A Recent Outbreak of Human Immunodeficiency Virus Type 1 Infection in Southern China Was Initiated by Two Highly Homogeneous, Geographically Separated Strains, Circulating Recombinant Form AE and a Novel BC Recombinant (original) (raw)
Abstract
New outbreaks of human immunodeficiency virus type 1 (HIV-1) among injecting drug users (IDUs) are spreading in China along heroin trafficking routes. Recently, two separate HIV-1 epidemics among IDUs were reported in Guangxi, Southern China, where partial sequencing of the env gene showed subtype C and circulating recombinant form (CRF) AE. We evaluated five virtually full-length HIV-1 genome sequences from IDUs in Guangxi to determine the genetic diversity and the presence of intersubtype recombinants. Sequence analysis showed two geographically separated, highly homogeneous HIV-1 strains. B/C intersubtype recombinants were found in three IDUs from Baise City, in a mountainous region near the Yunnan-Guangxi border. These were mostly subtype C, with portions of the capsid and reverse transcriptase (RT) genes from subtype B. The subtype B portion of the capsid was located in the N-terminal domain, which has been shown to influence virus core maturation, virus infectivity, and binding to cyclophilin A, whereas the subtype B portion of RT was located in the palm subdomain, which is the active site of the enzyme. These BC recombinants differed from a BC recombinant found in Xinjiang Province in northwestern China. CRF AE strains were found in IDUs from Nanning, the capital of Guangxi, and in IDUs from Pingxiang City near the China-Vietnam border. The AE and BC recombinants were both remarkable for their low interpatient diversity, less than 1% for the full genome. Rapid spread of HIV-1 among IDUs may foster the emergence of highly homogeneous strains, including novel recombinants in regions with multiple subtypes.
In China, human immunodeficiency virus type 1 (HIV-1) infection was first documented in 1985 in a tourist with AIDS and in four Chinese hemophiliac patients (35). The first HIV-1 epidemic occurred among injecting drug users (IDUs) in 1989 in Yunnan Province, a southwestern province of China that shares a border with Myanmar (Burma), one of the significant heroin-producing countries (27, 31, 36). Later, HIV-1 infection spread to IDUs outside Yunnan, accounting for more than 50% of new HIV-1 infections in China from 1995 to 1997 (21). During 1996 to 1997, an HIV-1 outbreak was reported among IDUs from two distinct regions of Guangxi Province: Baise City, which borders Yunnan, and Pingxiang City, which borders northern Vietnam (32, 33). Among the four transmission routes of HIV infection, injecting drug use, sexual and perinatal transmission, and blood transfusion, injecting drug use is a leading cause in China. By the end of 1997, a total of 9,333 HIV-1 infections were reported, and injecting drug use accounted for more than 70% of all infections (21). The rapid spread of HIV-1 infection in China among IDUs has been associated with needle sharing, which is commonly practiced among drug traders and users along routes of heroin trafficking (1, 32). Guangxi is a major transit area for heroin export from Myanmar and Laos to Hong Kong and, eventually, to the West (1, 32). Heroin transported by road from Yunnan must pass through the Yunnan-Guangxi border city of Baise and through Nanning City, the capital city of Guangxi (1, 32). Another route of heroin trafficking into Guangxi is from Myanmar and Laos to northern Vietnam and then across the China-Vietnam border to Pingxiang City (1, 32).
Multiple introductions of different HIV-1 subtypes have been reported in the southwest of China. The HIV-1 epidemic among IDUs in Yunnan was initiated in 1989 by subtype B strains, both North American-like subtype B strains and the Thailand variant of subtype B (B′ or Thai B) (31). Subsequently, subtype B′ began to predominate, from 20% of all subtype B in 1990 to 90% in 1996 (12, 28, 30). In 1992, subtype C strains similar to Indian subtype C (17) were first identified among IDUs in Yunnan (20). In late 1994, a circulating recombinant form (CRF) AE was found in women who had returned to Yunnan after commercial sex work in Thailand (6). Since 1995, HIV infection has rapidly spread beyond Yunnan (21); new cases had been reported in Guangxi, Sichuan, and Xinjiang provinces (5, 29; Y. Shao, P. Pan, X. Fan, Y. Feng, Y. Zhang, G. Qin, and Y. Zhang, Abstr. 5th Int. Congr. AIDS Asia Pacific, abstr. 428, 1999). A B/C intersubtype recombinant was found along the drug trafficking route from Yunnan to Sichuan, Gansu, Ninxia, and Xinjiang provinces in the west and far northwest of the country (1; L. Su, M. Graf, H. van Briesen, H. Xing, J. Kostler, H. Melzl, H. Wolf, Y. Shao, and R. Wagner, submitted for publication).
In 1996, subtype B′ was detected in commercial blood donors in Guangxi (5). More recently, based on partial envelope sequencing, subtype C and CRF AE detected among IDUs in Guangxi were segregated in two different geographic regions (32, 33). Only subtype C was found in Baise, a city in a mountainous subtropical area, whereas only CRF AE was found in Pingxiang on the plain (32, 33). The CRF AE strains found in Pingxiang were related to the HIV-1 epidemic in northern Vietnam (13, 32).
Intersubtype recombination in HIV-1 may occur in areas like Yunnan, where more than two subtypes of HIV-1 strains circulate in a high-risk population such as IDUs. In fact, B/C intersubtype recombinants were recently identified in Yunnan and other provinces in the west and northwest (L. Su, M. Graf, H. van Briesen, H. Xing, J. Kostler, H. Melzl, H. Wolf, Y. Shao, and R. Wagner, submitted for publication). However, whether similar recombinants are circulating in Guangxi, one of the major transit areas of drug trafficking from Yunnan, remains to be studied. New recombinants may complicate the development of effective vaccines to limit the HIV-1 epidemic.
To determine the genetic diversity of HIV-1 strains and the possibility that intersubtype recombinants are present among IDUs in Guangxi, China, we collected viral isolates from three cities located along the route of heroin trafficking in Guangxi and sequenced the full-length HIV-1 genome. We have found a new, unique BC recombinant different from the one recently found in Xinjiang and also CRF AE strains that are commonly found in southeast Asia. Both the BC and CRF AE strains are remarkable for their low interpatient diversity throughout the genome.
MATERIALS AND METHODS
Clinical samples.
Seventy-nine IDUs from Baise City (a city near the Yunnan-Guangxi border), 148 IDUs from Pingxiang City (a city bordering northern Vietnam), and 1 IDU from Nanning City (the capital city of Guangxi Province) were recruited from the city heroin detoxification centers between July 1996 and July 1997, as described previously (32). The protocols conformed to guidelines for human experimentation of the U.S. Department of Health and Human Services. All subjects provided informed consent and gave their blood for HIV testing. Heparinized blood samples were drawn and transported to the United States within 48 h of collection. Peripheral blood mononuclear cells (PBMCs) from HIV-1-positive individuals were separated on Ficoll-Hypaque (Pharmacia, Piscataway, N.J.) density gradient centrifugation. CD8+ T cells were depleted using Dynal beads (Dynal Inc., Lake Success, N.Y.). PBMCs from HIV-negative donors were stimulated with phytohemagglutinin (10 μg/ml) (Sigma-Aldrich Inc., St. Louis, Mo.) for 3 days. For virus isolation, CD8+ T-cell-depleted PBMCs from HIV-1-positive individuals were cocultured with phytohemagglutinin-stimulated PBMCs from HIV-negative donors in RPMI 1640 plus 20% fetal calf serum and interleukin-2 (10 U/ml) as previously described (34). Virus production was detected with an HIV-1 p24 capture enzyme-linked immunosorbent assay (Dupont, N. Billerica, Mass.). Five viral isolates were selected from samples of IDUs for full genetic analysis. Three samples were from Baise, one was from Pingxiang, and one was from Nanning.
