreth J E, Yu X F. Mutations in the matrix protein of human immunodeficiency virus type 1 inhibit surface expression and virion incorporation of viral envelope glycoproteins in CD4+ T lymphocytes. J Virol. 1997;71:1443–1452. doi: 10.1128/jvi.71.2.1443-1452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 17.Lu S, Santoro J C, Fuller D H, Haynes J R, Robinson H L. Use of DNAs expressing HIV-1 Env and noninfectious HIV-1 particles to raise antibody responses in mice. Virology. 1995;209:147–154. doi: 10.1006/viro.1995.1238. [DOI] [PubMed] [Google Scholar]
- 18.McAdam S, Kaleebu P, Krausa P, Goulder P, French N, Collin B, Blanchard T, Whitworth J, McMichael A, Gotch F. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. AIDS. 1998;12:571–579. doi: 10.1097/00002030-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath M J. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337:1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 20.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 21.Okuda K, Bukawa H, Hamajima K, Kawamoto S, Sekigawa K, Yamada Y, Tanaka S, Ishi N, Aoki I, Nakamura M. Induction of potent humoral and cell-mediated immune responses following direct injection of DNA encoding the HIV type 1 env and rev gene products. AIDS Res Hum Retroviruses. 1995;11:933–943. doi: 10.1089/aid.1995.11.933. [DOI] [PubMed] [Google Scholar]
- 22.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P, et al. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 23.Parslow T C. Post-transcriptional regulation of human retroviral gene expression. In: Cullen B R, editor. Human retroviruses. Durham, N.C: IRL Press at Oxford University Press; 1993. pp. 101–136. [Google Scholar]
- 24.Pavlakis G N. The molecular biology of human immunodeficiency virus type 1. In: DeVita J V T, Hellman S, Rosenberg S A, editors. AIDS: biology, diagnosis, treatment and prevention. 4th ed. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 25.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 26.Rizvi T A, Schmidt R D, Lew K A. Mason-Pfizer monkey virus (MPMV) constitutive transport element (CTE) functions in a position-dependent manner. Virology. 1997;236:118–129. doi: 10.1006/viro.1997.8728. [DOI] [PubMed] [Google Scholar]
- 27.Rizvi T A, Schmidt R D, Lew K A, Keeling M E. Rev/RRE-independent Mason-Pfizer monkey virus constitutive transport element-dependent propagation of SIVmac239 vectors using a single round of replication assay. Virology. 1996;222:457–463. doi: 10.1006/viro.1996.0444. [DOI] [PubMed] [Google Scholar]
- 28.Rowland-Jones S L, Nixon D F, Aldhous M C, Gotch F, Ariyoshi K, Hallam N, Kroll J S, Froebel K, McMichael A. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet. 1993;341:860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- 29.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabernero C, Zolotukhin A S, Bear J, Schneider R, Karsenty G, Felber B K. Identification of an RNA sequence within an intracisternal-A particle element able to replace Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Virol. 1997;71:95–101. doi: 10.1128/jvi.71.1.95-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townsend A, Bastin J, Gould K, Brownlee G, Andrew M, Coupar B, Boyle D, Chan S, Smith G. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J Exp Med. 1988;168:1211–1224. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trono D, Baltimore D. A human cell factor is essential for HIV-1 Rev action. EMBO J. 1990;9:4155–4160. doi: 10.1002/j.1460-2075.1990.tb07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, Ugen K E, Srikantan V, Agadjanyan M G, Dang K, Refaeli Y, Sato A I, Boyer J, Williams W V, Weiner D B. Gene inoculation generates immune responses against human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:4156–4160. doi: 10.1073/pnas.90.9.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X F, Matsuda Z, Yu Q C, Lee T H, Essex M. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J Gen Virol. 1995;76:3171–3179. doi: 10.1099/0022-1317-76-12-3171. [DOI] [PubMed] [Google Scholar]
- 36.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]