DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA (original) (raw)
Abstract
The Arabidopsis protein AINTEGUMENTA (ANT) is a member of a plant-specific family of transcription factors (AP2/EREBP) that share either one or two copies of an approximately 70 amino acid region called the AP2 repeat. DNA binding activity has been demonstrated previously for members of this family containing a single AP2 repeat. Using an in vitro selection procedure, the DNA binding specificity of the two AP2 repeat containing protein ANT was found to be 5′-gCAC(A/G)N(A/T)TcCC(a/g)ANG(c/t)-3′. This consensus site is much longer than sites recognized by proteins containing a single AP2 repeat and neither AP2 repeat of ANT was alone capable of binding to the selected sequences, suggesting that both AP2 repeats make DNA contacts. ANT binds to these DNA sequences as a monomer but a higher order complex is also observed at high protein concentrations. The ANT consensus site shows some similarity to the C-repeat/DRE elements bound by proteins that contain a single AP2 repeat, and we find that ANT binds weakly to such sites. We propose a model in which each AP2 repeat of ANT contacts adjacent sites within the consensus sequence. Our results suggest that the AP2 repeat can be utilized in different ways for DNA binding.
INTRODUCTION
The Arabidopsis protein AINTEGUMENTA (ANT) is a member of a large family of transcription factors that may be unique to plants. Members of the AP2/ethylene response element binding proteins (EREBP) family are involved in various aspects of plant growth and development (including flower development, hormone signal transduction and cellular differentiation) and in responses to biotic and abiotic stresses (1). These proteins contain either one (EREBP subfamily) or two (AP2 subfamily) copies of an approximately 70 amino acid domain termed the AP2 repeat because of its initial description in the floral homeotic protein APETALA2 (AP2) (2–4). In proteins containing a single AP2 repeat, this region has been shown to exhibit DNA binding activity. Proteins that have a single AP2 repeat include EREBPs/ethylene response factors (ERFs) (5–7), C-repeat/dehydration response element binding proteins (CBFs/DREBs) (8–10), ABI4 (11) and TINY (12). Proteins containing two AP2 repeats include AP2 (2,3), ANT (13,14) and the products of the maize indeterminate spikelet1 (ids1) (15) and Glossy15 (Gl15) (16) genes.
DNA binding specificity has been determined for three types of AP2/EREBP proteins that contain a single AP2 repeat. The first class are proteins which bind to ethylene response elements (ERE) or GCC boxes (tobacco EREBPs, Arabidopsis AtEBP and AtERF1-5, and tomato Pti4-6) found in the promoters of ethylene-inducible pathogenesis related genes (5–7,17). The GCC box is an 11 bp sequence (TAAGAGCCGCC) with a core GCCGCC sequence that is required for binding (5). The solution structure of the AP2 domain of AtERF1 bound to the GCC box has been determined (18). This protein domain folds into a novel DNA recognition structure consisting of a three-stranded β-sheet packed against an α-helix with residues in the β-sheet primarily responsible for DNA recognition.
The second class includes proteins that bind to the C-repeat or dehydration response element (DRE) in the promoters of genes that are turned on in response to low temperatures and/or water deficit (CBF1, CBF2, CBF3/DREB1A and DREB2A) (8–10). The C-repeat/DREs contain the core sequence CCGAC (19). The third class of AP2/EREBP-like proteins that have been shown to bind DNA consists of Arabidopsis RAV1 and RAV2. RAV1 and RAV2 are highly related proteins that contain two DNA binding motifs: a single AP2 repeat and a B3-like domain (20). RAV1 binds to a bipartite recognition sequence with the AP2 repeat binding to a CAACA motif and the B3 domain binding to a CACCTG sequence (20). Although either domain can bind DNA on its own, a much higher DNA binding affinity is achieved when both domains are present. The AP2 domains of RAV1 and RAV2 are more diverged from those of the EREBP-like proteins and may be considered a third subfamily of the AP2/EREBP family.
DNA binding has not yet been described for any proteins of the AP2 subfamily. These proteins are likely to possess DNA binding specificities that are distinct from those of the EREBP-like proteins and RAV1. ANT functions in several different processes in flower development including ovule development, floral organ initiation and growth, and petal development (13,14,21,22). To better understand how ANT functions in different aspects of reproductive development and to identify possible downstream targets of ANT regulation, we have determined the DNA binding specificity of ANT using an in vitro selection method. This procedure yielded the consensus sequence 5′-gCAC(A/G)N(A/T)TcCC(a/g)ANG(c/t)-3′ which is bound by ANT with high affinity. ANT binds as a monomer but at high protein concentrations a higher order complex is also observed. The length of the consensus site suggests that both AP2 repeats of ANT contact the DNA, and we show that neither AP2 repeat by itself is sufficient for binding to this DNA sequence. The ability of the second AP2 repeat of ANT to bind weakly to a site with similarity to part of the consensus site suggests that the two AP2 repeats may contact juxtaposed subsites within the consensus sequence. Our studies suggest that the AP2-like and EREBP-like proteins exhibit distinct mechanisms of DNA binding and that AP2 repeats can be utilized in different contexts for DNA recognition.