PCR amplification.
DNA was extracted from HIV-1-infected phytohemagglutinin-stimulated PBMCs by Capture Column (Gentra Systems Inc., Minneapolis, Minn.). The procedure for nested PCR amplification of virtually full-length HIV-1 genomes has been described (26). Briefly, the template DNA was limit diluted and amplified by using the Expand Long Template PCR system (Boehringer Mannheim GmbH, Mannheim, Germany). The outer primers annealing to the 5′ long terminal repeat (LTR) (msf12b: 5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′) and the 3′LTR (ofm-r1: 5′-TGAGGGATCTCTAGTTACCAGAGTC-3′) were used in the first-round PCR. The nested primers annealed to the 5′LTR (f2nst: 5′-GCGGAGGCTAGAAGGAGAGAGATGG-3′) and the 3′LTR (NL1/OFM19: 5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA-3′) were used in the second-round PCR to obtain one contiguous 9-kb-long segment. The nested primers are located at nucleotide positions 141 through 165 and 8957 through 8985, respectively, in HIV-1RL42.
DNA sequencing.
The PCR product served as a template for direct DNA sequencing with fluorescent dye terminators and an Applied Biosystems (Foster City, Calif.) model 373A automated DNA sequencer. Templates were fully sequenced on both strands, and all ambiguities were resolved. DNA sequences were assembled by Sequencher 3.1 software (Genecodes Inc., Ann Arbor, Mich.).
Phylogenetic and recombinant analyses.
The virtually full-length genome sequences of unknown isolates were aligned with reference sequences of HIV-1 subtypes A through J using Clustal V as implemented in the PHYLIP package (9) and gap stripped. The alignment was optimized by hand. Phylogenetic trees were constructed via the neighbor-joining method in the distance methods of the PHYLIP package (9), and the consistency of branching order was evaluated using bootstrapping (8). A bootstrap value equal to or greater than 70% was considered definitive. The reference isolates of subtype A (92UG037 and SE6594 from Uganda and Q2317 from Kenya), subtype B (MN, RF HAT3, and SF2 from the United States; OYI from Gabon; and CAM1 from the United Kingdom), subtype B′ (RL42 from China), subtype C (93IN301999, 93IN301904, 93IN301905, 94IN11246, and 95IN21068 from India; 92BR025 from Brazil; and ETH2220 from Ethiopia), subtype D (NDK, Z2Z6, and ELI from Zaire and 94UG1141 from Uganda), CRF AE (CM240 and 93TH253 from Thailand and 90CR402 from Central African Republic), subtype F (VI850 from Zaire and FIN9363 from Kenya), subtype G (SE6165 from Zaire and HH8793 from Kenya), subtype A/G (IBNG from Nigeria and DJ264 and DJ263 from Djibouti), subtype H (VI991 from Belgium), and subtype J (SE9173 and SE9280 from Zaire) were used (15).
Interpatient genetic distances were calculated using the Kimura two-parameter distance measurement in the DNA DIST part of the PHYLIP package (9) to determine the HIV-1 diversity and expressed as the average genetic distance for all pairwise combinations of nucleotide sequences from patients within each group. Genetic distances between groups were expressed as a mean. Two groups of reference strains were included in the analysis of interpatient HIV-1 diversity across the entire genome or individual genes: I included subtype C strains from eight patients with heterosexual transmission in Gaborone, Botswana (24) and II included subtype A strains from six Africans in Sweden who were infected in Uganda (n = 4), Somalia (n = 1), and Tanzania (n = 1) by heterosexual contact (3).
Distance scanning was performed to calculate genetic distances between the unknown sequence and the reference sequences using the algorithm of maximum likelihood with a sliding window of 300 nucleotides, overlapping by 50 nucleotides, and a transition-transversion ratio of 2.0 (4). The reference sequences for subtype A (92UG037), subtype B (MN), subtype B′ (RL42), subtype C (95IN21068), subtype D (NDK), CRF AE (CM240), subtype F (VI850), subtype G (SE6165), subtype H (VI991), and subtype J (SE9173) were used in the distance scanning (15).
Bootscanning (25) using the algorithm of maximum parsimony with a sliding window of 300 nucleotides overlapping by 50 nucleotides was performed to identify the putative recombination breakpoints. The reference sequences of subtype B′ (RL42), subtype C (95IN21068), CRF AE (CM240), and subtype J (SE9173) were used in the bootscanning (15). The nucleotide positions of breakpoints were designated with respect to reference strain HIVRL42 (GenBank accession number U71182) as previously described (12). The segments between the recombination breakpoints were used for building a phylogenetic tree via the neighbor-joining method with bootstrapping to confirm the disparate subtype origin of the different segments (8).
Identification of subtype signature sequences in the RT region of BC recombinants.
It appeared that the BC recombinants from Guangxi and Xinjiang (Su et al., submitted for publication) may have most of the subtype B segment in the coding region for reverse transcriptase (RT) in common. Subtype B or C signatures in the RT coding region were used to map the recombination breakpoints more precisely. We defined subtype signatures as those for which all reference strains of a given subtype have the same nucleotide in that position. Since we only used subtypes B and C as references, the presence of a subtype B or C signature in that position was mutually exclusive. The signatures were identified from 200 bp upstream and to 300 bp downstream of the putative recombination breakpoints in the RT coding region (from nucleotide positions 2044 to 2836 of HIV-1RL42). The reference strains of subtype B were MN, P896, WR27, LAI, RL42, 3202A12, OYI, CAM1, MANC, BCSG3, D31, RF HAT3, HAN, SF2, YU2, NY5CG, and JRCSF7 (HIV sequence database, http://hiv-web.lanl.gov). The reference strains of subtype C were 93IN301999, 93IN301904, 93IN301905, 94IN11246, 95IN21068, 92BR025, and ETH2220 (HIV sequence database website cited above).
Nucleotide sequence accession number.
The five virtually full-length sequences of our isolates (97CNGX-2F, -6F, -7F, -9F, and -11F) described in this article are available under GenBank accession no. AY008714 through AY008718.
RESULTS
Chinese isolates.
Chinese isolates of HIV-1, 97CNGX-2F, -6F, -7F, -9F, and -11F, were from five unrelated male IDUs (mean age, 26.4 years; range, 21 to 32 years). Three isolates (97CNGX-6F, -7F, and -9F) were from IDUs in Baise. Isolate 97CNGX-2F was from an IDU in Pingxiang. Isolate 97CNGX-11F was from an IDU in Nanning who had close contact with IDUs in Pingxiang. All subjects were in the asymptomatic phase of HIV-1 infection when samples were collected in July 1997. Since the Chinese surveillance system discovered that the epidemic of HIV-1 infection in Guangxi occurred during 1996 to 1997 (21), we estimated that the duration of infection in these five subjects was probably less than 2 years. All subjects had a history of needle sharing.
Phylogenetic relationships of Chinese isolates.
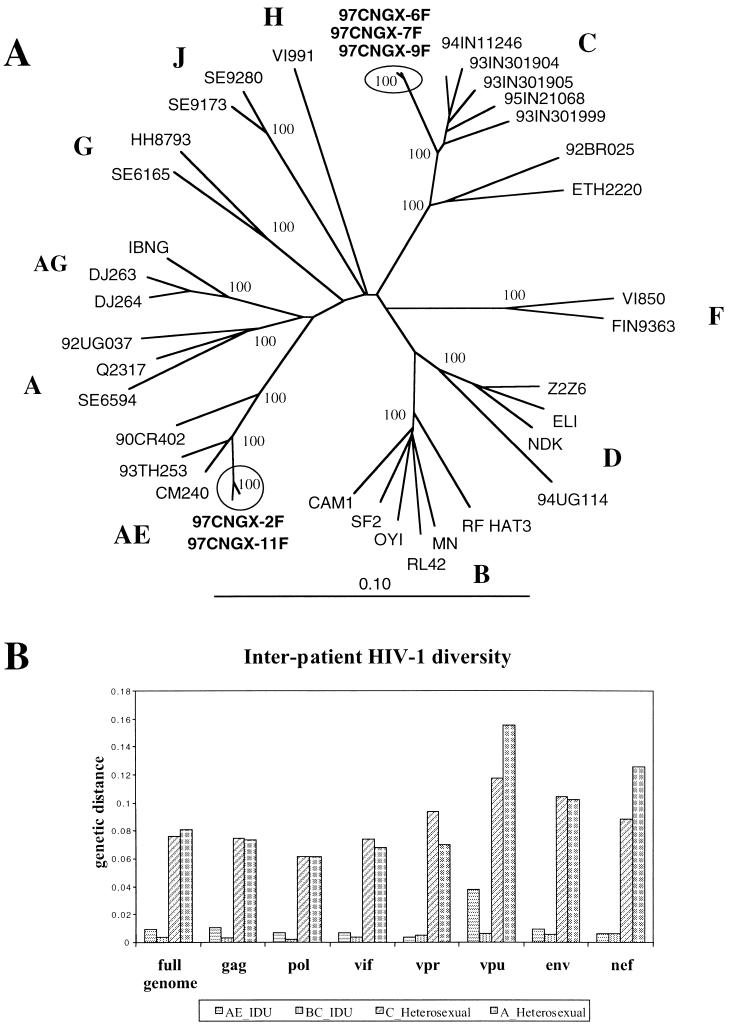
The five virtually full-length genome sequences of HIV-1 isolates were about 8.8 kb in length, extending from the beginning of gag to the early part (186 bp) of the 3′LTR. All isolates had intact open reading frames except 97CNGX-7F and 97CNGX-11F, which had stop codons in the middle of vpu. The sequences of the Chinese isolates were aligned with reference sequences from HIV-1 subtypes A to J, and phylogenetic trees were constructed by the neighbor-joining method with bootstrapping (Fig. 1A). Three isolates (97CNGX-6F, -7F, and -9F) from Baise were clustered in the subtype C strains, with a bootstrap value of 100% and a mean interpatient genetic distance of 0.37% (range, 0.28 to 0.45%). These isolates were only 4.75% divergent from subtype C strains found in India, whereas they were 8.50 and 9.28% divergent from subtype C strains 92BR025 from Brazil and ETH2220 from Ethiopia, respectively. Two isolates (97CNGX-2F and -11F) from Pingxiang and Nanning were grouped with the CRF AE, with a bootstrap value of 100% and an interpatient genetic distance of 0.89%. These isolates were grouped closer to CRF AE isolates CM240 and 93TH253 from Thailand (mean genetic distance between groups, 3.29%) than to CRF AE isolate 90CR402 from Central African Republic (mean genetic distance between groups, 6.14%).
FIG. 1.
(A) Phylogenetic analysis of virtually full-length genome sequences. The tree was constructed by the neighbor-joining method, and the number at each node indicates the percent bootstrap support from 100 bootstrap resamplings. The Chinese sequences (97CNGX-2F, -6F, -7F, -9F, and -11F) are shown in boldface. Subtype designations are shown in capital letters (A to J). The scale bar represents 10% difference. (B) Bar plot showing interpatient HIV-1 diversity across the entire genome or individual genes among Chinese isolates from IDUs compared with HIV-1 sequences from patients with heterosexual transmission. The interpatient genetic distances were calculated using the Kimura two-parameter distance measurement in the DNA DIST part of the PHYLIP package and are expressed as the average genetic distance for all pairwise combinations of nucleotide sequences from patients within each group. AE-IDU (horizontal) were Chinese CRF AE strains (97CNGX-2F and -11F) from two IDUs in Pingxiang City and Nanning City, and BC-IDU (vertical) were Chinese BC recombinants (97CNGX-6F, -7F, and -9F) from three IDUs in Baise City. Two groups of reference strains were included: C-Heterosexual (diagonal) were subtype C strains from eight patients with heterosexual transmission in Gaborone, Botswana (24), and A-Heterosexual (grid) were subtype A strains from six Africans in Sweden who were infected in Uganda, Somalia, and Tanzania by heterosexual contact (3).
Interpatient HIV-1 diversity.
We investigated the degree to which low interpatient diversity was maintained in different structural and regulatory genes. The interpatient genetic distances across the entire genome or individual genes among Chinese IDU isolates were compared with HIV-1 sequences from patients with heterosexual transmission (Fig. 1B). While both Chinese BC and CRF AE strains were remarkable for their low interpatient diversity in all structural and regulatory genes (0.37 and 0.89%, respectively), the reference strains of subtype C from Botswana (23) and subtype A from Uganda, Somalia, and Tanzania (3) had a very high degree of interpatient diversity (7.6 and 8.1%, respectively).
Detection of a novel BC recombinant.