MATERIALS AND METHODS
Plasmid construction and protein expression
ANT protein constructs were made by inserting PCR amplified DNA fragments into the _Bam_HI site of pQE12 or pQE-8 (Qiagen). The c-myc tag (EQKLISEEDLN) was added by annealing two complementary oligonucleotides that created _Bgl_II sticky ends. All plasmids were sequenced on an ABI 377. Proteins were expressed by induction with 1 mM IPTG in either XL1-Blue MRF′ Tet (Stratagene) or M15(pREP4) (Qiagen) cells. The cells were harvested after growth at 25–30°C for 4–8 h. In the case of ANT-AP2R1R2 and ANT-AP2R2, cells were lysed by sonication and purified using Ni-NTA (Qiagen) according to the manufacturer’s instructions. Escherichia coli cells expressing ANT-AP2R1 were lysed by stirring in 8 M urea, 100 mM Na-phosphate, 10 mM Tris–HCl pH 8.
Selection of ANT binding sequences from random oligonucleotides
A labeled pool of random sequences was generated by a Klenow fill-in reaction using R3 (5′-CTCGTCCAGTTACGTCTAGACT-3′) and R1 (5′-CTAGACTAGGTGCAGAATTCACTTG(N)20ACCAGTCTAGACGTAACTGGACGAG-3′), where N indicates bases of mixed A,C,G,T identity. Purified ANT-AP2R1R2 (0.8 µg) was incubated with 0.25 µg of labeled DNA in binding buffer (20 mM Tris pH 8, 100 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, 100 µg/ml poly dI–dC) for 30 min at 25°C. An aliquot of 0.125 µg of anti-cmyc Ab (PharMingen) was added and the binding reactions were incubated for 1 h at room temperature. Protein A–Sepharose (100 µg) was added and the binding reactions were incubated for an additional hour. The Sepharose was washed twice with 500 µl binding buffer without DTT and dI–dC. Bound DNA was eluted from the Sepharose after 30 min of incubation in 200 µl of 50 mM Tris pH 8, 5 mM EDTA, 100 mM NaOAc, 0.5% SDS at 45°C. The recovered DNA was extracted with phenol, precipitated, and used as a template in a PCR reaction with R2 (5′-CTAGACTAGGTGCAGAATTCAC-3′) and R3. The radiolabeled PCR product was purified on a 3% NuSieve gel and treated with GELase (Epicentre Technologies). This selection procedure was repeated eight times before switching to gel mobility shift assays. Binding reactions for the gel shifts were performed in 20 mM Tris pH 8, 100 mM KCl, 12% glycerol, 1 mM EDTA, 1 mM DTT, 20 ng/µl dI–dC, 20 ng/µl calf thymus DNA and 0.3 mg/ml BSA. The binding reactions were separated on a 1.8% SeaPlaque agarose gel (FMC) in 1× TBE at 4°C. Bound DNA was cut out of the gel, rehydrated in 100 µl TE, treated with GELase and used for PCR as described above. After 10 rounds of selection with gel shifts, the recovered DNA was PCR amplified and cloned into pCR-Script (Stratagene). Sequence analysis was performed on 34 clones using an ABI 377.
Gel mobility shift assays
Gel mobility shift assays for characterizing ANT binding were similar to those described above for the selection experiments except that the protein–DNA complexes were separated on 5% acrylamide (29:1 polyacrylamide:bisacrylamide) gels in 1× TBE at 4°C. Quantitative gel shift assays were performed at room temperature and the binding reactions were incubated for 4 h prior to loading. Quantitation of bound and free DNA was performed using a Molecular Dynamics Storm 860 phosphoimager. Data were fit to the equation Y = [Pt]/([Pt]+Kd), where Y = fractional saturation, using Kaleidagraph Software (Synergy Software, Reading, PA). The protein concentrations used in the experiment were in large excess compared to the DNA such that the concentration of free protein corresponds essentially to total protein.
DNA binding site probes
Binding site 15 and mutated versions (M1, M2, M3 and M4) were cloned into pGEM3Z (Promega) by annealing two oligos that created _Bam_HI sticky ends. Probes for these binding sites were prepared by digestion with _Eco_RI and _Hin_dIII, fill-in labeling reaction, purification on a 3% NuSieve gel, recovery by GELase digestion and precipitation. The COR15a, COR78-1 and mCOR15a C-repeat/DRE probes were prepared similarly by digestion of plasmids pEJS44, pEJS57 and pEJS210, respectively, with _Eco_RI and _Hin_dIII. For DNase I footprinting and methylation interference the plasmids were cut with either _Eco_RI or _Hin_dIII, labeled and then cut with the other enzyme.
DNase I footprinting
Footprinting reactions were performed in 25 mM Tris–HCl pH 8, 5 mM MgCl2, 1 mM CaCl2, 100 mM KCl, 2 mM DTT, 5 µg/ml dI–dC and 100 µg/ml BSA. Proteins were incubated with ∼10 000 c.p.m. of singly end-labeled probe for 20 min at room temperature. An aliquot of 5 U of DNase I (Pharmacia) was added to a 200 µl reaction volume and incubated for 1 min. The reaction was stopped by the addition of 700 µl of DNase stop solution containing 73 µg/ml tRNA and 0.73 M NH4OAc. The DNA was precipitated by incubation in a dry ice/EtOH bath for 15 min followed by centrifugation. The pellets were resuspended in 3–5 µl of formamide buffer, incubated at 90°C for 5 min and loaded onto an 8% acrylamide gel (40:2 polyacrylamide:bisacrylamide).