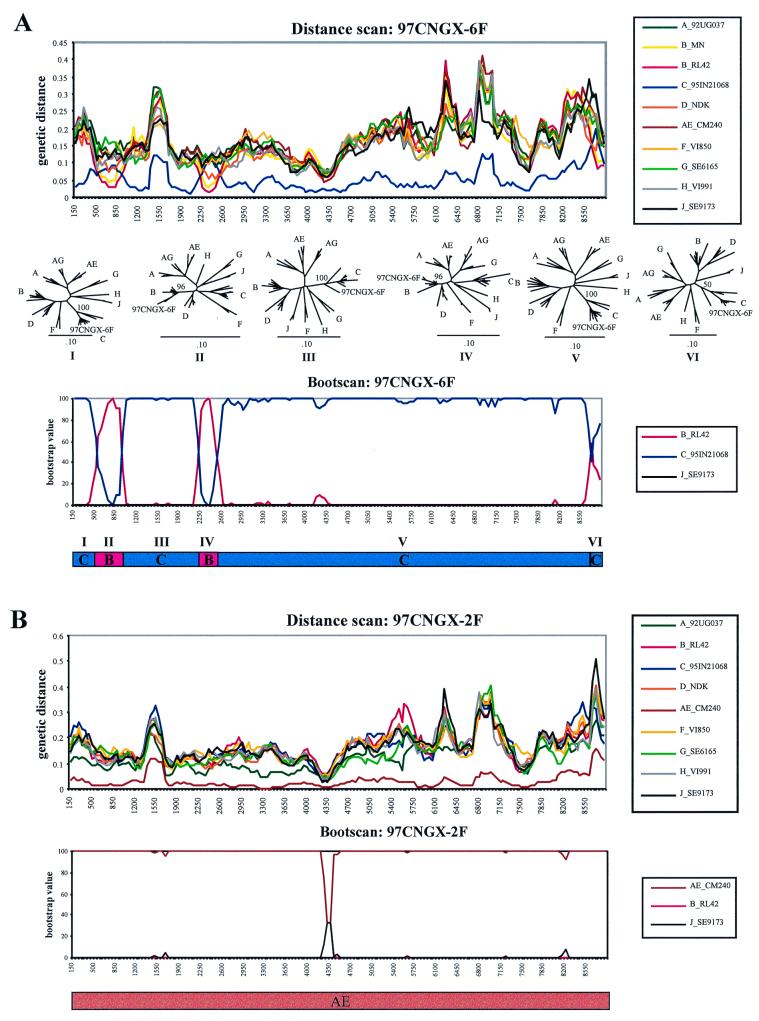
Since subtypes B and C have been detected in Guangxi (5, 32, 33), we examined these strains for evidence of recombination. The genetic distances between isolate 97CNGX-6F and each subtype were calculated in a sliding window of 300 bp overlapping by 50 bp using the maximum-likelihood distances (Fig. 2A). The distance scan showed alternating regions close to subtypes B and C. The isolate was mostly subtype C, accounting for its initial classification as subtype C, but three regions were far from subtype C and closer to subtype B: these regions were located in the p24 domain of gag, the RT domain of pol, and the nef gene. We investigated whether isolate 97CNGX-6F was a B/C intersubtype recombinant. Since isolates 97CNGX-6F, -7F, and -9F were nearly identical, the distance scans between isolates 97CNGX-7F or -9F and each reference subtype were similar to that shown Fig. 2A (data not shown).
FIG. 2.
Detection of (A) novel BC recombinant 97CNGX-6F and (B) CRF AE 97CNGX-2F by distance scanning and bootscanning. The genetic distances between unknown isolate and each reference subtype were calculated for distance scanning with maximum likelihood. The x axis is the nucleotide position in the multiple alignment of the full-length HIV-1 sequences, and the y axis is the genetic distance. The distances between the unknown and the reference subtype A (green), subtype B (yellow), subtype B′ (red), subtype C (blue), subtype D (orange), CRF AE (brown), subtype F (light orange), subtype G (light green), subtype H (gray), and subtype J (black) were plotted. The reference subtypes are indicated in the box. The bootscanning of the unknown isolate and the reference subtypes was performed using the algorithm of maximum parsimony. The x axis is the nucleotide position in the multiple alignment of the full-length HIV-1 sequences, and the y axis is the bootstrap value (percent). The reference subtype B′ (red), subtype C (blue), CRF AE (brown), and subtype J (black) are indicated in the box. The recombination breakpoints were used to deduce the subtype structure of the virus as shown below the bootscan. The phylogenetic trees of the segments between the breakpoints (designated segments I to VI) of 97CNGX-6F were constructed by the neighbor-joining method with bootstrap values (percent) at the nodes with the unknown isolate.
Next, bootscanning was performed to further investigate the possibility of recombination. Bootscanning using maximum parsimony showed results similar to distance scanning; there were three B/C intersubtype recombination sites at the p24 domain of gag, RT domain of pol, and nef gene (Fig. 2A). The phylogenetic trees constructed from subregions defined by these breakpoints confirmed the shifted subtype between subtype B and subtype C in gag and pol (bootstrap values = 96 and 96%, respectively). Due to the short segment and low bootstrap value of 50%, we could not definitely assign the nef region to subtype B. The positions of the recombination sites were approximately at nucleotides 475 to 925 and 2200 to 2500 of the multiple sequence alignment (Fig. 2A) or at nucleotide positions 604 to 1054 and 2241 to 2538 (corresponding to amino acid residues 17 to 165 of p24 and 102 to 200 of RT) of reference strain HIV-1RL42 (12), respectively. In all of these analyses, the subtype C portions were closest to the Indian isolates 93IN301904, 93IN301905, 93IN301999, 94IN11246, and 95IN21068 (17), and the subtype B portions were closest to isolate RL42 from Yunnan, China (12).
Analysis of CRF AE.
Two Guangxi strains clustered with the AE recombinant strains, and we evaluated their relationship to CRF AE throughout the genome. The genetic distances between isolate 97CNGX-2F and each subtype were calculated by using distance scanning (Fig. 2B). Since the distance between 97CNGX-2F and CRF AE (CM240) was shortest, this suggested that 97CNGX-2F grouped with CRF AE throughout the genome. The bootscanning (Fig. 2B) and phylogenetic analysis of subregion (data not shown) also confirmed that 97CNGX-2F was a CRF AE strain. Since isolates 97CNGX-2F and -11F were nearly identical, the distance scanning and bootscanning results between 97CNGX-11F and other reference subtypes were similar to those in Fig. 2B (data not shown). Phylogenetic analysis of the envelope gene also found that our CRF AE strains from Pingxiang and Nanning were very close to the CRF AE strains from Vietnam (data not shown).
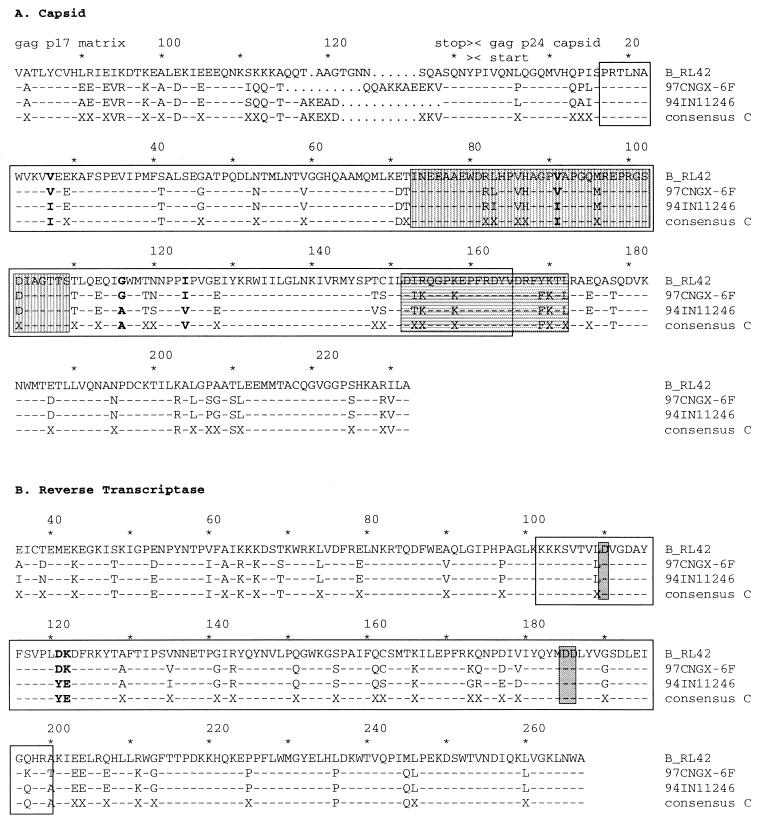
Comparison of amino acid alignments of subtype B, subtype C, and BC recombinant.
The BC recombinant 97CNGX-6F contained two segments from subtype B inserted into the backbone of subtype C. The first subtype B segment was located in the N-terminal domain of p24, which included a cyclophilin A binding motif and a major homology region (Fig. 3A). The N-terminal domain (residues 1 to 151) of p24 is important for virus core maturation and influences virus infectivity, while the C-terminal domain (residues 152 to 231) is important for protein-protein interaction and critical for virus assembly (7, 11). The second subtype B segment was located in the palm subdomain of RT (Fig. 3B), which contained three conserved aspartate residues at positions 110, 185, and 186 that form the active site of the enzyme (14). Thus, we determined the effect of the B/C intersubtype recombination on the protein sequence of 97CNGX-6F. We compared amino acid alignments of BC recombinant 97CNGX-6F, subtype B RL42 (12), and subtypes C from India (17) in the region of p24 (Fig. 3A) and RT (Fig. 3B). The comparison was done in the fragments extending from 200 bp upstream and to 200 bp downstream of the putative recombination breakpoints of 97CNGX-6F in the gag and pol genes (from nucleotide positions 404 to 1254 and 2040 to 2738 of HIV-1RL42, corresponding to amino acid residue 82 of p17 matrix to residue 232 of p24 capsid and residues 36 to 267 of RT, respectively). The reference strain RL42 is the subtype B′ prevalent in southern China (12). The consensus sequence of subtype C was derived from Indian subtype C strains (93IN301904, 93IN301905, 93IN301999, 94IN11246, and 95IN21068) using the Sequence Consensus IUPAC method in the PHYLIP package (9, 17). 94IN11246 was randomly chosen to represent Indian subtypes C (17). 97CNGX-6F had four nonconservative mutations in p24 and two mutations in RT segments that were similar to subtype B but different from subtype C. One of the four mutations in p24 was in the cyclophilin A binding motif.
FIG. 3.
Alignments of the deduced amino acid sequences of a BC recombinant (97CNGX-6F), subtype B, and subtype C in the region of (A) p24 capsid and (B) RT. The comparison was done for fragments extending from 200 bp upstream to 200 bp downstream of the putative recombination breakpoints of 97CNGX-6F in the gag and pol genes (from nucleotide positions 404 to 1254 and 2040 to 2738 of HIV-1RL42, corresponding to amino acid residue 82 of p17 matrix to residue 232 of p24 capsid and residues 36 to 267 of RT, respectively). RL42 is the subtype B′ prevalent in southern China (12). The consensus sequence of subtype C was derived from Indian subtype C strains (93IN301904, 93IN301905, 93IN301999, 94IN11246, and 95IN21068) (17). 94IN11246 was randomly chosen to represent Indian subtypes C (17). Dashes represent identity with the consensus sequences of subtype C. Dots represent gaps. X indicates the nonconsensus amino acid of Indian subtype C. Amino acids of 97CNGX-6F that were similar to RL42 but different from consensus subtype C are shown in boldface. Amino acid positions are shown above the protein sequences. Empty boxes indicate subtype B segments between the recombination breakpoints of 97CNGX-6F estimated by bootscanning. The cyclophilin A binding motif (vertical gray box) and major homology region (horizontal gray box) of p24 capsid and highly conserved aspartate residues 110, 185, and 186 of RT (gray box) are shown.
It is still an open question whether recombinant viruses offer any advantages over their parental viruses during transmission. The fact that our BC recombinant contained the N-terminal domain of capsid from subtype B and C-terminal domain from subtype C is consistent with the idea that these domains are functionally distinct and rather flexible (7). Two nonconservative mutations near a highly conserved aspartate residue (position 110) of RT were aspartate and lysine residues (positions 121 and 122). This may influence the efficiency and/or accuracy of reverse transcription.
Comparison of BC recombinants from Guangxi and Xinjiang provinces.
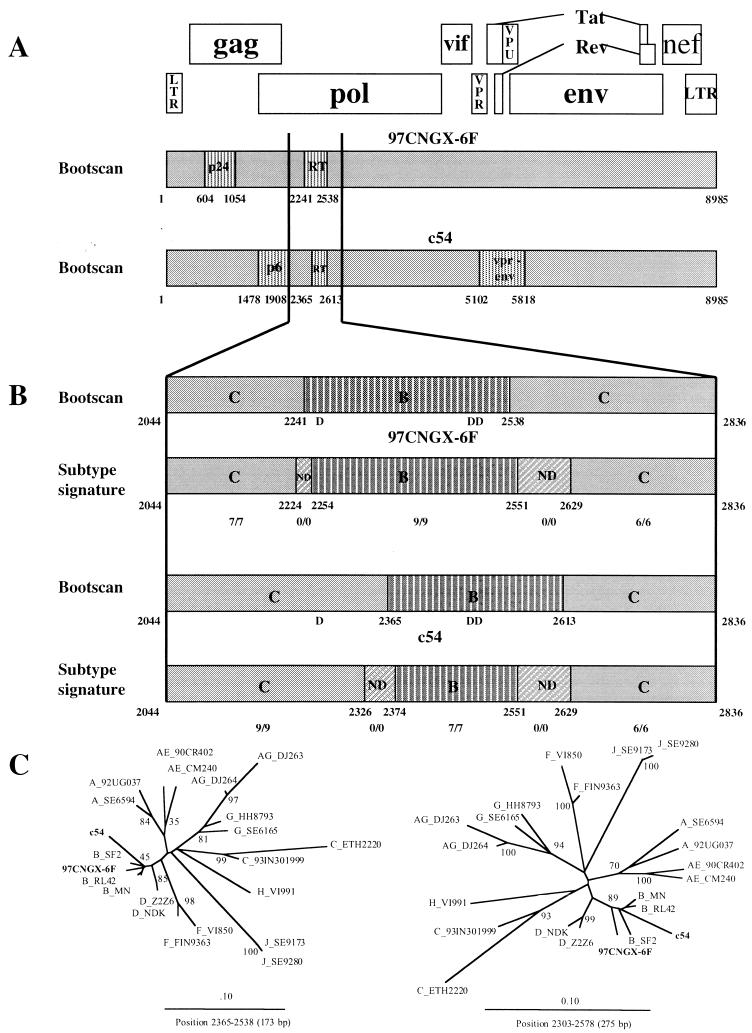
Su et al. (submitted for publication) recently found recombinant HIV-1 between subtypes B and C in five provinces located in the southwest, northwest, and far northwest parts of China. By full genome sequencing, it was determined that isolate c54, found in Xinjiang Province, was a BC recombinant and was mostly subtype C with portions of gag (p6 region), pol (RT), and vpr-env from subtype B. The positions of the recombination sites in c54 were approximately at nucleotide positions 1478 to 1908, 2365 to 2613, and 5102 to 5818 of reference strain HIV-1RL42 (Fig. 4A). The BC recombinant from Xinjiang was different from the BC recombinant found in Guangxi in that our recombinant was mostly subtype C with portions of gag (p24) and pol (RT) from subtype B.