Methylation interference
100 000–300 000 c.p.m. of probe in 200 µl of 50 mM sodium cacodylate, 1 mM EDTA pH 8 was methylated by incubation with 1 µl dimethylsulfate (DMS) for 5 min at room temperature. The reaction was terminated by the addition of 40 µl of DMS stop buffer (1.5 M NaOAc, 1 M β-mercaptoethanol), 250 µg/ml tRNA and 600 µl EtOH. The samples were incubated in a dry ice/EtOH bath for 10 min and centrifuged for 10 min. The DNA pellet was resuspended in 250 µl of 0.3 M NaOAc, 1 mM EDTA. The DNA was precipitated with the addition of 750 µl EtOH and incubated in a dry ice/EtOH bath for 10 min. After centrifugation, the recovered DNA was resuspended and again precipitated. The methylated DNA was used as a probe in a gel shift experiment. Bands corresponding to the bound DNA and free DNA were cut out of the gel, rehydrated in 200 µl of TE, treated with GELase and precipitated. The recovered DNA was cleaved by incubation with 100 µl of 1 M piperidine for 30 min at 90–95°C. The DNA was lyophilized, resuspended in 100 µl of H2O, lyophilized, resuspended once more in 100 µl of H2O and lyophilized. The samples were resuspended in formamide loading buffer and separated on an 8% acrylamide gel.
RESULTS
Determination of optimal DNA binding sequences for ANT
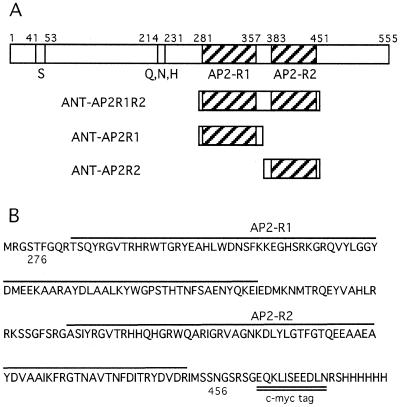
To determine whether ANT has DNA binding abilities, and if so, the nature of its DNA recognition sequence, a random oligonucleotide in vitro selection method (SELEX) was used (23). Labeled oligonucleotides containing a 20 bp randomized central sequence (flanked by defined sequences on either side) were incubated with a modified ANT protein consisting of amino acids 276–456 fused to a c-myc epitope tag. This portion of ANT corresponds to the two AP2 repeats and the linker region between them and will be referred to as ANT-AP2R1R2 (Fig. 1). DNA sequences bound by ANT-AP2R1R2 were initially separated by immunoprecipitation using an antibody against the c-myc tag and later by gel mobility shift assays. Sequences bound by ANT-AP2R1R2 were recovered, amplified by PCR and subjected to additional rounds of selection. After 18 rounds of selection (eight by immunoprecipitation and 10 by gel shift), PCR products were cloned and sequenced. These sequences were aligned (Table 1) and a consensus binding site determined (Table 2). The consensus binding site has the sequence 5′-gCAC(A/G)N(A/T)TcCC(a/g)ANG(c/t)-3′ where the uppercase letters indicate the most highly conserved positions (present in >90% of the selected sites) and lowercase letters indicate somewhat less conserved positions (present in at least 65% of the selected sites). N indicates positions for which no particular base appeared to be preferred. The binding site is a 16 bp sequence containing 14 conserved positions.
Figure 1.
ANT protein constructs used in DNA binding assays. (A) Schematic diagram of full-length ANT (top), ANT-AP2R1R2, ANT-AP2R1 and ANT-AP2R2. S and Q, N, H indicate Ser- and Gln, Arg, His-rich regions respectively, while AP2-R1 and AP2-R2 refer to the first and second AP2 repeats. (B) Amino acid sequence of ANT-AP2R1R2 which corresponds to amino acids 276–456 of full-length ANT. The two AP2 repeats are indicated with lines while the c-myc tag is double underlined.