FIG. 4.
(A) Deduced subtype structure of the full-length HIV-1 genome of BC recombinants from Guangxi and Xinjiang provinces. 97CNGX-6F from Guangxi was mostly subtype C (gray) with portions of gag (p24 capsid) and pol (RT) genes from subtype B (vertical), whereas c54 from Xinjiang was subtype C with portions of gag (p6-protease), pol (RT), and vpr-env genes from subtype B. (B) Recombination breakpoints of BC recombinants in the region of RT estimated by bootscanning and subtype signature identification. B, subtype B (vertical); C, subtype C (gray); ND, undetermined fragment indicating the subtype switching zone (diagonal). Nucleotide positions are shown below the deduced subtype structure and refer to the position in reference strain HIV-1RL42 (12). Conserved aspartate residues 110, 185, and 186 of RT are marked D. The numbers of the subtype B or C signatures over the total signature site in that fragment are shown below the nucleotide positions. (C) Phylogenetic trees of RT segments from BC recombinants and reference strains were constructed by the neighbor-joining method with bootstrap values at the nodes. The 173-bp segment (between nucleotide positions 2365 and 2538 of HIV-1RL42) was the minimal overlapping region between 97CNGX-6F and c54 in the RT region. The longer 275-bp segment (between nucleotide positions 2303 and 2578 of HIV-1RL42) was derived from the midpoint of upstream recombination breakpoints (position 2241 of 97CNGX-6F and position 2365 of c54) and the midpoint of downstream breakpoints (position 2538 of 97CNGX-6F and position 2613 of c54). Isolates 97CNGX-6F and c54 are shown in boldface.
Although different, we investigated whether the BC recombinants from Guangxi and Xinjiang could have a subtype B segment in RT in common (Fig. 4B). We compared the apparent breakpoints determined by bootscanning with patterns of subtype signatures in the region. The number of subtype B or C signatures over the total signature sites in that segment is shown (Fig. 4B). Isolate 97CNGX-6F had the upstream recombination breakpoint between nucleotide positions 2224 and 2254 and the downstream breakpoint between nucleotide positions 2551 and 2629. Isolate c54 had the upstream recombination breakpoint between nucleotide positions 2326 and 2374, which was different from 97CNGX-6F, and the downstream breakpoint between nucleotide positions 2551 and 2629, which was the same as in 97CNGX-6F. Because of the overall differences in structure and the different upstream breakpoints in the RT region, it appeared that 97CNGX-6F and c54 represent independent BC recombinations. However, they had a common subtype B RT sequence of 173 to 275 bp. Phylogenetic trees of the RT segments (173-bp segment from nucleotide positions 2365 to 2538 and 275-bp segment from nucleotide positions 2303 to 2578 of HIV-1RL42) were constructed (Fig. 4C). It showed that the BC recombinants found in Guangxi and Xinjiang were grouped with subtype B with bootstrap values of 45 and 89%, respectively. The common RT segment in the BC recombinants would encode amino acid positions 144 to 200 of RT, which include aspartate residues at positions 185 and 186, thought to form the active site of the enzyme with the aspartate residue at position 110 (Fig. 4B).
DISCUSSION
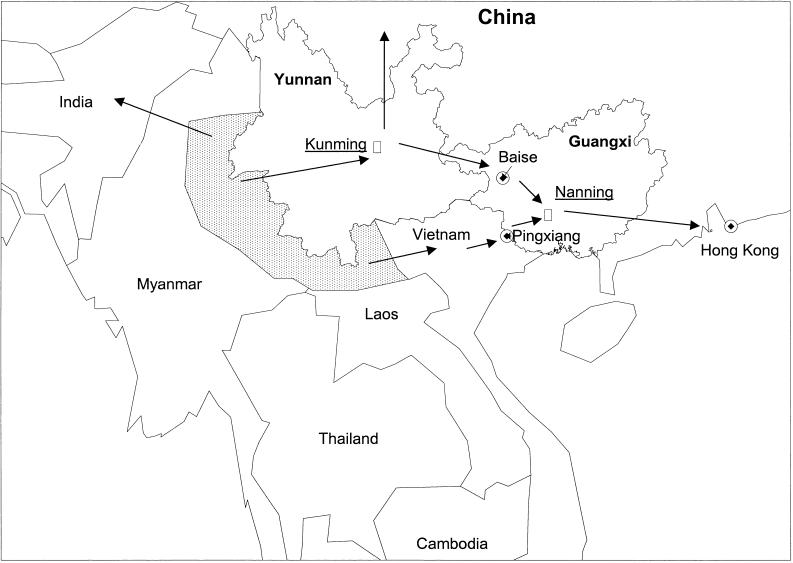
HIV-1 infection continues to spread in China primarily through the IDU networks. The rate of infection has increased about 80% per year over the past several years (21). Unlike other HIV-1 epidemics which have spread from urban to rural areas, infection began in rural areas of Yunnan in 1989 and is spreading to urban sites along the route of drug trafficking (1, 29, 31). At least three outbreaks of HIV-1 infection among IDUs in China are related to the activity of drug transportation (Fig. 5), suggesting that both local traffickers and users may play an important role in the expanding HIV epidemic (1). Drug users test heroin purity through self-injection and share needles with traders as part of their drug-purchasing behavior.
FIG. 5.
Heroin trafficking routes in southeast Asia. The dotted area indicates the opium-growing areas in Myanmar and Laos. The arrows indicate potential drug-trafficking routes. At least three drug-trafficking routes were associated with HIV-1 infection in China: from Yunnan through the Yunnan-Guangxi border city of Baise and through Nanning City, the capital city of Guangxi; from Myanmar and Laos to northern Vietnam, and then across the China-Vietnam border to Pingxiang City; and from Yunnan to west and northwest China.
New outbreaks of HIV infections with subtype C and CRF AE have been reported in Guangxi (5, 32, 33), and here we report five virtually full-length genome sequences from these IDUs. Phylogenetic analysis showed two geographically separated, highly homogeneous HIV-1 strains. The BC recombinants were found in Baise in the west of Guangxi. CRF AE strains were found in Pingxiang near the southern border. Meanwhile, both subtype C (5) and CRF AE strains were found in Nanning, the capital of Guangxi, located halfway between Baise and Pingxiang. The BC recombinants isolated in Guangxi had two small segments of subtype B inserted in the backbone of subtype C and were different from a BC recombinant, c54, found in Xinjiang (Su et al., submitted for publication). Rapid spread of multiple subtypes of HIV-1 including novel recombinants among IDUs may complicate the vaccine effort in China.