Table 1. Sequences selected from pool of random oligonucleotides.
| Clone number | Sequence | ANT binding |
|---|---|---|
| 1 | gtTCGGCGCAATTCCCGATGTTcagg | ** |
| 2 | ggCGAGCACGGTTCCCAAAGCAcagt | *** |
| 3 | agaattcacgGTTCCCGAGGGCTTTG | ** |
| 4 | CGAGAGCACAGTTCCCcaggtgaatt | * |
| 5 | TCAGCGCACAAATCCCcaggtgaatt | * |
| 6 | CATCAGCACGACTCCCcaggtgaatt | * |
| 8 | agaattcacgGTTCCCGAGGGCTTTG | ** |
| 9 | ctgtGGCACAAACTCCGATGTCCAcg | ** |
| 10 | CCACTGCACAGATCCCcaagtgaatt | * |
| 11 | CCCAGGCACGCATCCcaagtgaattc | *** |
| 12 | CGAGAGCTCAGTTCCCcaggtgaatt | ** |
| 13 | AACACACACGGATCCCAAGGcagtga | *** |
| 14 | AACACACACGGATCCCGAGGcagtga | *** |
| 15 | ttgGTGCACATATCCCGATGCTTaca | *** |
| 16 | cactgACACACTTACCGAGGTGTGTa | *** |
| 17 | CAATAGCACGTTTCCCcaggtgaatt | ** |
| 19 | cctgGGTGCGTTTCCCAAAGCAGGac | *** |
| 21 | gGGTCGCACAAATCCCAATGTacagt | |
| 22 | cactgACACACTTACCGAGGTGTGTa | |
| 23 | ctgtGGCACAAACTCCGATGTCCAcg | |
| 24 | cctgTACACGGTTCCCGATACTCTac | |
| 25 | cttgGGCACAGTTCCCCAAGCTACac | |
| 26 | ctggtACACACTTCCCAAGAAGCCCc | |
| 28 | CCTTGGCACAGTTACCcaagtgaatt | |
| 29 | TGATGACACGCTTTCCAaccagtcta | |
| 30 | ttgGTGCACATATCCCGATGCTTaca | |
| 31 | gGGTCGCACAAATCCCAATGTacagt | |
| 33 | ACNGTGCACAGATTCTcaagtgaatt | |
| 34 | ctgtGGCACAAAGTCCGATGTCCAcg | |
| 36 | agactgCACATATCCCTAGGTTCCTC | |
| 37 | GAGGCGCACAGGTCCCcaggtgaatt | |
| 38 | gactggtACGCTTCCCAATACCTTGG | |
| 39 | CCTTGGCACAGTTACCcaagtgaatt | |
| 40 | AACACACACGGATCCCGAGGcagtga |
Table 2. ANT consensus binding site.
| | –3 | –2 | –1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | | | --------- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | | A | 9 | 6 | 5 | 8 | 0 | 31 | 0 | 21 | 8 | 15 | 0 | 4 | 0 | 0 | 9 | 34 | 7 | 3 | 2 | 8 | 12 | 19 | | C | 8 | 4 | 7 | 0 | 32 | 0 | 34 | 0 | 6 | 1 | 3 | 25 | 34 | 33 | 11 | 0 | 1 | 1 | 10 | 6 | 10 | 4 | | G | 9 | 12 | 13 | 24 | 0 | 2 | 0 | 13 | 15 | 1 | 0 | 0 | 0 | 0 | 13 | 0 | 16 | 29 | 3 | 14 | 4 | 3 | | T | 7 | 12 | 9 | 2 | 2 | 1 | 0 | 0 | 5 | 17 | 31 | 5 | 0 | 1 | 1 | 0 | 10 | 1 | 19 | 6 | 8 | 8 | | consensus | | | | g | C | A | C | A | N | A | T | c | C | C | a | A | N | G | c | | | | | | | | | | | | | G | | T | | | | | g | | | | T | | | | |
To confirm that ANT-AP2R1R2 bound to these sequences, individual gel mobility shifts were performed with 17 of the 34 sites (Table 1). All 17 of these sequences bound to ANT-AP2R1R2 but with somewhat different affinities as judged by comparing the intensity of the shifted band in reactions containing equal amounts of proteins. Binding sites 4, 5, 6 and 10 exhibited the lowest affinity while binding sites 2, 11, 13–16 and 19 bound with the highest affinity. The relative affinities did not necessarily correlate with the number of positions matching the consensus site. Binding sites 2 and 15 with matches at all 14 positions exhibited high affinity binding. However several sequences (binding sites 16 and 19), which matched the consensus site in 12 of the 14 positions, exhibited higher binding affinity than some sites with 13 matches (binding sites 4, 5 and 10). One similarity noted among the weakest binding sites was the presence of a C at position 12 of the recognition site. While 11 of the 34 sites have a C in this position, in all but one of these sequences the C corresponds to the first position of the flanking sequence and not from a randomized position. The presence of the sequence AAGT following this C in the flanking region matches positions 13–16 in the consensus binding site and may have contributed to the presence of such sequences in the selected pool. All 34 of the sequenced sites match the consensus site in at least 10 of the 14 positions.
Characterization of ANT binding to the consensus site
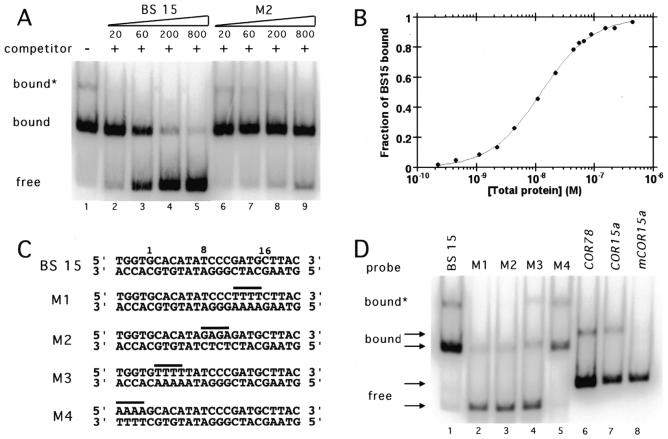
Binding site 15 and ANT-AP2R1R2 without a c-myc epitope tag were used for further characterization of the DNA binding properties of ANT. Competitive gel shifts performed in the presence of increasing amounts of unlabeled binding site 15 or a mutated version of site 15 that binds with significantly reduced affinity (M2, see below) showed that the binding was specific. Binding to the labeled 15 was competed by unlabeled 15 but not by equivalent concentrations of the mutated version of 15 (Fig. 2A). Note that a faint slower migrating band is present in lanes 1 and 6–8. The presence of this band was protein concentration dependent (data not shown) and presumably corresponds to a higher order ANT complex. Gel shift experiments using a larger fragment of ANT confirmed that the predominant shifted band corresponds to a monomer (data not shown). Binding isotherms of ANT-AP2R1R2 for binding site 15 were generated using gel mobility shifts and fit to a dissociation constant of 1.3 × 10–8 M (Fig. 2B). This value is in the range expected for a transcription factor that binds DNA specifically.
Figure 2.
ANT-AP2R1R2 binds specifically and with high affinity to binding site 15 (BS 15). (A) DNA binding by ANT to BS 15 was competed by unlabeled BS 15 (lanes 2–5) but not by similar amounts of an unlabeled mutated site M2 (lanes 6–9). Note the presence of a small amount of a slower migrating complex (bound*) corresponding to a higher order ANT protein complex in lane 1. 20, 60, 200 and 800 indicate the ratio of unlabeled DNA to labeled DNA. (B) Binding isotherm of ANT-AP2R1R2 for binding site 15. The data were fit with an equilibrium dissociation constant of 1.3 × 10–8 M. (C) Sequences of BS 15 and the mutated sites M1, M2, M3 and M4. The bases altered in each mutated site are indicated by a line. Positions 1, 8 and 16 of the consensus site are indicated by numbers. (D) ANT-AP2R1R2 binds weakly to M1 (lane 2), M2 (lane 3) and M3 (lane 4) and strongly to M4 (lane 5). ANT-AP2R1R2 also binds weakly to the C repeat/DRE elements present in COR78 (lane 6) and COR15a (lane 7) but does not bind to a mutated COR15a (m_COR15a_) (lane 8). The COR probes were shorter in length than those of BS 15 and M1–M4. Equivalent amounts of ANT-AP2R1R2 were present in each lane.
Mutated versions of binding site 15 were created to determine the relative importance of different sequences within the consensus site. In each of these mutated sites, the identity of four adjacent nucleotides was altered from that of site 15. These alterations correspond to the bases at positions 12–15 (M1), 8–11 (M2), 2–5 (M3) and –4 to –1 (M4) (Fig. 2C). ANT-AP2R1R2 bound weakly to M1, M2 and M3 but bound M4 with an affinity similar to that of the wild-type 15 site (Fig. 2D). Thus, mutations in any part of the consensus sequence severely reduced DNA binding while mutation of sequences outside of the consensus sequence had only a slight affect.
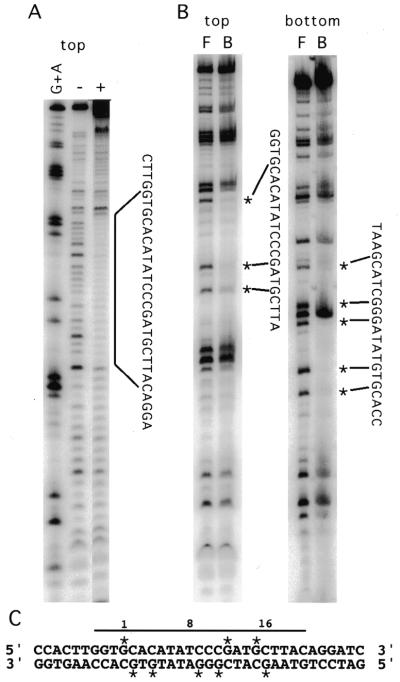
To confirm that ANT-AP2R1R2 interacts with the conserved nucleotides of binding site 15, DNase I footprinting and methylation interference were performed. The region of site 15 that ANT-AP2R1R2 protects from DNase I cleavage corresponds to the consensus site and some flanking nucleotides (Fig. 3A and C). Methylation of the Gs at positions 1, 12 and 15 of the top strand and positions 2, 4, 9, 11 and 16 of the bottom strand severely interfered with DNA binding (Fig. 3B and C). Interestingly, methylation of the G at position 10 of the bottom strand, which corresponds to the middle G of a GGG sequence, did not interfere with binding. Methylation of Gs outside of the consensus sequence did not interfere with binding.
Figure 3.