Yunnan borders the Golden Triangle region of southeast Asia, composed of northern and eastern Myanmar and western Laos. Yunnan is very vulnerable to HIV spread from eastern Myanmar, across the border into Yunnan, then through Guangxi, Hong Kong, and to Western countries (1, 32). Our BC recombinants were detected in the city of Baise, located along this major route shared with Yunnan. Su et al. reported another BC recombinant (c54) found along the northwestern route of drug trafficking. This heroin route begins in eastern Myanmar, leads to Yunnan, and goes northwest to Sichuan and Xinjiang provinces, and then across the Chinese border to Kazakhstan (1). Since c54 had recombination breakpoints different from those that occurred in our BC recombinants, these two recombinant strains may be generated in different groups of IDUs, probably in Yunnan. The heroin traders and users harboring these two different BC recombinant HIV-1 strains may live in geographically separated areas and may not interact socially. The third route of drug trafficking into China is from Myanmar and Laos eastward, through northern Vietnam, then to China at the China-Vietnam border city of Pingxiang (1, 32). The CRF AE from Pingxiang and Nanning were very close to the CRF AE strains from Vietnam, confirming that this route is an important gateway of HIV entry into southern Guangxi (13, 32). Injecting drug use plays a key role in the spread of HIV-1 in this area. By combining data from this report with other sources (1, 5, 32, 33; Su et al., submitted for publication), it appears that such a drug trafficking route has its own unique HIV-1 strain. Our discovery that each drug trafficking route is associated with a different and sometimes unique HIV-1 recombinant supports the association between heroin trafficking and HIV spread in China. These findings have important implications for preventive interventions.
An interesting finding of our study is that the new BC recombinant HIV-1 strains detected among drug users in Guangxi have low heterogeneity. It is unlikely that this result was due to PCR contamination. The original patient PBMC samples were divided into two parts. One part was used for extracting DNA, PCR, and HIV-1 env sequencing, which was published previously (32). The other part was used to isolate HIV-1 viruses. These virus isolates were then used to infect phytohemagglutinin-stimulated HIV-negative PBMCs. These PBMC samples were used to generate the three full-length BC recombinant HIV-1 sequences. When these sequences were compared to HIV-1 env sequences derived from uncultured PBMCs from the same three individuals, it was clear that the full-length sequence from each individual viral isolate matched that from the uncultured PBMC-derived env sequence from the same individual. Furthermore, although the three BC recombinant full-length sequences are highly related, they are not identical. Each full-length sequence has its own unique pattern in the variable V4 loop of the Env sequences (data not shown). These results clearly argue against the possibility of PCR contamination.
HIV-1 infection among IDUs has another unique characteristic: a very low genetic diversity among patients. The study of HIV outbreaks among IDUs in countries of the former Soviet Union, including Belarus, Ukraine, Kaliningrad, Latvia, and Russia, showed that the mean interpatient genetic distances in the C2-V3 region of the envelope were less than 2%, but a study from Nepal showed two components, one of low diversity (2, 10, 16, 18, 19, 22, 23). Meanwhile, the interpatient genetic distances among HIV-1 patients with heterosexual transmission were much higher, about 8% (3, 23). Why these HIV-1 isolates from IDUs have low diversity remains unclear, but some hypotheses can be advanced. (i) The virus circulating among patients infected by heterosexual contact may be highly selected for minor variants during mucosal transmission, while virus circulating among IDUs has direct transmission into the bloodstream, and the most abundant form may repeatedly establish itself. (ii) A single introduction of HIV-1 into a cluster of IDUs with a high transmission rate may cause a strong founder effect, resulting in low genetic diversity. Our study showed that BC recombinant and CRF AE from Guangxi were highly homogeneous, with an interpatient diversity of 0.4 and 0.9%, respectively, and that this genetic restriction applied to all structural and regulatory genes. This finding suggests that BC recombinants from three IDUs in Baise may have a common ancestor in the very recent past. We speculate that this ancestor might be in Yunnan, because subtypes B and C, based on envelope sequencing, commonly circulated among IDUs in Yunnan during the early 1990s (20, 30, 31), while subtype C was first introduced into IDUs in Guangxi around 1996 to 1997 (5, 32, 33).
Although the BC recombinants from Guangxi and Xinjiang had different parts of subtype B inserted into the backbone of subtype C, they had most of a subtype B segment in the RT region in common. However, the bootscanning and subtype signature identification of these recombinants showed that the recombination breakpoints in the RT region were not the same, supporting the independent origin of the BC recombinants and concerted selection for the subtype B RT segment. Why these two strains picked up the same region of RT remains to be explained, but possibly this subtype B RT segment may influence the error rate of reverse transcription, leading to a slower accumulation of mutations per replication cycle.
The reason for the low HIV-1 diversity in IDU epidemics must be pursued. Further studies should focus on the serial follow-up of IDUs with these unique BC recombinant strains. Maintenance of a low within-patient diversity over time would favor our hypothesis of a low RT error rate, while normal rates of quasi-species diversification would suggest the transmission mode as a key factor in limiting diversity. Comparative studies with different subtypes and recombinants would be highly recommended in this regard.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants DA12326 and DA12327 (X.-F. Yu), in part by a fellowship/grant from Fogarty International Center/USNIH (2D43TW000010-AITRP), and by a cooperative agreement between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense.
We thank Deborah L. Birx at the U.S. Military HIV Research Program, Walter Reed Army Institute of Research, for critical review of the manuscript.