ANT-AP2R1R2 interacts with the conserved nucleotides of BS 15. (A) DNase I footprinting shows that ANT-AP2R1R2 protects the consensus site and some flanking sequence from cleavage by DNase I. (B) Methylation interference of BS 15. Bands corresponding to G residues that when methylated interfered with DNA binding by ANT-AP2R1R2 are indicated by *. (C) Summary of the results from DNase I footprinting and methylation interference. Positions 1, 8 and 16 of the consensus site are indicated by numbers. The protected region is indicated by a line while stars mark G residues important for binding.
ANT binds to C-repeat/DREs present in COR15a and COR78 and to genomic sites with similarity to the consensus site
One part of the consensus site (positions 10–13) shows similarity to the C-repeat/DRE bound by several EREBP-like proteins. The C-repeat/DREs from the COR78 promoter (5′-ACTACCGACATGA-3′) matches the consensus site in six of nine positions (underlined bases) while the C-repeat/DRE from the COR15a promoter (5′-ATGGCCGACCTGC-3′) matches the consensus site in five of nine positions (underlined bases). The ability of ANT-AP2R1R2 to bind either of these sites was investigated by gel mobility shift assays. ANT-AP2R1R2 binds weakly to both the COR78 and COR15a sites but does not show binding to a mutant COR15a site (mCOR15a) (5′-ATGGAATCACTGC-3′) that lacks the CCGAC core (Fig. 2D).
To identify possible targets of ANT regulation, we searched the Arabidopsis genome for sites that match the consensus binding sequence using the program PatMatch (http://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl ). Only two exact matches were found and both of these sites were located in the introns of putative transposon proteins. However, over 200 sequences were identified that matched the consensus site in 15 of the 16 positions. At least nine of these sequences were present within 1.7 kb of the start of a known or predicted gene and are shown in Table 3. Four of these sites were assayed for ANT binding and all four were shown to bind ANT (data not shown).
Table 3. Genomic sequences that lie within 1.7 kb of the start of a gene and that exhibit similarity to the ANT consensus site.
| Gene | Sequence | ANT binding |
|---|---|---|
| β-9 tubulin | GCACATATCACAAAGC | ND |
| Similarity to cytochrome P450 | ACACGTTTCCCAAAGT | + |
| Similarity to ARF1 | GCACGCTTTCCAAAGC | + |
| Putative | TCACACTTCCCGAAGT | + |
| Putative | GCACACTCCCCGAGGT | ND |
| Putative | GCACACTTCCTAAGGT | ND |
| Putative | GCACGTTTCCCAATTC | + |
| Phosphoenol pyruate carboxy kinase | GCACGCATCTCAAAGT | ND |
| Putative | ACACACTTCCCAAAGT | ND |
Both AP2 repeats of ANT are required for DNA binding to the consensus site
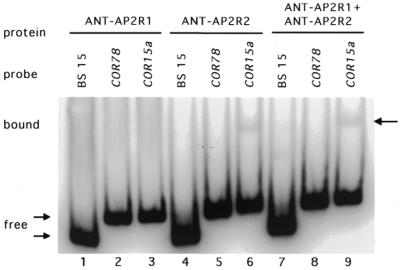
Since the single AP2 domain in several EREBP-like proteins has been shown to be sufficient for DNA binding, we investigated whether both AP2 repeats of ANT are required for binding to site 15. Proteins corresponding to the first and second repeat, ANT-AP2R1 and ANT-AP2R2, respectively, were expressed in E.coli and purified. ANT-AP2R2 was obtained in a soluble form but ANT-AP2R1 had to be purified in a denatured form. Neither ANT-AP2R1 nor ANT-AP2R2 was able to bind site 15 as assayed by gel mobility shifts but very weak binding of ANT-AP2R2 to the COR15a site was detected (Fig. 4). Furthermore, a mixture of equal molar amounts of ANT-AP2R1 and ANT-AP2R2 showed no binding to site 15 (Fig. 4). Although the ANT-AP2R1 protein could only be isolated in an insoluble form, ANT-AP2R1R2 isolated under the same denaturing conditions was capable of DNA binding (data not shown).
Figure 4.
ANT-AP2R2 binds weakly to COR15a. ANT-AP2R1 did not bind to BS 15 (lane 1), COR78 (lane 2) or COR15a (lane 3). ANT-AP2R2 did not bind to BS 15 (lane 4) or COR78 (lane 5) but did bind weakly to COR15a (lane 6). Incubation of an equimolar mixture of ANT-AP2R1 and ANT-AP2R2 with the BS15 (lane 7), COR78 (lane 8) and COR15a (lane 9) probes showed only the shifted band corresponding to the ANT-AP2R2 complex with COR15a (lane 9).
DISCUSSION
ANT binds to DNA using both AP2 repeats
The data presented here are the first report of DNA binding by a member of the AP2 subfamily of AP2/EREBP transcription factors. Using an in vitro selection method, we have isolated DNA sequences that are bound with high affinity by a truncated ANT protein containing the two AP2 repeats of ANT. Therefore, AP2 repeats are capable of DNA binding when present as a single domain (in EREBP-like proteins) or as tandem copies within a protein (in AP2-like proteins). The DNA sequence to which ANT binds [5′-gCAC(A/G)N(A/T)TcCC(a/g)ANG(c/t)-3′] is much longer than the binding sites of the EREBP-like proteins, suggesting that both AP2 repeats contact the DNA. Both AP2 repeats are required for DNA binding to this consensus site.