REFERENCES
- 1.Beyrer C, Razak M H, Lisam K, Chen J, Lui W, Yu X-F. Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. AIDS. 2000;14:1–9. doi: 10.1097/00002030-200001070-00009. [DOI] [PubMed] [Google Scholar]
- 2.Bobkov A, Kazennova E, Selimova L, Bobkova M, Khanina T, Ladnaya N, Kravchenko A, Pokrovsky V, Cheingsong-Popov R, Weber J. A sudden epidemic of HIV type 1 among injecting drug users in the former Soviet Union: identification of subtype A, subtype B, and novel gagA/envB recombinants. AIDS Res Hum Retroviruses. 1998;14:669–676. doi: 10.1089/aid.1998.14.669. [DOI] [PubMed] [Google Scholar]
- 3.Carr J K, Laukkanen T, Salminen M O, Albert J, Alaeus A, Kim B, Sanders-Buell E, Birx D L, McCutchan F E. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS. 1999;13:1819–1826. doi: 10.1097/00002030-199910010-00003. [DOI] [PubMed] [Google Scholar]
- 4.Carr J K, Salminen M O, Koch C, Gotte D, Artenstein A W, Hegerich P A, St Louis D, Burke D S, McCutchan F E. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Young N L, Subbarao S, Warachit P, Saguanwongse S, Wongsheree S, Jayavasu C, Luo C C, Mastro T D. HIV type 1 subtypes in Guangxi Province, China, 1996. AIDS Res Hum Retroviruses. 1999;15:81–84. doi: 10.1089/088922299311754. [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Zhang J, Capizzi J, Young N L, Mastro T D. HIV-1 subtype E in Yunnan, China. Lancet. 1994;344:953–954. doi: 10.1016/s0140-6736(94)92304-3. [DOI] [PubMed] [Google Scholar]
- 7.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. Confidence limits of phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. PHYLIP: phylogenetic inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 10.Ferdats A, Konicheva V, Dievberna I, Lilja E, Albert J. An HIV type 1 subtype A outbreak among injecting drug users in Latvia. AIDS Res Hum Retroviruses. 1999;15:1487–1490. doi: 10.1089/088922299310007. [DOI] [PubMed] [Google Scholar]
- 11.Frankel A D, Young J A. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Graf M, Shao Y, Zhao Q, Seidl T, Kostler J, Wolf H, Wagner R. Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res Hum Retroviruses. 1998;14:285–288. doi: 10.1089/aid.1998.14.285. [DOI] [PubMed] [Google Scholar]
- 13.Kato K, Shiino T, Kusagawa S, Sato H, Nohtomi K, Shibamura K, Nguyen T H, Pham K C, Truong X L, Mai H A, Hoang T L, Bunyaraksyotin G, Fukushima Y, Honda M, Wasi C, Yamazaki S, Nagai Y, Takebe Y. Genetic similarity of HIV type 1 subtype E in a recent outbreak among injecting drug users in northern Vietnam to strains in Guangxi Province of southern China. AIDS Res Hum Retroviruses. 1999;15:1157–1168. doi: 10.1089/088922299310250. [DOI] [PubMed] [Google Scholar]
- 14.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 15.Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J, Sodroski J. Human retroviruses and AIDS. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Library; 1998. [Google Scholar]
- 16.Liitsola K, Tashkinova I, Laukkanen T, Korovina G, Smolskaja T, Momot O, Mashkilleyson N, Chaplinskas S, Brummer-Korvenkontio H, Vanhatalo J, Leinikki P, Salminen M O. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS. 1998;12:1907–1919. doi: 10.1097/00002030-199814000-00023. [DOI] [PubMed] [Google Scholar]
- 17.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconvertors in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukashov V V, Huismans R, Rakhmanova A G, Lisitsina Z N, Akhtyrskaya N A, Vlasov N N, Melnick O B, Goudsmit J. Circulation of subtype A and gagA/envB recombinant HIV type 1 strains among injecting drug users in St. Petersburg, Russia, correlates with geographical origin of infections. AIDS Res Hum Retroviruses. 1999;15:1577–1583. doi: 10.1089/088922299309874. [DOI] [PubMed] [Google Scholar]
- 19.Lukashov V V, Karamov E V, Eremin V F, Titov L P, Goudsmit J. Extreme founder effect in an HIV type 1 subtype A epidemic among drug users in Svetlogorsk, Belarus. AIDS Res Hum Retroviruses. 1998;14:1299–1303. doi: 10.1089/aid.1998.14.1299. [DOI] [PubMed] [Google Scholar]
- 20.Luo C C, Tian C, Hu D J, Kai M, Dondero T, Zheng X. HIV-1 subtype C in China. Lancet. 1995;345:1051–1052. doi: 10.1016/s0140-6736(95)90792-0. [DOI] [PubMed] [Google Scholar]
- 21.Ministry of Health, People's Republic of China; United Nations Theme Group on HIV/AIDS in China. China responds to AIDS: HIV/AIDS situation and needs assessment report. Beijing, China: Chinese Ministry of Health; 1997. [Google Scholar]
- 22.Novitsky V A, Montano M A, Essex M. Molecular epidemiology of an HIV-1 subtype A subcluster among injection drug users in the Southern Ukraine. AIDS Res Hum Retroviruses. 1998;14:1079–1085. doi: 10.1089/aid.1998.14.1079. [DOI] [PubMed] [Google Scholar]
- 23.Novitsky V A, Montano M A, McLane M F, Renjifo B, Vannberg F, Foley B T, Ndung'U T P, Rahman M, Makhema M J, Marlink R, Essex M. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J Virol. 1999;73:4427–4432. doi: 10.1128/jvi.73.5.4427-4432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oelrichs R B, Shrestha I L, Anderson D A, Deacon N J. The explosive human immunodeficiency virus type 1 epidemic among injecting drug users of Kathmandu, Nepal, is caused by a subtype C virus of restricted genetic diversity. J Virol. 2000;74:1149–1157. doi: 10.1128/jvi.74.3.1149-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salminen M O, Carr J K, Burke D S, McCutchan F E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 26.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Nan J, Guo Q. AIDS and HIV infection in China. AIDS. 1994;8(Suppl. 2):S55–S59. [PubMed] [Google Scholar]
- 28.Wagner R, Deml L, Teeuwsen V, Heeney J, Yiming S, Wolf H. A recombinant HIV-1 virus-like particle vaccine: from concepts to a field study. Antibiot Chemother. 1996;48:68–83. doi: 10.1159/000425160. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe M. China faces increased spread of HIV. Nat Med. 1998;4:1216. doi: 10.1038/3188. [DOI] [PubMed] [Google Scholar]
- 30.Weniger B G, Takebe Y, Ou C Y, Yamazaki S. The molecular epidemiology of HIV in Asia. AIDS. 1994;8(Suppl. 2):S13–S28. [PubMed] [Google Scholar]
- 31.Xia M, Kreiss J K, Holmes K K. Risk factors for HIV infection among drug users in Yunnan province, China: association with intravenous drug use and protective effect of boiling reusable needles and syringes. AIDS. 1994;8:1701–1706. [PubMed] [Google Scholar]
- 32.Yu X-F, Chen J, Shao Y, Beyrer C, Liu B, Wang Z, Liu W, Yang J, Liang S, Viscidi R P, Gu J, Gurri-Glass G, Lai S. Emerging HIV infections with distinct subtypes of HIV-1 infection among injection drug users from geographically separate locations in Guangxi Province, China. J AIDS. 1999;22:180–188. doi: 10.1097/00126334-199910010-00011. [DOI] [PubMed] [Google Scholar]
- 33.Yu X F, Chen J, Shao Y, Beyrer C, Lai S. Two subtypes of HIV-1 among injection-drug users in southern China. Lancet. 1998;351:1250. doi: 10.1016/S0140-6736(05)79316-8. [DOI] [PubMed] [Google Scholar]
- 34.Yu X F, Wang Z, Beyrer C, Celentano D D, Khamboonruang C, Allen E, Nelson K. Phenotypic and genotypic characteristics of human immunodeficiency virus type 1 from patients with AIDS in northern Thailand. J Virol. 1995;69:4649–4655. doi: 10.1128/jvi.69.8.4649-4655.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Y, Fan J, Zhang Q, Wang P C, Tang D J, Zhon S C, Zheng X W, Liu D P. Detection of antibody to LAV/HTLV-III in sera from hemophiliacs in China. AIDS Res. 1986;2(Suppl. 1):S147–S149. [PubMed] [Google Scholar]
- 36.Zheng X, Tian C, Choi K H, Zhang J, Cheng H, Yang X, Li D, Lin J, Qu S, Sun X, et al. Injecting drug use and HIV infection in southwest China. AIDS. 1994;8:1141–1147. doi: 10.1097/00002030-199408000-00017. [DOI] [PubMed] [Google Scholar]