Located within this consensus site is a sequence CC(a/g)AN (positions 10–13) that is similar to the core binding element (CCGAC) of the C-repeat/DRE sequences bound by members of the EREBP-like subfamily. We find that ANT-AP2R1R2 as well as the second AP2 repeat (ANT-AP2R2) are capable of weak binding to the C-repeat sequence in the COR15a promoter. Since ANT-AP2R1R2 and ANT-AP2R2 exhibit similar weak affinity for this site, it is possible that only one AP2 repeat of ANT-AP2R1R2 is actually binding to the COR15a site. Thus, although single AP2 repeats may be capable of DNA binding, both AP2 repeats appear to be required for high affinity DNA binding by AP2-like proteins. High affinity DNA binding by single AP2 repeats of ANT cannot be ruled out however, since it is possible that these single repeats have DNA binding specificities distinct from that of the two repeat containing protein. This is supported by the ability of ANT-AP2R2 to bind to COR15a but not the consensus site. Such a situation has previously been described for the POU-specific domain and POU-type homeodomain of POU domain proteins (24).
ANT binds DNA in a manner distinct from the EREBP-like proteins
The EREBP-like proteins use a single AP2 domain to bind DNA with very tight affinity. AtERF has been reported to bind GCC boxes with a dissociation constant as low as 5 pM (25). In contrast, the two repeats of ANT together do not achieve as high a binding affinity suggesting that their interaction with DNA is significantly different. Despite these differences, ANT recognizes a sequence that contains a CC(a/g) motif that is similar to the CCG cores of both the EREBP and CBF/DREB proteins. This is surprising, considering that the three arginines shown to contact the guanines of this motif (18) are not conserved between the AP2-like class and the EREBP-like class.
The weak DNA binding by ANT-AP2R2 is similar to that observed for the single AP2 repeat of RAV1/2. In both cases a second DNA binding motif is required for high affinity binding. The arrangement of these two motifs in RAV1 results in extreme flexibility in DNA recognition in that the DNA sequences bound by the AP2 and B3 domains can be found in different spacings and orientations (20). It is not yet clear whether such flexibility will be a feature of AP2-like proteins. The conserved nature of the linker between the two AP2 repeats might suggest that this is not the case. In addition, no such variability was seen in the sequences selected in vitro although their presence may have been precluded by the small number of random positions in the starting oligonucleotides compared to the size of the binding site (20 versus 16). It is possible that the length of the initial random oligo provided an artificial constraint on the types of sites that could be selected. Further studies using a longer random sequence will be necessary to determine whether this is the case.
One possible model for the interaction between ANT and the consensus site is that each AP2 repeat contacts juxtaposed subsites within the consensus site. Because ANT-AP2R2 but not ANT-AP2R1 binds weakly to the COR15a site, we propose that the first AP2 repeat of ANT binds to the 5′ part of the consensus site [gCAC(A/G)] and that the second repeat contacts the more 3′ part of the consensus site [cCC(a/g)A] (Fig. 5). Each repeat is likely to bind in the major groove as demonstrated for the AP2 repeat of ERF1 (18), with the linker bridging the distance between these major groove sites. The conservation of the linker among all AP2-like proteins indicates that this region serves an important function, perhaps in orienting the two AP2 repeats for DNA binding or alternatively in contacting the DNA itself.
Figure 5.
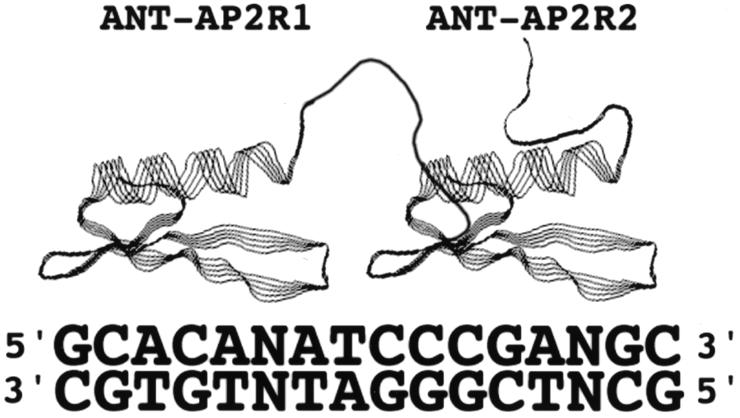
Model for interaction of ANT with the consensus site. The first AP2 repeat (ANT-AP2R1) is proposed to contact the 5′ half of the consensus site while the second AP2 repeat (ANT-AP2R2) is proposed to contact the more 3′ half of the consensus site. The individual domains are predicted to fold into domains similar in structure to that of AtERF (18). The depicted linker connecting the two AP2 repeats is not based on any structural information.
Possible targets of ANT regulation
The identification of DNA sequences bound by ANT has allowed us to identify possible targets of ANT regulation. One means of doing this was to search the Arabidopsis genome for sequences with high similarity to the ANT consensus site. We have shown that ANT binds to several of these sequences. One of these genomic matches is particularly intriguing as it is found upstream of a putative cytochrome P450. Overexpression of the P450 gene, ROTUNDIFOLIA3 (ROT3), causes increased elongation of leaves and floral organs (26), a phenotype that is similar to that found in plants ectopically expressing ANT (27,28).
Another possible target of ANT regulation is the floral homeotic gene AGAMOUS (AG) (22). We have found that ANT binds to a region within the AG second intron that is also bound by AP2 (G.Gocal and D.Weigel, personal communication). The AG fragment used in these DNA binding studies contained two partially overlapping sequences that show some similarity to the ANT consensus site. Further work is necessary to determine whether ANT is binding to either or both of these sequences. It is impossible to predict how the in vitro and in vivo DNA binding specificities of ANT will compare. In at least some cases ANT DNA binding specificity is likely to be modified in vivo by interactions with other factors. The results described here serve as an important first step in addressing such questions and in understanding how ANT functions in different aspects of reproductive development.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Sandra Snyder for technical assistance, Mike Thomashow and Eric Stockinger for providing plasmids containing wild-type and mutant C-repeat/DREs, Detlef Weigel and Greg Gocal for sharing their unpublished observations, and Jose Luis Riechmann for information on the AP2/EREBP family and comments on the manuscript. This work was supported by Department of Energy Grant 98ER20312.
REFERENCES
- 1.Riechmann J.L. and Meyerowitz,E.M. (1998) Biol. Chem., 379, 633–646. [DOI] [PubMed] [Google Scholar]
- 2.Jofuku K.D., den Boer,B.G.W., Van Montagu,M. and Okamuro,J.K. (1994) Plant Cell, 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigel D. (1995) Plant Cell, 7, 388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamuro J.K., Caster,B., Villarroel,R., Montagu,M.V. and Jofuku,K.D. (1997) Proc. Natl Acad. Sci. USA, 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohme-Takagi M. and Shinshi,H. (1995) Plant Cell, 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buttner M. and Singh,K.B. (1997) Proc. Natl Acad. Sci. USA, 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimoto S.Y., Ohta,M., Usui,A., Shinshi,H. and Ohme-Takagi,M. (2000) Plant Cell, 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stockinger E.J., Gilmour,S.J. and Thomashow,M.F. (1997) Proc. Natl Acad. Sci. USA, 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmour S.J., Zarke,D.G., Stockinger,E.J., Salazar,M.P., Houghton,J.M. and Thomashow,M.F. (1998) Plant J., 16, 433–442. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q., Kasuga,M., Sakuma,Y., Abe,H., Miura,S., Yamaguchi-Shinozaki,K. and Shinozaki,K. (1998) Plant Cell, 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein R.R., Wang,M.L., Lynch,T.J., Rao,S. and Goodman,H.M. (1998) Plant Cell, 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson K., Long,D., Swinburne,J. and Coupland,G. (1996) Plant Cell, 8, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott R.C., Betzner,A.S., Huttner,E., Oakes,M.P., Tucker,W.Q.J., Gerentes,D., Perez,P. and Smyth,D.R. (1996) Plant Cell, 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klucher K.M., Chow,H., Reiser,L. and Fischer,R.L. (1996) Plant Cell, 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuck G., Meeley,R.B. and Hake,S. (1998) Genes Dev., 12, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moose S.P. and Sisco,P.H. (1996) Genes Dev., 10, 3018–3027. [DOI] [PubMed] [Google Scholar]
- 17.Zhou J., Tang,X. and Martin,G.B. (1997) EMBO J., 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen M.D., Yamasaki,K., Ohme-Takagi,M., Tateno,M. and Suzuki,M. (1998) EMBO J., 17, 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker S.S., Wilhelm,K.S. and Thomashow,M.F. (1994) Plant Mol. Biol., 24, 701–713. [DOI] [PubMed] [Google Scholar]
- 20.Kagaya Y., Ohmiya,K. and Hattori,T. (1999) Nucleic Acids Res., 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneitz K., Baker,S.C., Gasser,C.S. and Redweik,A. (1998) Development, 125, 2555–2563. [DOI] [PubMed] [Google Scholar]
- 22.Krizek B.A., Prost,V. and Macias,A. (2000) Plant Cell, 12, 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klug S.J. and Famulok,M. (1994) Mol. Biol. Rep., 20, 97–107. [DOI] [PubMed] [Google Scholar]
- 24.Verrijzer C.P., Alkema,M.J., van Weperen,W.W., Van Leeuwen,H.C., Strating,M.J.J. and van der Vliet,P.C. (1992) EMBO J., 11, 4993–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao D., Ohme-Takagi,M. and Sarai,A. (1998) J. Biol. Chem., 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- 26.Kim G.-T., Tsukaya,H., Saito,Y. and Uchimiya,H. (1999) Proc. Natl Acad. Sci. USA, 96, 9433–9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krizek B.A. (1999) Dev. Genet., 25, 224–236. [DOI] [PubMed] [Google Scholar]
- 28.Mizukami Y. and Fischer,R.L. (2000) Proc. Natl Acad. Sci. USA, 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]